Abstract
The molecular profiling of peripheral tissues, including circulating leukocytes, may hold promise in the discovery of biomarkers for diagnosing and treating neurodegenerative diseases, including Alzheimer’s disease (AD). As a proof-of-concept, we performed a proteomics study on peripheral leukocytes from patients with AD both before and during treatment with divalproex sodium. Using two-dimensional gel electrophoresis and MALDI-TOF mass spectrometry, we identified ten differentially expressed proteins: two up-regulated proteins, 14-3-3 protein ε and peroxiredoxin 2; and eight down-regulated proteins, actin-interacting protein, mitogen activated protein kinase 1, beta actin, annexin A1, glyceraldehyde 3-phosphate dehydrogenase, transforming protein RhoA, acidic leucine-rich nuclear phosphoprotein 32 family member B, and a currently unidentified protein. A subset was validated on both the transcript and protein levels in normal human peripheral blood mononuclear cell cultures treated with valproic acid. These proteins comprise a number of functional classes that may be important to the biology of AD and to the therapeutic action of valproate. These data also suggest the potential of using peripheral leukocytes to monitor pharmaceutical action for neurodegenerative diseases.
Keywords: 2D electrophoresis, Alzheimer’s disease, biomarkers, glycogen synthase kinase-3, GSK-3, histone deacetylase inhibitor, HDAC, PBMC, proteomics, qRT-PCR, valproic acid, VPA
1. Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder that results in the death of specific neuronal populations in a temporally and spatially distinct pattern and subsequent cognitive, functional, and behavioral impairments. Current FDA approved treatment options for AD are limited to two classes of drugs, those that enhance cholinergic function via inhibition of acetylcholinesterase and those that alter glutamatergic synaptic function via uncompetitive, low-to-moderate affinity NMDA-receptor antagonism. Neither drug class has been shown to alter disease progression. However, disease-modifying treatments are in preclinical or clinical development that target a variety of AD-related pathologies, including amyloid beta (Aβ) plaques, hyperphosphorylated tau-containing neurofibrillary tangles (NFT), oxidative stress, and inflammation.
Other therapeutics are also widely prescribed to help alleviate the neuropsychiatric and behavioral features of AD, which are estimated to affect greater than 80% of patients with dementia (Lyketsos et al., 2002; Weiner et al., 2005) and often lead to a number of adverse outcomes, including increased caregiver burden, earlier institutionalization, and decreased quality of life. One such treatment is the short-chain branched fatty acid, valproic acid (2-propylpentanoic acid, valproate; VPA), which is often administered to patients as the divalproex sodium formulation. Some studies suggest that VPA may afford symptomatic relief of agitation in people with dementia (Porsteinsson et al., 1997; Porsteinsson et al., 2003; Sival et al., 2002; Tariot et al., 2002), although the most recent study found no benefit (Tariot et al., 2005). A multicenter clinical trial is currently being conducted by the Alzheimer’s Disease Cooperative Study (ADCS) that is investigating the potential for VPA to attenuate progression of the clinical features of AD (Tariot et al., 2002).
While its mechanisms of action remain unclear, a number of studies suggest that VPA inhibits the activity of the histone deacetylases (HDACs) (Bradbury et al., 2005; Gottlicher et al., 2001; Gurvich et al., 2004; Phiel et al., 2001; Yildirim et al., 2003), which in turn leads to derepression of gene transcription. Other studies have also shown that VPA inhibits a major signaling enzyme, glycogen synthase kinase-3 beta [GSK3β; (Chen et al., 1999; Kim et al., 2005)], although this effect may be an indirect function of more global changes resulting from HDAC inhibition [(De Sarno et al., 2002); reviewed in (Gurvich and Klein, 2002)]. Regardless, VPA appears to exert complex effects on diverse molecular signaling pathways and a number of studies have demonstrated its neuroprotective potential (Dou et al., 2003; Hashimoto et al., 2002; Jeong et al., 2003; Mora et al., 1999; Morland et al., 2004; Wang et al., 2003). Thus, while additional research is necessary, these findings suggest the possibility that VPA and similar drugs could alter the course of AD [reviewed in (Chuang, 2005; Loy and Tariot, 2002; Tariot et al., 2002)].
In addition to alterations within the central nervous system (CNS) AD has both systemic manifestations and resultant compensatory responses that affect a number of peripheral tissues. This suggests the potential use of these tissues in understanding disease biology, progression, and therapeutic actions. Studies in fibroblasts from people with AD have found alterations in proteins related to the disease compared to controls (Mazzola and Sirover, 2001; Scali et al., 2002; Takeda et al., 1992; Zhao et al., 2003; Zhao et al., 2002) and Nagasaka et al. (Nagasaka et al., 2005) recently reported a unique gene expression profile in people with one of three familial AD mutations (APPSWE, APPARC, or PSEN H163Y) compared to their unaffected (wild-type) siblings. Other studies have found AD-specific changes in peripheral leukocytes that mirror changes in neurons, including alterations in glucose utilization (Blum-Degen et al., 1995; Urcelay et al., 2001), increased sensitivity to apoptotic stimuli (Blandini et al., 2006; Eckert et al., 1998; Mecocci et al., 2002; Morocz et al., 2002; Velez-Pardo et al., 2002), cell cycle reentry (de las Cuevas et al., 2003; Marx et al., 1999; Nagy et al., 2002; Urcelay et al., 2001), increased oxidation (Cecchi et al., 1999; Kadioglu et al., 2004; Leutner et al., 2005; Straface et al., 2005), altered calcium signaling (de las Cuevas et al., 2003; Eckert et al., 1996; Mattson et al., 2001; Palotas et al., 2002), and a variety of specific proteomic (Hye et al., 2005; Jabbour et al., 1992; Jung et al., 1999; Mirinics et al., 2002; Tacconi et al., 2004; Tayebati et al., 2001) and transcriptomic changes (Coleman et al., Submitted; Ebstein et al., 1996; Kalman et al., 2005). Peripheral leukocytes have also been used to study molecular changes in response to therapy in AD (Casademont et al., 2003; Gambi et al., 2004; Palotas et al., 2004a; Palotas et al., 2004b; Reale et al., 2004; Reale et al., 2005; Wong et al., 2004). Finally, a recently published microarray study demonstrated the utility of biomolecular profiling of peripheral leukocytes in patients with Huntington’s disease (HD) (Borovecki et al., 2005). A number of significantly elevated transcripts were identified in leukocytes from patients compared to control subjects, which were also found to be altered within the disease-affected caudate region in brains of HD patients and whose expression levels within leukocytes were abrogated following treatment with the HDAC inhibitor, sodium butyrate. Taken in aggregate, these studies illustrate a rationale for the use of peripheral tissues, including leukocytes, as reporters of central nervous system (CNS) diseases and as potential monitors of therapy.
We have developed a working hypothesis that VPA-induced alterations in genes and gene products in peripheral leukocytes are reflective of generalized molecular and cellular alterations and may provide important information regarding the effects of VPA on the CNS. As an initial proof-of-concept, we set out to determine differential protein expression profiles in peripheral leukocytes in AD patients on VPA therapy. Using both unbiased discovery as well as validation approaches, we demonstrate VPA-dependent alterations in leukocyte transcripts and proteins and illustrate the potential of peripheral leukocytes to serve as biological surrogates for CNS disease and therapy.
2. Materials and Methods
2.1 Chemicals
All chemicals were obtained from Sigma-Aldrich Corp. (St. Louis, MO) and were of molecular biology grade or better unless otherwise noted. Cell culture reagents (e.g. RPMI - 1640) were obtained from GIBCO/Invitrogen (Carlsbad, CA); standard laboratory solutions were obtained from Mediatech, Inc. (Herndon, VA); fetal bovine serum (FBS) was obtained from Hyclone (Logan, UT); Ficoll-Paque PLUS for leukocyte separations was obtained from GE Healthcare / Amersham Biosciences (Piscataway, NJ).
2.2 Study design, participants, and VPA dosing
This study was primarily an adjunct to a placebo-controlled clinical trial addressing the safety and tolerability of divalproex sodium in outpatients with moderate AD who lacked agitation or psychotic symptoms (Profenno et al., 2005). The purpose of the parent study was to inform the design of a multicenter ADCS trial to alter the clinical progression of AD (Tariot et al., 2002). This adjunct study was designed to identify potential biomarkers of therapeutic potential in peripheral leukocytes obtained from these patients. In addition, as part of an ongoing biomarkers project, we obtained blood samples from three patients with AD who were treated clinically with divalproex sodium and we collected blood samples from non-demented control subjects in order to perform validation studies. All studies were reviewed and approved by the University of Rochester Institutional Review Board. Informed consents were obtained from the family members of each person with dementia and, where relevant, consent/assent was obtained from each participant.
A total of six male and nine female participants were recruited between June and December 2002 at the University of Rochester Medical Center. The clinical trial involved outpatients with probable AD who met clinical criteria for the current ADCS trial (Profenno et al., 2005). Participants were 50–90 years old, inclusive (77.2±5.6 years; mean ± SEM), weighed at least 44 kg at baseline (68.7±11.6 kg; mean ± SEM), and lived within the community. Acute medical conditions were ruled out by history, physical examination, and laboratory testing. Standard therapies were permitted if used at stable doses. Subjects participated in a 10-week randomized, double-blind, placebo-controlled study was conducted assessing the safety and tolerability of two dosages (1000 mg/day and 1500 mg/day) of divalproex sodium delayed-release tablets versus placebo (Abbott Laboratories, Abbott Park, IL) over eight weeks, followed by a switch at week eight to divalproex sodium extended-release tablets. The 15 subjects in the present study included 12 of the 16 participants in the above clinical trial from whom suitable samples were obtained for purposes of this study, as well as three inpatients who were also treated with divalproex sodium.
2.3 Sample collection and preparation
Peripheral blood samples were collected at baseline and 3–4 weeks (25.9 ± 2.7 days) following initiation of VPA treatment into 10 mL K3EDTA-containing Vacutainer lavender top tubes (BD, Franklin Lakes, NJ) using standard venipuncture. Samples were processed for leukocyte proteins by selective lysis of red blood cells using RBC Lysis Buffer (Gentra Systems, Minneapolis, MN), followed by a single wash in PBS, centrifugation of the sample, retention of the leukocyte pellet, and storage at −80°C. Prior to two-dimensional electrophoresis (2DE) gel analysis, samples were quickly thawed on ice and lysed using a standard solubilization buffer: 20 mM Tris, pH 7.5; 0.5% Nonidet; 1 mM EDTA; 0.1 mM NaCl; 1 mM phenylmethanesulfonyl fluoride; 1:100 each Sigma Protease Inhibitor Cocktail 1 and Phosphatase Inhibitor Cocktails 1 and 2. Protein concentrations were determined using the BioRad DC Protein Assay (Hercules, CA). Protein aliquots were frozen (−80°C) and later subjected to 2DE gel analysis (Kendrick Laboratories, Inc, Madison, WI).
2.4 2D gels and analysis
2DE and spot analysis were performed by Kendrick Laboratories, Inc. Electrophoresis was performed according to the method of O’Farrell (O'Farrell, 1975). Briefly, isoelectric focusing (IEF) was carried out in glass tubes of inner diameter 2.0 mm and using 2% pH 3.5–10 ampholines for 9600 volt-hours. Following equilibration in SDS buffer, the second dimension was run on a 10% acrylamide slab gel for ~4 hours at 12.5 mA. Fifty µg of total sample protein was loaded per gel, along with an IEF (tropomyosin) standard and molecular weight (MW; myosin, 200 kDa; phophorylase A, 94 kDa; catalase, 60 kDa; actin, 43 kDa; carbonic anhydrase, 29 kDa; lysozyme, 14 kDa) standards. MW and pI measurements are approximate and are derived from the standards and a pH gradient plot using an algorithm during spot analysis.
Gels were silver stained, dried, and scanned with a laser densitometer (Molecular Dynamics, Inc., Sunnyvale, CA). Gels were analyzed using the Progenesis software system (Ver. 2002.01, NonLinear Dynamics, Newcastle upon Tyne, UK), which included automatic spot finding and quantification, automatic background subtraction (mode of non-spot), and automatic spot matching in conjunction with detailed manual checking of the spot finding and matching functions. Spot intensities were expressed as the percentage of the integrated spot density (volume) over the total density of all measured spots. Differential spot comparisons were made between a subject’s own four week VPA and baseline blood draws and were expressed as fold changes ([4 week spot value]/[baseline spot value]). Spots that were differentially expressed between baseline and 4 weeks on VPA were then sent for identification (see below).
2.5 Matrix assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF MS) and protein identification
Following spot analysis, separate preparative 2DE gels were run as above, using 200 µg of total sample protein (Kendrick Laboratories, Inc.). These gels were then Coomassie blue-stained and protein spots of interest were excised from the gels, trypsin-digested, and analyzed using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, Protein Chemistry Core, Howard Hughes Medical Institute/Columbia University, New York, NY). Briefly, an excised gel spot was transferred to clean microfuge tubes, water was added to completely hydrate the gel, and the plastic gel coating was removed with clean tweezers. Spots were prepared for digestion by washing twice with 100 µL 0.05 M Tris, pH 8.5/30% acetonitrile for 20 minutes with shaking, then with 100% acetonitrile for one to two minutes. After removing the washes, the gel pieces were dried for 30 minutes in a Speed-Vac concentrator. Gels were then digested by adding 0.06 µg modified trypsin (sequencing grade, Roche Applied Science, Indianapolis, IN) in 13–15 µL 0.025 M Tris, pH 8.5. Tubes were heated overnight at 32°C. Peptides were then extracted with 2X 50 µL 50% acetonitrile/2% trifluoroacetic acid (TFA) and combined extracts were dried and resuspended in matrix solution.
Matrix solution for the MALDI-TOF MS was prepared from a 10 mg/mL solution of 4-hydroxy-α-cyanocinnamic acid in 50% acetonitrile/ 0.1% TFA with the addition of two internal standards, angiotensin and adrenocorticotropic hormone (ACTH) 7–38 peptide. The dried digest was dissolved in 3 µL matrix/standard solution and 0.5 mL was spotted onto the sample plate. Following complete drying, the spot was washed twice with water. MALDI mass spectrometric analysis was then performed on the digest using an Applied Biosystems Voyager DE Pro mass spectrometer in the linear mode. Peptide masses were entered into the ProFound (http://129.85.19.192/profound-bin/WebProFound.exe) and the MS-Fit (http://prospector.ucsf.edu) search programs to search the NCBI and/or GenPept databases for a protein match. Average masses were used for searching with the following parameters: Taxonomy: Mammals; Protein Mass Range: +/− 30% of the expected mass of the protein; Partial Modifications: Cysteine modified by acrylamide; Mass tolerance: +/− 0.5 Da; Charge State: MH+; Enzyme: trypsin; Maximum missed cleavages: 1. More complete mass spectra data, including percent coverage, are provided in Supplemental Data Table 1.
2.6 In vitro validation studies
2.6.1 Peripheral blood mononuclear cell cultures
Peripheral blood mononuclear cell (PBMC) cultures treated with VPA were utilized for validation of the VPA-responsive targets identified in the biomarker study described above. Briefly, cultures were prepared from freshly drawn human peripheral blood following standard venipuncture. Blood samples were drawn from four control lab volunteers over the course of the studies who had no known medical or psychiatric conditions (55.8±2.4 years; mean ± SEM). PBMCs (lymphocytes and monocytes) were isolated using Ficoll-Paque PLUS gradient centrifugation and plated in RPMI-1640 with 10% fetal bovine serum (FBS) at a final density of 1×106 cells / mL (1.5 mL plated per well of a 6-well tissue culture dish or 4.5 mL per 60-mm dish). Cultures were treated immediately following plating in triplicate wells with 0–5 mM VPA (valproic acid sodium salt; Sigma), diluted in water, and were maintained in a standard incubator for 2 days in vitro (DIV). Following treatment, cells were harvested, rinsed briefly, lysed, and protein concentrations determined as described above. Prior to cell lysis, an aliquot of each well’s collection was counted for cell viability using the Trypan blue exclusion assay (Sigma) following the manufacturer’s protocol. For RNA studies cells were harvested, rinsed briefly, counted, and RNA collected as described below.
2.6.2 Quantitative real-time PCR (qRT-PCR)
Following cell harvest, total RNA was collected from each culture well using the RNeasy Mini Kit (Qiagen, Valencia, CA) per the manufacturer’s directions. Samples were treated on-column using RNase-Free DNase (Promega, Madison, WI), followed by quality and quantity assessment using standard UV spectrophotmetry and storage at −80°C until further analysis. First strand cDNA for each sample was produced from 200 ng total RNA using the SuperScript™ III First-Strand synthesis kit (Invitrogen), following the manufacturer’s protocol using random hexamer primers, and stored at −20°C until qRT-PCR analysis.
We used the commercially available Assays-on-Demand™ gene expression products (Applied Biosystems, Foster City, CA) for qRT-PCR, which utilize unlabeled PCR primers and a TaqMan® MGB probe (FAM™ dye-labeled). The following human specific Assays-on-Demand™ primer/probe sets were used: 18s rRNA (Hs99999901_s1), 14-3-3 ε (YWHAE, Hs00356749_g1), beta actin (ACTB, Hs99999903_m1), annexin A1 (ANXA1, Hs00167549_m1), acidic leucine-rich nuclear phosphoprotein 32 family member B (ANP32B or APRIL, Hs00742713_s1), glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Hs99999905_m1), peroxiredoxin 2 (PRDX2, Hs00853603_m1), and Rho A (RHOA, Hs00357608_m1). Each qRT-PCR reaction contained the following components per 25 µL: 2.5 µL high-quality cDNA, 1.25 µL Assays-on-Demand™ 20x primer/probe, 12.5 µL 2x qPCR Master Mix Plus (Eurogentec, San Diego, CA), 8.75 µL nuclease-free water (Invitrogen). Standard curves using target specific constructs were included for each qRT-PCR run. Reactions were run, fluorescence signals detected, and data analyzed using the Perkin-Elmer Applied Biosystems Sequence Detector 7700 or 7300 Real-Time PCR System with the appropriate detection software (Ver. 1.9.1, 7700; Ver. 1.3, 7300). (All samples for a given primer/probe set were run and analyzed on only one system.) Briefly, the thermocycler run included a 2 min incubation (50°C), a 10 min denaturation step (95°C), and 40 PCR cycles (15 sec at 95°C and 1 min at 60°C). All data were normalized to a separate 18S rRNA run for each sample. Data from each individual sample were generated from duplicate measures from either triplicate culture wells (n=5 individual culture experiments) or from single culture wells from one culture experiment (n=5–6 per target per treatment group).
2.6.3 Western blot analysis and antibodies
Standard denaturing SDS-PAGE and Western blot analysis on PVDF membranes (GE / Amersham) were performed on a small subset of the patient samples (n=11 of 15 subjects) and on cultured lymphocyte samples treated with VPA (10–20 µg/lane; n=3 separate culture experiments) to probe for ACTB, ANXA1, APRIL, GAPDH, and PRDX2. A subset of cultured samples was collected and nuclear extracts were prepared and probed for acetylated H3 and H4 histones (n=1 culture experiment). Plated cells were trypsinized, washed in PBS, and resuspended in ice-cold buffer containing: 10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCl; 50 µM DTT; 2 µM PMSF for 10 min to lyse the cells. Lysates were mixed by vortex for 30 sec and centrifuged. The supernatant was retained for the cytoplasmic fraction and the pellet resuspended in ice-cold buffer containing: 10 mM HEPES, pH 7.9; 25% glycerol; 420 mM NaCl; 1.5 mM MgCl2; 200 µM EDTA; and 50 µM DTT for 15 min to lyse the nuclei. The lysate was centrifuged and the supernatant fraction stored at −80°C and later assayed for nuclear protein. Total and phosphorylated GSK3 expression levels, as well as identified target proteins, were probed using total cellular lysates as described previously. Equal sample loading and protein transfer were visualized using Ponceau S stain (Sigma-Aldrich) of each membrane prior to Western blot analysis. Antibody binding was visualized using SuperSignal West or West Dura ECL reagents (Pierce Biotechnology, Inc., Rockford, IL) and the ECL signal was detected and quantified using the UVP LabWorks system (Upland, CA) or, alternatively, using film (Biomax XAR, Eastman Kodak Company, Rochester, NY; Hyperfilm ECL, GE / Amersham) that was scanned and quantified using the UVP system. The following antibodies and concentrations were used in this study: rabbit anti-ACTB (1:5000; Sigma-Aldrich); rabbit anti-ANXA1 (1:40,000; Zymed Laboratories, Inc., San Francisco, CA); rabbit anti-APRIL (1:5000; kind gift of Dr. Joan Steitz, Howard Hughes Medical Institute, Yale University); mouse anti-GAPDH (1:1000; Chemicon International, Inc., Temecula, CA); rabbit anti-PRDX2 (1:1000; LabFrontier Co., Seoul, South Korea); rabbit anti-acetyl-histone H3 (1:500; Upstate, Charlottesville, VA); rabbit anti-acetyl-histone H4 (1:500; Upstate); rabbit anti-phospho-GSK3α/β (1:1000; Cell Signaling Technology, Danvers, MA); mouse anti-GSK3α/β (1;1000; Assay Designs & Stressgen Bioreagents, Ann Arbor, MI); horseradish peroxidase (HRP) conjugated goat anti-rabbit (1:40,000 ACTB; 1:80,000, ANXA1; 1:5000, APRIL; 1:10,000; phospho-GSK3α/β; 1:2000; acetylated histone H3, H4; PRDX2; 1:5000; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA); HRP conjugated goat anti-mouse (1:80,000, GSK3α/β; 1:5000, GAPDH, Sigma-Aldrich). Data from each individual sample were generated from either triplicate culture wells (n=2–3 individual culture experiments) or from single culture wells from one culture experiment (n=3–4 per target per treatment group).
2.7 Data analysis and statistics
All data were transferred to Excel (Microsoft, Redmond, WA) for general data handling (normalization, etc.). A total of sixteen spots were chosen for mass spectrometry: eight spots were chosen based upon a qualitative trend analysis where ≥7 of 15 subjects demonstrated at least a 1-fold change in the same direction following VPA treatment (three of these spots also had a p-value of ≤0.05 following a single-sample t-test). The remaining eight spots were chosen based upon a single-sample t-test with significance determined at p<0.05. Correlation coefficients for 2DE and Western blot comparisons were calculated using the Pearson Product-Moment run in the SAS statistical program (Ver. 9.1, SAS Institute Inc., Cary, NC). Statistical analyses for the protein and mRNA validation studies were performed with the StatView for Windows statistical program (Ver. 5.0, SAS Institute, Inc.) using analysis of variance (ANOVA) and are reported at the 5% confidence interval using Fisher's Protected Least Significant Difference (PLSD).
3. Results
Thirty peripheral blood samples were collected from 15 patients with AD prior to and following four weeks (25.9±2.7 days) on divalproex sodium therapy. Subjects had mean Mini Mental Status Examination scores of 15.5±1.3 and Clinical Dementia Rating scores of 1.3±0.2 at the time of study enrollment. The mean 4-week sodium divalproex dose was 832±50 mg/day (12.3±0.9 mg/kg), resulting in a mean plasma valproate level of 64.9±9.3 mcg/mL. All data are expressed as mean ± SEM. The majority of patients were concurrently taking cholinesterase inhibitors (11 of 15) and vitamin C (11) as well as other concomitant medications, as is common in these populations [see (Profenno et al., 2005)].
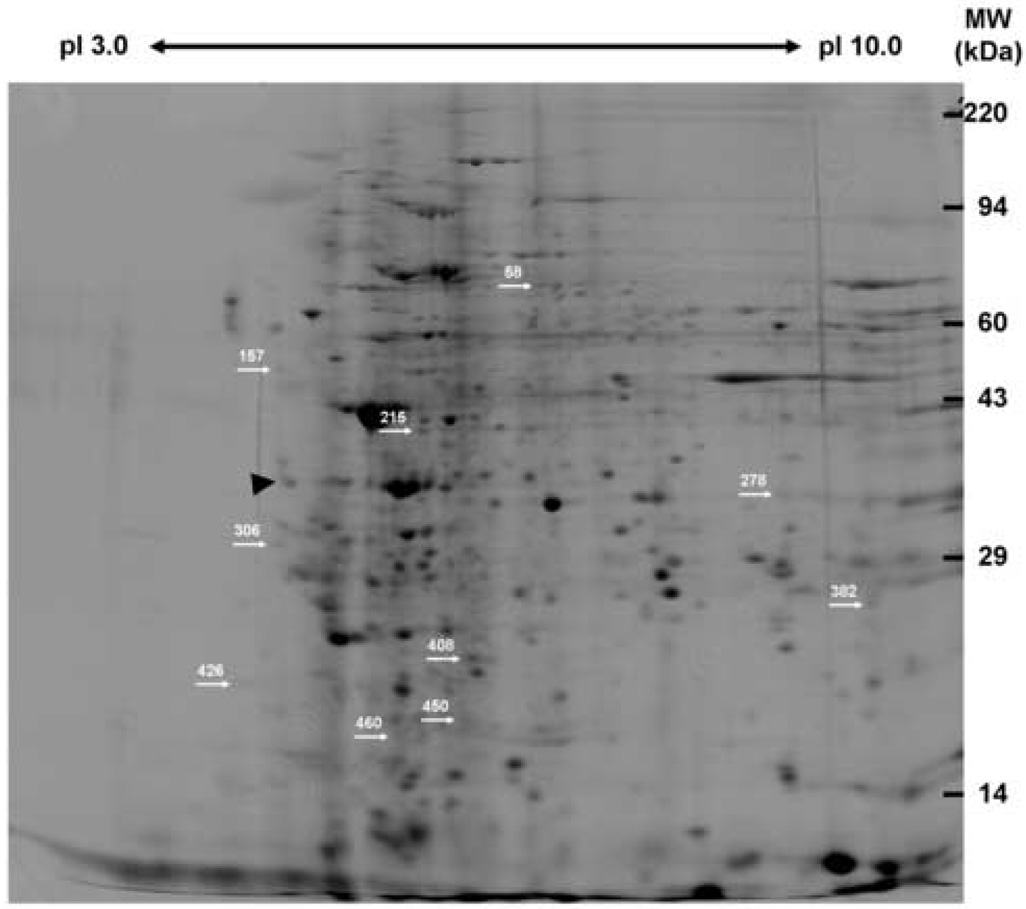
Peripheral leukocyte protein lysates were prepared subjected to 2DE and computer-aided densitometric analysis of the silver stained gels (Figure 1). From this leukocyte proteome 978 unique peptide spots were identified on at least one of the 2D gels, while 457 spots were matched on at least two gels. A total of 253 spots were matched between all 30 gels with an additional 78 spots matched on 28 gels and 41 more spots matched on 26 gels. Thus, approximately 81% (372 of 457 spots) of these spots that were matched on at least two gels were also matched in at least 13 of the 15 subjects’ samples. Aggregate comparisons of the 2D gel spot pattern were made between each subject’s baseline sample and 4-week sample and identified 16 spots that were differentially expressed in subjects following VPA treatment. Using separate preparative gels these spots were localized, excised, and subjected to MALDI-TOF MS for protein identification. Nine of these spots were successfully identified based upon an average ~43% amino acid coverage (range: 21.4%–54.5%) and an average of nine peptides matched (range: 5–16) for each protein (Table 1; Supplemental Table 1). A tenth spot (#450, mean fold difference between 4-week and baseline, −1.368±0.297, mean ± SEM, p<0.0035) had a good mass spectrum, but was not identified in any of the databases. The remaining spots were identified as either keratin or hemoglobin contaminants (three of six spots), an actin fragment (one of six spots), or were unidentifiable due to poor spectra (three of six spots). The nine identified proteins belong to a number of functional classes and two of these proteins were significantly up-regulated following VPA treatment: 14-3-3 protein ε (YWHAE) and peroxiredoxin 2 (PRDX2). The remaining seven proteins were significantly down-regulated after four weeks of VPA therapy and included: WD-repeat protein 1/actin-interacting protein 1 (AIP1, WDR1), mitogen activated protein kinase 1/extracellular signal-regulated kinase 2 (MAPK1, ERK2), beta actin (ACTB), annexin A1 (ANXA1), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), transforming protein RhoA (RHOA), and acidic leucine-rich nuclear phosphoprotein 32 family member B/acidic protein rich in leucines (ANP32B, APRIL).
Figure 1. Representative silver-stained 2D gel of human peripheral leukocytes.
Protein lysates from human peripheral leukocytes were prepared and analyzed via 2DE. Individual spot intensities were normalized to total spot intensity for each gel and differences were expressed as a ratio of 4 week VPA treatment to baseline. Nine spots were significantly differentially expressed following VPA treatment (p<0.05) and were identified via MALDI-TOF MS (see Table 1; Supplementary Table 1). A tenth spot (#450) that was significantly differentially expressed was not identifiable using MS. The differentially expressed proteins are indicated by spot number and arrows. This figure was taken from an original scanned gel that was contrast enhanced, despeckled, and sharpened using Adobe ImageReady CS2 for illustration purposes only. Gels were analyzed using the Progenesis software system as detailed in Materials and Methods with detailed manual checking of the spot finding and matching functions.
Table 1.
Differentially Expressed 2DE Gel Spots Following 4-Week VPA Treatment
| Increased | ||||||
|---|---|---|---|---|---|---|
| Spot # | Protein1 | Accession #2 | pI3 | MW4 | Change Following VPA5 | Protein Function |
| #306 | 14-3-3 Protein ε (YWHAE) | P62258 | 5.10 (4.63) |
32.6 (29.2) |
1.292 ± 0.396 (12/15) |
Signal transduction (apoptosis, transcription, cell cycle) |
| #460 | Peroxiredoxin 2 (PRDX2) | P32119 | 5.81 (5.66) |
20.7 (21.9) |
1.017 ± 0.350 (11/15) |
Antioxidant enzyme; anti-apoptotic |
| Decreased | ||||||
| Spot # | Protein1 | Accession #2 | pI3 | MW4 | Change Following VPA5 | Protein Function |
| #58 | WD-repeat Protein 1 (WDR1, Actin-Interacting Protein 1) | O75083 | 6.87 (6.17) |
72.5 (66.2) |
−0.874 ± 0.338 (11/15) |
Cytoskeletal; modulates actin dynamics |
| #157* | Mitogen Activated Protein Kinase 1 (MAPK1, ERK2) | P28482 | 5.13 (6.67) |
53.8 (40.4) |
−0.510 ± 0.505 (8/15) |
Signal transduction (proliferation, differentiation, survival) |
| #215 | Beta-actin (ACTB) | P60709 | 5.95 (5.29) |
42.4 (41.6) |
−0.735 ± 0.356 (10/15) |
Cytoskeletal; cellular structure; motility |
| #278* | Annexin A1 (ANXA1) | P04083 | 8.48 (6.64) |
36.0 (38.6) |
−0.519 ± 0.437 (9/15) |
Regulates phospholipase A2 activity; involved in diverse cellular processes (exocytosis, differentiation, apoptosis) |
| #382* | Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) | P04406 | 9.07 (8.58) |
27.2 (35.9) |
−1.002 ± 0.468 (9/15) |
Glycolytic enzyme; involved in diverse cellular processes (apoptosis, interaction with cytoskeletal elements) |
| #408¥ | Transforming Protein RhoA (RHOA) | P61586 | 6.31 (5.83) |
25.4 (21.4) |
−1.267 ± 0.507 (10/15) |
GDP/GTP binding protein; involved in diverse cellular processes (maintenance of focal adhesions, cell cycle progression, gene expression) |
| #426¥ | Acidic Leucine-Rich Nuclear Phosphoprotein 32 Family Member B (ANP32B, APRIL) | Q92688 | 4.94 (4.19) |
23.4 (22.3) |
−1.045 ± 0.405 (7/15) |
Nucleocytoplasmic shuttling protein; involved in diverse cellular processes (cell cycle progression, mRNA stability, anti-apoptotic?) |
Protein name with gene name in parentheses
Accession # from UniProt KB/Swiss-Prot Entry (http://us.expasy.org/sprot/)
pI from 2D gels; in parentheses, predicted pI from Swiss-Prot
MW (kDa) from 2DE gels; in parentheses, average MW from Swiss-Prot
Mean fold change in spot intensity ± SEM (data in parentheses are the total number of subjects with a change in the same direction as the mean change over the total number of subjects).
Spots were chosen based upon a qualitative trend analysis
Spots were chosen for analysis based upon log normalization of the data, followed by single-sample t-test (p<0.05); All other spots were chosen based upon a single-sample t-test (p<0.05). Spot #450 (21.8 kDa; 6.29 pI) gave a good mass spectrum, but was not identifiable.
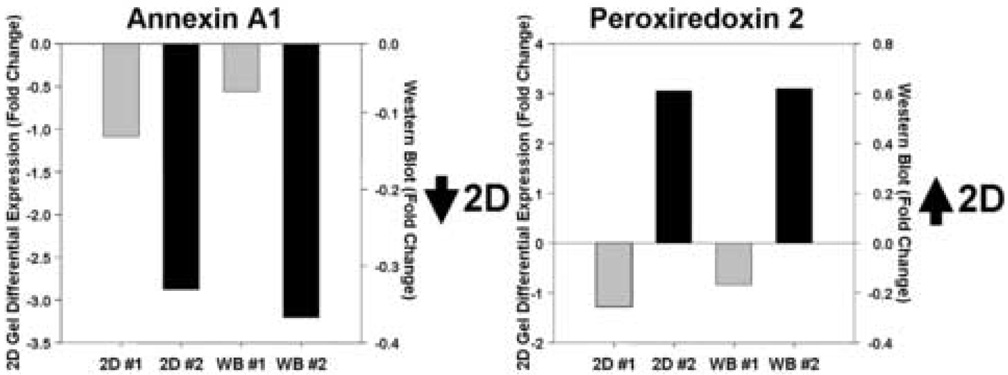
We next performed a limited validation of the VPA-responsive targets both in patient samples and in a series of in vitro experiments. In a demonstration of the sensitivity and specificity of the 2DE gel studies we compared relative expression levels of identified ANXA1 and PRDX2 silver-stained spots with Western immunoblotting data using specific antibodies to these two proteins (Figure 2). While the relative fold changes are different, there was good correspondence of expression levels in specific subjects using both techniques. Furthermore, in an examination of 11 of the 15 patient samples we found good correspondence and correlation between the 2DE data and Western blot analyses for PRDX2 (7 of 11 show same change; r=0.59, p=0.055) and GAPDH (8 of 11 show same change; r=0.70, p=0.016), but a non-significant correlation in the ANXA1 data (6 of 11 subjects show same change in direction; r=0.31, p=0.364). We did not undertake further proteomic or transcriptomic studies from this patient population because of a limited supply of samples.
Figure 2. Correspondence of relative expression levels of two proteins between 2DE analysis and Western immunoblot analysis following VPA treatment.
Examination of annexin a1 (Spot #278) and peroxiredoxin 2 (Spot #460) expression in two study patients using 2DE differential spot analysis and immunoblot analysis demonstrate similar relative levels of expression between the two methodologies. Expression levels are indicated as fold-level changes between the 4-week treatment and baseline samples. 2D, 2D gel analysis; WB, Western blot analysis; #1, subject #1; #2, subject #2.
As an additional set of validation studies we examined alterations in transcript and protein expression in cultured human peripheral blood mononuclear cells (PBMCs; lymphocytes and monocytes) treated with VPA. We hypothesized that VPA-induced alterations identified in the in vivo studies would be generalizable to these culture experiments. PBMCs were isolated from healthy lab volunteers and cultured for two days in vitro (2 DIV) in the presence of varying concentrations of VPA (0–5 mM). This range of concentrations comports with other studies in a variety of cell types (Bradbury et al., 2005; Fraser et al., 1999; Fuller et al., 2002; Gurvich et al., 2004; Marchion et al., 2005; Shen et al., 2005) and has been shown to substantially inhibit GSK3β [0.5 mM, see (Chen et al., 1999; Kim et al., 2005)] and HDAC activities [0.1–20 mM, see (Gottlicher et al., 2001; Gurvich et al., 2004; Phiel et al., 2001)]. Cell viability as assayed by Trypan blue exclusion was significantly reduced at the highest doses in cultures following 2 DIV VPA treatment (data not shown). Next, we examined the effects of in vitro treatment on two putative VPA targets, the HDACs and GSK3β. In our culture system we found increased acetylation by Western blot analysis of the histones H3 and H4, which is an indirect measure of HDAC inhibition (data not shown). Likewise, we also observed an increase in phosphorylated GSK3β in cultures treated with VPA compared to controls with no observed increase in total GSK3β levels (data not shown). While we did not detect a dose-response relationship in these measures, these data are consistent with VPA inhibiting both HDACs and GSK3β in cultured human PBMCs.
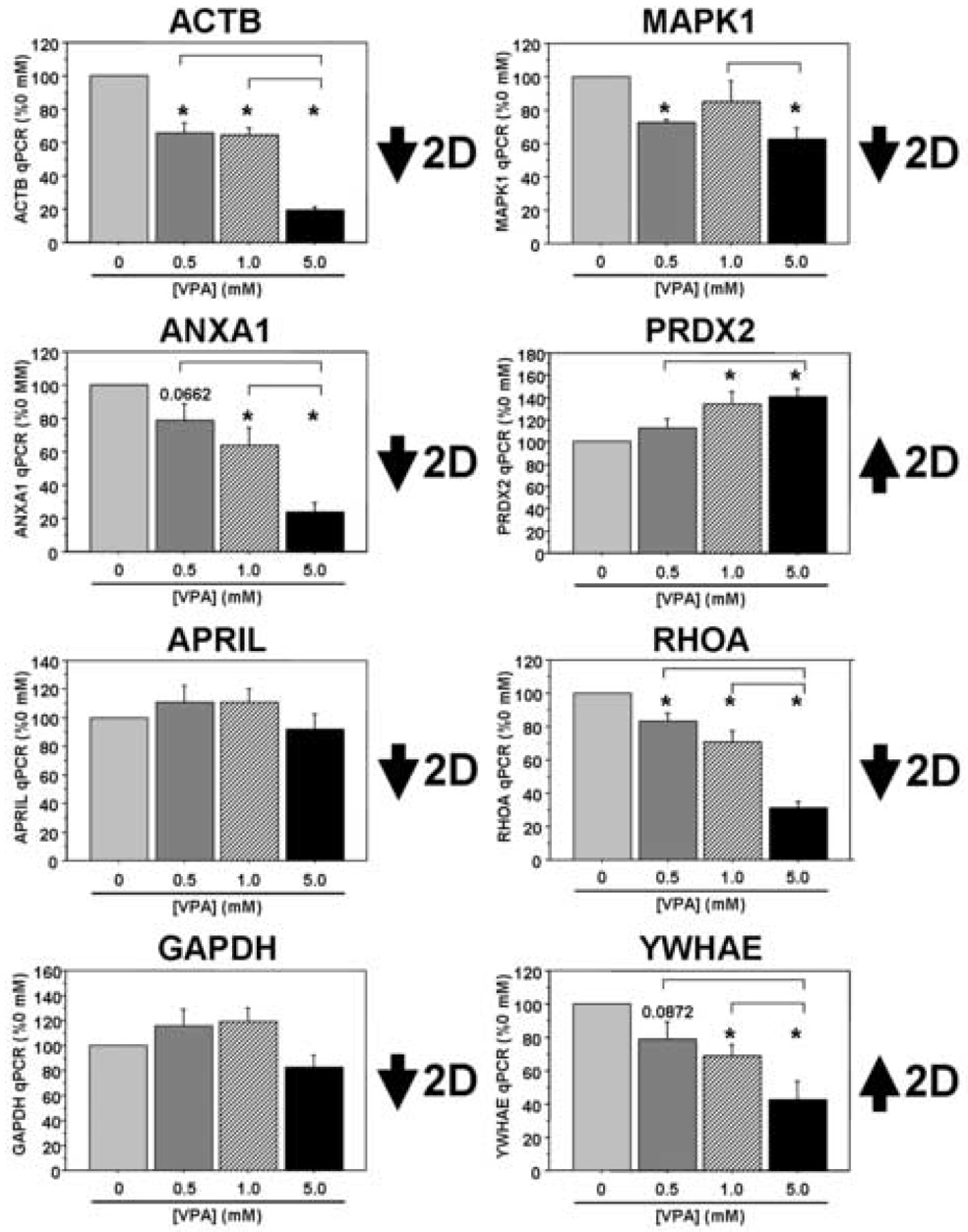
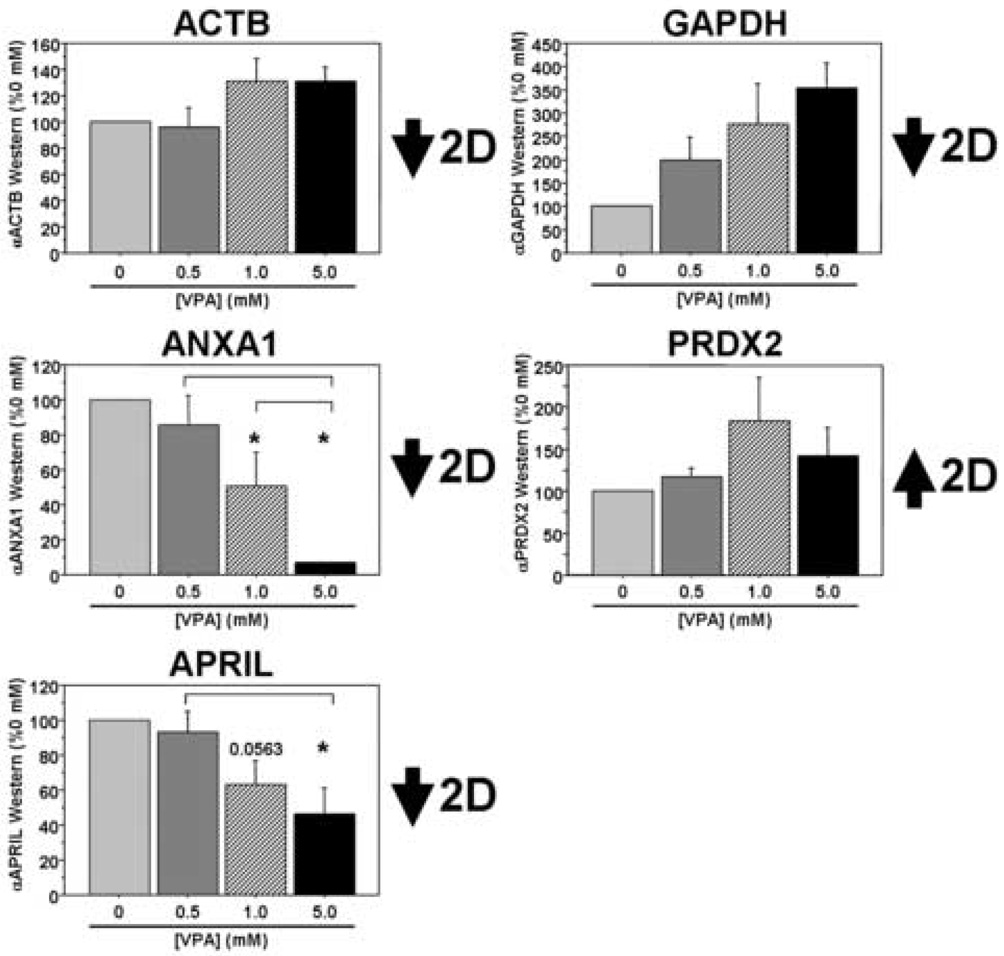
Finally, we investigated a number of potential VPA targets identified from the 2DE studies both on the transcript and the protein levels in our PBMC cultures following VPA treatment. On the transcript level we found good correlation between five of the eight targets examined: ACTB, ANXA1, MAPK1, PRDX2, and RHOA (Figure 3). The GAPDH data were more complex and trended toward decreased expression levels between the two middle VPA concentrations and the highest concentration (overall p=0.0709). We found no significant change in APRIL transcript levels in these cultures, while YWHAE transcript levels showed changes in the opposite direction compared to the proteomic data from our initial patient cohort. In separate sets of cultures, there was a significant dose-dependent decrease in ANXA1 and APRIL at the protein level and a trend toward increased PRDX2 protein expression in PBMC cultures treated with VPA, mimicking the data from our in vivo patient study (Figure 4). Interestingly, ACTB protein expression was relatively stable across the range of VPA concentration, while GAPDH expression levels were in the opposite direction from those observed in the patient study (overall p=0.0624). Thus, three of the five targets surveyed in the in vitro study demonstrate consistent changes on the protein level with those observed in the initial patient study. While we acknowledge differences in these cultures, both in terms of complexity and the origins of these samples (e.g. normal volunteers versus patients with AD), these results suggest commonalities in VPA-dependent biochemical alterations between this culture system and the patient population we studied.
Figure 3. VPA-responsive targets identified in patients on valproate therapy are also differentially regulated at the transcript level in cultured human PBMCs following VPA treatment.
PBMCs were isolated from healthy lab volunteers and cultured in the presence of varying concentrations of VPA for two days in vitro. Eight of the nine targets identified in the 2DE experiments were analyzed using qRT-PCR and all data were normalized to 18S rRNA (n=5 separate experiments for MAPK1; n=6 separate experiments all other targets). All data are expressed as a percent of the 0 mM (control) treatment condition (mean ± SEM). *, significantly different between “0 mM VPA”, p<0.05; inverted brackets, significantly different between the two bracketed treatments, p<0.05.
Figure 4. VPA-responsive targets identified in patients on valproate therapy are also differentially regulated at the protein level in cultured human PBMCs following VPA treatment.
PBMCs were isolated from healthy lab volunteers and cultured in the presence of varying concentrations of VPA for two days in vitro. Five of the nine targets identified in the 2DE experiments were analyzed using Western blots (n=3 separate experiments). All data are expressed as a percent of the 0 mM (control) treatment condition (mean ± SEM). *, significantly different between “0 mM VPA”, p<0.05; inverted brackets, significantly different between the two bracketed treatments, p<0.05.
4. Discussion
We lack rapid and minimally invasive techniques to monitor therapeutic actions at the molecular and cellular levels in vivo. Methodologies that incorporate these into testing strategies may be valuable in clinical trials, as well as in clinical monitoring of approved therapies. Molecular profiling of peripheral tissues, including circulating leukocytes, is one such method. The critical role of inflammation in disease pathogenesis and the demonstration of shared alterations among a number of classes of molecules in AD brain and peripheral leukocytes (Coleman et al., Submitted) have led us to consider this cellular population as a potentially useful surrogate for diagnosis, prognosis, and therapeutic monitoring in AD. In our discovery and validation proteomics study on peripheral leukocytes from AD patients at baseline and following four weeks of VPA treatment we identified ten differentially expressed proteins: one unidentified protein and nine others representing functional classes related to the cytoskeleton, cell signaling, apoptosis, and redox status. A subset of these were validated on the initial patient samples and in cultured human leukocytes treated with VPA.
In addition to its efficacy in treating epilepsy, bipolar disorder, and migraine, VPA and related agents have recently gained attention as potential therapeutics for cancer (Yoo and Jones, 2006), human immunodeficiency virus (HIV) infection (DiCenzo et al., 2004; Lehrman et al., 2005), and neurodegenerative disorders (Ren et al., 2004; Sugai et al., 2004; Sumner et al., 2003). While we did not evaluate HDAC or GSK3β in our patient samples, we did find an increase in both acetylated histones and GSK3β phosphorylation in PBMC cultures treated with VPA at therapeutic concentrations, suggesting its inhibitory effects on these molecules in this reduced system. Although it remains to be determined whether or not these effects are generalizable in vivo, it appears that VPA can induce an overall change in transcriptional status of a cellular population, either via inhibition of HDACs and/or GSK3β, that would result in predictable patterns of differential protein expression related to its therapeutic effects.
Limitations and caveats of the current study
While the present study demonstrates the potential use of proteomic profiling of peripheral tissues to ascertain biochemical signatures in the treatment of CNS disease, a number of caveats are warranted. A major limitation of this current 2DE study is the small fraction of the leukocyte proteome actually examined. Certain 2DE studies routinely identify thousands of spots in a given sample (2000–10,000; reviewed in (Choe et al., 2006; Gorg et al., 2004)) and while we identified nearly 1000 unique spots in aggregate within our samples, only 253 spots were identified and matched between all 30 gels. Furthermore, the differentially expressed spots identified by MS analysis are abundant within the cytosolic compartment, consistent with the sample preparation protocol that we followed. Thus, the small fraction of the proteome examined is a function of both the method by which these samples were prepared (e.g. enriched for cytosolic proteins versus proteins from other intracellular compartments) and the inherent limitation of 2DE studies, which tend to overlook proteins of low abundance, proteins at the extremes of molecular weight (e.g. very high or very low MW proteins), and highly hydrophobic proteins (Gorg et al., 2004; Lubec et al., 2003). Issues related to technical variability between gels, including gel streaking and areas of poor sample resolution, also make matching between samples difficult and reduce the sensitivity of this method. A final, potentially limiting factor is the heterogeneous nature of the tissue examined, which consists of at least five distinct cell types and numerous cellular subtypes, complicating identification of more subtle, cell-specific changes in protein expression. Future proteomic discovery efforts beyond proof-of-concept should include considerations of intra-subject proteome variation, cellular enrichment, subcellular fractionation, protein enrichment or depletion, and the use of other, complementary proteomic profiling technologies (e.g. LC-MS-MS; ICAT).
While the majority of data were consistent between the discovery and validation studies (5 of 8 targets on the transcript level; 3 of 5 targets on the protein level), we did find that a small number of targets did not validate either on the transcript level or on the protein level. A number of explanations may account for these discrepancies, including the common divergence often seen between transcript and protein levels in biological studies (Gygi et al., 1999), as well differences between the in vivo and in vitro environments (VPA dose, duration of dose, subpopulations of cells examined in culture, individual variation in drug response, etc.). Differences between the clinical population samples and the in vitro culture system may also be a factor since cultured leukocytes from normal volunteers may not mimic the complexity of disease processes and compensatory mechanisms that may be ongoing in the original patient population. Ideally, validation of these targets would have been performed in a separate cohort of AD patients on VPA therapy. However, the commonalities between the in vivo and the in vitro data are promising in the sense that these may be generalizable targets of VPA. Importantly, these types of in vitro studies may also help inform future, reciprocal sets of experiments that examine the biological effects of VPA treatment both in peripheral and CNS tissues. Finally, these data do not necessarily point to a particular set of biomarkers for VPA in AD patients. More rigorous studies, such as the current ADCS trial, and follow up validation studies will be necessary to examine the ability of the peripheral leukocyte proteome to report clinical efficacy or predict individual responsiveness to therapy. Taken in aggregate, however, the data do offer a proof-of-concept for the biomolecular profiling of peripheral tissues in CNS disease and may suggest pathways that are significant to the therapeutic or potentially neuroprotective actions of VPA and to the biology of AD, which mandate further study.
Summary
We have identified a set of differentially expressed leukocyte proteins obtained from patients with AD following VPA therapy within a dose range and for a duration that is expected to induce cellular neuroprotective changes within the CNS and that is tolerable for long-term daily usage. These proteins comprise a number of functional classes related to cytoskeletal dynamics, intracellular signaling, gene expression, apoptosis, immune function, and intracellular redox status. The precise nature of these changes on any specific molecular, cellular, or clinical outcome regarding VPA therapy is currently unknown. We speculate that these changes identified in peripheral tissues are generalizable to the CNS and may in part define a VPA-dependent molecular program that would lead to altered synaptic, antioxidant, and anti-apoptotic properties within neurons. Alternatively, these changes may be more closely related to the immunological compartment (leukocytes, microglia) and instead reflect VPA-dependent alterations in inflammation and immune response. While caution is currently necessary in interpreting these results, these data nonetheless support the importance of peripheral tissues in these types of studies. Given the intimate relationship between the peripheral leukocyte and CNS compartments in neurodegenerative diseases, both in terms of shared intracellular signaling pathways and the role of inflammation in disease, it appears that peripheral leukocytes may provide important molecular and cellular information regarding disease diagnosis, pathogenesis, and therapeutic potential. A more complete understanding of these relationships is part of ongoing studies within our laboratories.
Supplementary Material
Acknowledgements
This work was supported by in part by the National Institutes of Health Grant AG10483 (The Alzheimer’s Disease Cooperative Study, National Institute on Aging) and by an investigator-initiated grant from Abbott Laboratories. Study medications were also provided by Abbott Laboratories. The rabbit anti-APRIL antibody was a kind gift of Dr. Joan Steitz, Howard Hughes Medical Institute, Yale University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement for Authors.
(a) A portion of these studies was supported by an investigator initiated grant from Abbot Laboratories, which also provided all study medications. PDC, HF, RL, KM-Z, TRM, LAP, and PT have received support in the past or currently receive support from the Abbott Laboratories grant. These authors have also filed a U.S. Provisional Patent Application (“Biomarkers of Alzheimer ’s Disease”) based upon a portion of this work. DZ does not have any actual or potential conflicts of interest.
HF discloses the following: “I am a founder and equity owner of AmpliVex and MedGenesis. I have received consulting honoraria from the High Q Foundation, Spinal Muscular Atrophy NINDS Advisory Board, and speakers’ honoraria from non-academic entities Amgen, GE Healthcare, Sirtris Pharmaceuticals, the Ernst Scherring Foundation and the Institute on the Study of Aging. I’ve received research support from the NINDS, NIA, DoD and Abbott Laboratories.“
RL has received investigator-initiated grants, speaker fees and travel support from Abbott Laboratories.
TM has received consulting honoraria from MedGenesis.
PT also discloses the following: “I have received consulting fees from Memory Pharmaceuticals, Inc., and Novartis AG; consulting fees and research support from Abbott Laboratories, Bristol-Myers Squibb, Eisai Inc., GlaxoSmithKline, Janssen Pharmaceutica, Eli Lilly and Company, Merck and Company, Myriad Pharmaceuticals, Pfizer Inc., Sanofi-Synthelabo, Schwabe, and Takeda Pharmaceuticals North America Inc.; educational fees from Lundbeck; consulting fees, research support, and educational fees from AstraZeneca, Eisai Inc, Forest Laboratories, Pfizer Inc.; research support from Elan, Mitsubishi Pharma Corporation, Neurochem, Ono Pharmaceuticals, and Wyeth Laboratories. Other research support: NIA, NIMH, Alzheimer’s Association, Arizona Department of Health Services, and the Institute for Mental Health Research. I have no investments to disclose. I am listed as a contributor to a provisional patent application, “Biomarkers of Alzheimer’s Disease.” I am not on any speakers’ bureaus but was in 2005: AstraZeneca; Eisai Inc; Forest Pharmaceuticals Inc; Pfizer Inc.”
(b) All study protocols were reviewed and approved by the University of Rochester Institutional Review Board and informed consent was obtained from each participant or from a responsible study partner.
References
- Blandini F, Sinforiani E, Pacchetti C, Samuele A, Bazzini E, Zangaglia R, Nappi G, Martignoni E. Peripheral proteasome and caspase activity in Parkinson disease and Alzheimer disease. Neurology. 2006;66:529–534. doi: 10.1212/01.wnl.0000198511.09968.b3. [DOI] [PubMed] [Google Scholar]
- Blum-Degen D, Frolich L, Hoyer S, Riederer P. Altered regulation of brain glucose metabolism as a cause of neurodegenerative disorders? 46. 1995:139–147. [PubMed] [Google Scholar]
- Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV, Krainc D. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci U S A. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury CA, Khanim FL, Hayden R, Bunce CM, White DA, Drayson MT, Craddock C, Turner BM. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19:1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- Casademont J, Miro O, Rodriguez-Santiago B, Viedma P, Blesa R, Cardellach F. Cholinesterase inhibitor rivastigmine enhance the mitochondrial electron transport chain in lymphocytes of patients with Alzheimer's disease. J Neurol Sci. 2003;206:23–26. doi: 10.1016/s0022-510x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Cecchi C, Latorraca S, Sorbi S, Iantomasi T, Favilli F, Vincenzini MT, Liguri G. Gluthatione level is altered in lymphoblasts from patients with familial Alzheimer's disease. Neurosci Lett. 1999;275:152–154. doi: 10.1016/s0304-3940(99)00751-x. [DOI] [PubMed] [Google Scholar]
- Chen G, Huang LD, Jiang YM, Manji HK. The mood-stabilizing agent valproate inhibits the activity of glycogen synthase kinase-3. J Neurochem. 1999;72:1327–1330. doi: 10.1046/j.1471-4159.2000.0721327.x. [DOI] [PubMed] [Google Scholar]
- Choe LH, Werner BG, Lee KH. Two-dimensional protein electrophoresis: from molecular pathway discovery to biomarker discovery in neurological disorders. NeuroRx. 2006;3:327–335. doi: 10.1016/j.nurx.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang DM. The antiapoptotic actions of mood stabilizers: molecular mechanisms and therapeutic potentials. Ann N Y Acad Sci. 2005;1053:195–204. doi: 10.1196/annals.1344.018. [DOI] [PubMed] [Google Scholar]
- Coleman PD, Cox C, Weimer J, Xiao Y, Federoff HJ, Marshall FJ, Chow N. Inflammatory and stress related transcripts distinguish early Alzheimer’s disease peripheral blood leukocytes. (Submitted) [Google Scholar]
- de las Cuevas N, Urcelay E, Hermida OG, Saiz-Diaz RA, Bermejo F, Ayuso MS, Martin-Requero A. Ca2+/calmodulin-dependent modulation of cell cycle elements pRb and p27kip1 involved in the enhanced proliferation of lymphoblasts from patients with Alzheimer dementia. Neurobiol Dis. 2003;13:254–263. doi: 10.1016/s0969-9961(03)00040-8. [DOI] [PubMed] [Google Scholar]
- De Sarno P, Li X, Jope RS. Regulation of Akt and glycogen synthase kinase-3 beta phosphorylation by sodium valproate and lithium. Neuropharmacology. 2002;43:1158–1164. doi: 10.1016/s0028-3908(02)00215-0. [DOI] [PubMed] [Google Scholar]
- DiCenzo R, Peterson D, Cruttenden K, Morse G, Riggs G, Gelbard H, Schifitto G. Effects of valproic acid coadministration on plasma efavirenz and lopinavir concentrations in human immunodeficiency virus-infected adults. Antimicrob Agents Chemother. 2004;48:4328–4331. doi: 10.1128/AAC.48.11.4328-4331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Birusingh K, Faraci J, Gorantla S, Poluektova LY, Maggirwar SB, Dewhurst S, Gelbard HA, Gendelman HE. Neuroprotective activities of sodium valproate in a murine model of human immunodeficiency virus-1 encephalitis. J Neurosci. 2003;23:9162–9170. doi: 10.1523/JNEUROSCI.23-27-09162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebstein RP, Nemanov L, Lubarski G, Dano M, Trevis T, Korczyn AD. Changes in expression of lymphocyte amyloid precursor protein mRNA isoforms in normal aging and Alzheimer's disease. Brain Res Mol Brain Res. 1996;35:260–268. doi: 10.1016/0169-328x(95)00227-j. [DOI] [PubMed] [Google Scholar]
- Eckert A, Cotman CW, Zerfass R, Hennerici M, Muller WE. Lymphocytes as cell model to study apoptosis in Alzheimer's disease: vulnerability to programmed cell death appears to be altered. 1998;54:259–267. doi: 10.1007/978-3-7091-7508-8_25. [DOI] [PubMed] [Google Scholar]
- Eckert A, Forstl H, Zerfass R, Hartmann H, Muller WE. Lymphocytes and neutrophils as peripheral models to study the effect of beta-amyloid on cellular calcium signalling in Alzheimer's disease. Life Sci. 1996;59:499–510. doi: 10.1016/0024-3205(96)00329-3. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Sills GJ, Butler E, Thompson GG, Lindsay K, Duncan R, Howatson A, Brodie MJ. Effects of valproate, vigabatrin and tiagabine on GABA uptake into human astrocytes cultured from foetal and adult brain tissue. Epileptic Disord. 1999;1:153–157. [PubMed] [Google Scholar]
- Fuller LC, Cornelius SK, Murphy CW, Wiens DJ. Neural crest cell motility in valproic acid. Reprod Toxicol. 2002;16:825–839. doi: 10.1016/s0890-6238(02)00059-x. [DOI] [PubMed] [Google Scholar]
- Gambi F, Reale M, Iarlori C, Salone A, Toma L, Paladini C, De Luca G, Feliciani C, Salvatore M, Salerno RM, et al. Alzheimer patients treated with an AchE inhibitor show higher IL-4 and lower IL-1 beta levels and expression in peripheral blood mononuclear cells. J Clin Psychopharmacol. 2004;24:314–321. doi: 10.1097/01.jcp.0000125683.74595.2f. [DOI] [PubMed] [Google Scholar]
- Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665–3685. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, Sleeman JP, Lo CF, Nervi C, Pelicci PG, Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. 20. 2001:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvich N, Klein PS. Lithium and valproic acid: parallels and contrasts in diverse signaling contexts. Pharmacol Ther. 2002;96:45–66. doi: 10.1016/s0163-7258(02)00299-1. [DOI] [PubMed] [Google Scholar]
- Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004;64:1079–1086. doi: 10.1158/0008-5472.can-03-0799. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Hough C, Nakazawa T, Yamamoto T, Chuang DM. Lithium protection against glutamate excitotoxicity in rat cerebral cortical neurons: involvement of NMDA receptor inhibition possibly by decreasing NR2B tyrosine phosphorylation. J Neurochem. 2002;80:589–597. doi: 10.1046/j.0022-3042.2001.00728.x. [DOI] [PubMed] [Google Scholar]
- Hye A, Kerr F, Archer N, Foy C, Poppe M, Brown R, Hamilton G, Powell J, Anderton B, Lovestone S. Glycogen synthase kinase-3 is increased in white cells early in Alzheimer's disease. Neurosci Lett. 2005;373:1–4. doi: 10.1016/j.neulet.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Jabbour W, Pouplard-Barthelaix A, Houlgatte R, Emile J. Abnormal expression of actin in lymphocytes of Alzheimer's disease and Down's syndrome patients. J Neuroimmunol. 1992;38:199–208. doi: 10.1016/0165-5728(92)90013-b. [DOI] [PubMed] [Google Scholar]
- Jeong MR, Hashimoto R, Senatorov V, Fujimaki K, Ren M, Lee MS, Chuang DM. Valproic acid, a mood stabilizer and anticonvulsant, protects rat cerebral cortical neurons from spontaneous cell death: a role of histone deacetylase inhibition. FEBS Letters. 2003;542:74–78. doi: 10.1016/s0014-5793(03)00350-8. [DOI] [PubMed] [Google Scholar]
- Jung SS, Gauthier S, Cashman NR. Beta-amyloid precursor protein is detectable on monocytes and is increased in Alzheimer's disease. Neurobiol Aging. 1999;20:249–257. doi: 10.1016/s0197-4580(99)00051-2. [DOI] [PubMed] [Google Scholar]
- Kadioglu E, Sardas S, Aslan S, Isik E, Esat Karakaya A. Detection of oxidative DNA damage in lymphocytes of patients with Alzheimer's disease. Biomarkers. 2004;9:203–209. doi: 10.1080/13547500410001728390. [DOI] [PubMed] [Google Scholar]
- Kalman J, Kitajka K, Pakaski M, Zvara A, Juhasz A, Vincze G, Janka Z, Puskas LG. Gene expression profile analysis of lymphocytes from Alzheimer's patients. Psychiatr Genet. 2005;15:1–6. doi: 10.1097/00041444-200503000-00001. [DOI] [PubMed] [Google Scholar]
- Kim AJ, Shi Y, Austin RC, Werstuck GH. Valproate protects cells from ER stress-induced lipid accumulation and apoptosis by inhibiting glycogen synthase kinase-3. J Cell Sci. 2005;118:89–99. doi: 10.1242/jcs.01562. [DOI] [PubMed] [Google Scholar]
- Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366:549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutner S, Schindowski K, Frolich L, Maurer K, Kratzsch T, Eckert A, Muller WE. Enhanced ROS-generation in lymphocytes from Alzheimer's patients. Pharmacopsychiatry. 2005;38:312–315. doi: 10.1055/s-2005-916186. [DOI] [PubMed] [Google Scholar]
- Loy R, Tariot PN. Neuroprotective properties of valproate: potential benefit for AD and tauopathies. J Mol Neurosci. 2002;19:303–307. doi: 10.1385/jmn:19:3:301. [DOI] [PubMed] [Google Scholar]
- Lubec G, Krapfenbauer K, Fountoulakis M. Proteomics in brain research: potentials and limitations. Prog Neurobiol. 2003;69:193–211. doi: 10.1016/s0301-0082(03)00036-4. [DOI] [PubMed] [Google Scholar]
- Lyketsos CG, Lopez O, Jones B, Fitzpatrick AL, Breitner J, DeKosky S. Prevalence of neuropsychiatric symptoms in dementia and mild cognitive impairment: results from the cardiovascular health study. Jama. 2002;288:1475–1483. doi: 10.1001/jama.288.12.1475. [DOI] [PubMed] [Google Scholar]
- Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res. 2005;65:3815–3822. doi: 10.1158/0008-5472.CAN-04-2478. [DOI] [PubMed] [Google Scholar]
- Marx F, Blasko I, Zisterer K, Grubeck-Loebenstein B. Transfected human B cells: a new model to study the functional and immunostimulatory consequences of APP production. 1999;34:783–795. doi: 10.1016/s0531-5565(99)00049-2. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Chan SL, Camandola S. Presenilin mutations and calcium signaling defects in the nervous and immune systems. Bioessays. 2001;23:733–744. doi: 10.1002/bies.1103. [DOI] [PubMed] [Google Scholar]
- Mazzola JL, Sirover MA. Reduction of glyceraldehyde-3-phosphate dehydrogenase activity in Alzheimer's disease and in Huntington's disease fibroblasts. J Neurochem. 2001;76:442–449. doi: 10.1046/j.1471-4159.2001.00033.x. [DOI] [PubMed] [Google Scholar]
- Mecocci P, Polidori MC, Cherubini A, Ingegni T, Mattioli P, Catani M, Rinaldi P, Cecchetti R, Stahl W, Senin U, Beal MF. Lymphocyte oxidative DNA damage and plasma antioxidants in Alzheimer disease. 59. 2002:794–798. doi: 10.1001/archneur.59.5.794. [DOI] [PubMed] [Google Scholar]
- Mirinics ZK, Calafat J, Udby L, Lovelock J, Kjeldsen L, Rothermund K, Sisodia SS, Borregaard N, Corey SJ. Identification of the presenilins in hematopoietic cells with localization of presenilin 1 to neutrophil and platelet granules. Blood Cells Mol Dis. 2002;28:28–38. doi: 10.1006/bcmd.2002.0486. [DOI] [PubMed] [Google Scholar]
- Mora A, Gonzalez-Polo RA, Fuentes JM, Soler G, Centeno F. Different mechanisms of protection against apoptosis by valproate and Li+ Eur J Biochem. 1999;266:886–891. doi: 10.1046/j.1432-1327.1999.00919.x. [DOI] [PubMed] [Google Scholar]
- Morland C, Boldingh KA, Iversen EG, Hassel B. Valproate is neuroprotective against malonate toxicity in rat striatum: an association with augmentation of high-affinity glutamate uptake. J Cereb Blood Flow Metab. 2004;24:1226–1234. doi: 10.1097/01.WCB.0000138666.25305.A7. [DOI] [PubMed] [Google Scholar]
- Morocz M, Kalman J, Juhasz A, Sinko I, McGlynn AP, Downes CS, Janka Z, Rasko I. Elevated levels of oxidative DNA damage in lymphocytes from patients with Alzheimer's disease. 2002;23:47–53. doi: 10.1016/s0197-4580(01)00257-3. [DOI] [PubMed] [Google Scholar]
- Nagasaka Y, Dillner K, Ebise H, Teramoto R, Nakagawa H, Lilius L, Axelman K, Forsell C, Ito A, Winblad B, et al. A unique gene expression signature discriminates familial Alzheimer's disease mutation carriers from their wild-type siblings. Proc Natl Acad Sci U S A. 2005;102:14854–14859. doi: 10.1073/pnas.0504178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Combrinck M, Budge M, McShane R. Cell cycle kinesis in lymphocytes in the diagnosis of Alzheimer's disease. 2002;317:81–84. doi: 10.1016/s0304-3940(01)02442-9. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Palotas A, Kalman J, Palotas M, Juhasz A, Janka Z, Penke B. Beta-amyloid-induced increase in the resting intracellular calcium concentration gives support to tell Alzheimer lymphocytes from control ones. Brain Res Bull. 2002;58:203–205. doi: 10.1016/s0361-9230(02)00773-6. [DOI] [PubMed] [Google Scholar]
- Palotas A, Puskas LG, Kitajka K, Palotas M, Molnar J, Pakaski M, Janka Z, Penke B, Kalman J. Altered response to mirtazapine on gene expression profile of lymphocytes from Alzheimer's patients. Eur J Pharmacol. 2004a;497:247–254. doi: 10.1016/j.ejphar.2004.06.059. [DOI] [PubMed] [Google Scholar]
- Palotas A, Puskas LG, Kitajka K, Palotas M, Molnar J, Pakaski M, Janka Z, Penke B, Kalman J. The effect of citalopram on gene expression profile of Alzheimer lymphocytes. Neurochem Res. 2004b;29:1563–1570. doi: 10.1023/b:nere.0000029570.57903.74. [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- Porsteinsson AP, Tariot PN, Erb R, Gaile S. An open trial of valproate for agitation in geriatric neuropsychiatric disorders. Am J Geriatr Psychiatry. 1997;5:344–351. doi: 10.1097/00019442-199700540-00010. [DOI] [PubMed] [Google Scholar]
- Porsteinsson AP, Tariot PN, Jakimovich LJ, Kowalski N, Holt C, Erb R, Cox C. Valproate therapy for agitation in dementia: open-label extension of a double-blind trial. Am J Geriatr Psychiatry. 2003;11:434–440. [PubMed] [Google Scholar]
- Profenno LA, Jakimovich L, Holt CJ, Porsteinsson A, Tariot PN. A randomized, double-blind, placebo-controlled pilot trial of safety and tolerability of two doses of divalproex sodium in outpatients with probable Alzheimer's disease. Curr Alzheimer Res. 2005;2:553–558. doi: 10.2174/156720505774932205. [DOI] [PubMed] [Google Scholar]
- Reale M, Iarlori C, Gambi F, Feliciani C, Salone A, Toma L, DeLuca G, Salvatore M, Conti P, Gambi D. Treatment with an acetylcholinesterase inhibitor in Alzheimer patients modulates the expression and production of the pro-inflammatory and anti-inflammatory cytokines. J Neuroimmunol. 2004;148:162–171. doi: 10.1016/j.jneuroim.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Reale M, Iarlori C, Gambi F, Lucci I, Salvatore M, Gambi D. Acetylcholinesterase inhibitors effects on oncostatin-M, interleukin-1 beta and interleukin-6 release from lymphocytes of Alzheimer's disease patients. Exp Gerontol. 2005;40:165–171. doi: 10.1016/j.exger.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Ren M, Leng Y, Jeong M, Leeds PR, Chuang DM. Valproic acid reduces brain damage induced by transient focal cerebral ischemia in rats: potential roles of histone deacetylase inhibition and heat shock protein induction. J Neurochem. 2004;89:1358–1367. doi: 10.1111/j.1471-4159.2004.02406.x. [DOI] [PubMed] [Google Scholar]
- Scali C, Prosperi C, Bracco L, Piccini C, Baronti R, Ginestroni A, Sorbi S, Pepeu G, Casamenti F. Neutrophils CD11b and fibroblasts PGE(2) are elevated in Alzheimer's disease. Neurobiol Aging. 2002;23:523–530. doi: 10.1016/s0197-4580(01)00346-3. [DOI] [PubMed] [Google Scholar]
- Shen WT, Wong TS, Chung WY, Wong MG, Kebebew E, Duh QY, Clark OH. Valproic acid inhibits growth, induces apoptosis, and modulates apoptosis-regulatory and differentiation gene expression in human thyroid cancer cells. Surgery. 2005;138:979–984. doi: 10.1016/j.surg.2005.09.019. discussion 984-975. [DOI] [PubMed] [Google Scholar]
- Sival RC, Haffmans PM, Jansen PA, Duursma SA, Eikelenboom P. Sodium valproate in the treatment of aggressive behavior in patients with dementia--a randomized placebo controlled clinical trial. 2002;17:579–585. doi: 10.1002/gps.653. [DOI] [PubMed] [Google Scholar]
- Straface E, Matarrese P, Gambardella L, Vona R, Sgadari A, Silveri MC, Malorni W. Oxidative imbalance and cathepsin D changes as peripheral blood biomarkers of Alzheimer disease: a pilot study. FEBS Lett. 2005;579:2759–2766. doi: 10.1016/j.febslet.2005.03.094. [DOI] [PubMed] [Google Scholar]
- Sugai F, Yamamoto Y, Miyaguchi K, Zhou Z, Sumi H, Hamasaki T, Goto M, Sakoda S. Benefit of valproic acid in suppressing disease progression of ALS model mice. Eur J Neurosci. 2004;20:3179–3183. doi: 10.1111/j.1460-9568.2004.03765.x. [DOI] [PubMed] [Google Scholar]
- Sumner CJ, Huynh TN, Markowitz JA, Perhac JS, Hill B, Coovert DD, Schussler K, Chen X, Jarecki J, Burghes AH, et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- Tacconi S, Perri R, Balestrieri E, Grelli S, Bernardini S, Annichiarico R, Mastino A, Caltagirone C, Macchi B. Increased caspase activation in peripheral blood mononuclear cells of patients with Alzheimer's disease. Exp Neurol. 2004;190:254–262. doi: 10.1016/j.expneurol.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Takeda M, Tatebayashi Y, Nishimura T. Change in the cytoskeletal system in fibroblasts from patients with familial Alzheimer's disease. Prog Neuropsychopharmacol Biol Psychiatry. 1992;16:317–328. doi: 10.1016/0278-5846(92)90083-q. [DOI] [PubMed] [Google Scholar]
- Tariot PN, Loy R, Ryan JM, Porsteinsson A, Ismail S. Mood stabilizers in Alzheimer's disease: symptomatic and neuroprotective rationales. Adv Drug Deliv Rev. 2002;54:1567–1577. doi: 10.1016/s0169-409x(02)00153-9. [DOI] [PubMed] [Google Scholar]
- Tariot PN, Raman R, Jakimovich L, Schneider L, Porsteinsson A, Thomas R, Mintzer J, Brenner R, Schafer K, Thal L. Divalproex sodium in nursing home residents with possible or probable Alzheimer Disease complicated by agitation: a randomized, controlled trial. Am J Geriatr Psychiatry. 2005;13:942–949. doi: 10.1176/appi.ajgp.13.11.942. [DOI] [PubMed] [Google Scholar]
- Tayebati SK, Amenta F, Amici S, El Assouad D, Gallai V, Ricci A, Parnetti L. Peripheral blood lymphocytes muscarinic cholinergic receptor subtypes in Alzheimer's disease: a marker of cholinergic dysfunction? 2001;121:126–131. doi: 10.1016/s0165-5728(01)00435-0. [DOI] [PubMed] [Google Scholar]
- Urcelay E, Ibarreta D, Parrilla R, Ayuso MS, Martin-Requero A. Enhanced proliferation of lymphoblasts from patients with Alzheimer dementia associated with calmodulin-dependent activation of the na+/H+ exchanger. 2001;8:289–298. doi: 10.1006/nbdi.2000.0381. [DOI] [PubMed] [Google Scholar]
- Velez-Pardo C, Ospina GG, Jimenez del Rio M. Abeta[25–35] peptide and iron promote apoptosis in lymphocytes by an oxidative stress mechanism: involvement of H2O2, caspase-3, NF-kappaB, p53 and c-Jun. Neurotoxicology. 2002;23:351–365. doi: 10.1016/s0161-813x(02)00081-5. [DOI] [PubMed] [Google Scholar]
- Wang JF, Azzam JE, Young LT. Valproate inhibits oxidative damage to lipid and protein in primary cultured rat cerebrocortical cells. Neuroscience. 2003;116:485–489. doi: 10.1016/s0306-4522(02)00655-3. [DOI] [PubMed] [Google Scholar]
- Weiner MF, Hynan LS, Bret ME, White C., 3rd Early behavioral symptoms and course of Alzheimer's disease. Acta Psychiatr Scand. 2005;111:367–371. doi: 10.1111/j.1600-0447.2004.00472.x. [DOI] [PubMed] [Google Scholar]
- Wong GT, Manfra D, Poulet FM, Zhang Q, Josien H, Bara T, Engstrom L, Pinzon-Ortiz M, Fine JS, Lee HJ, et al. Chronic treatment with the gamma-secretase inhibitor LY-411,575 inhibits beta-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J Biol Chem. 2004;279:12876–12882. doi: 10.1074/jbc.M311652200. [DOI] [PubMed] [Google Scholar]
- Yildirim E, Zhang Z, Uz T, Chen CQ, Manev R, Manev H. Valproate administration to mice increases histone acetylation and 5-lipoxygenase content in the hippocampus. Neurosci Lett. 2003;345:141–143. doi: 10.1016/s0304-3940(03)00490-7. [DOI] [PubMed] [Google Scholar]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- Zhao WQ, Feng C, Alkon DL. Impairment of phosphatase 2A contributes to the prolonged MAP kinase phosphorylation in Alzheimer's disease fibroblasts. Neurobiol Dis. 2003;14:458–469. doi: 10.1016/s0969-9961(03)00124-4. [DOI] [PubMed] [Google Scholar]
- Zhao WQ, Ravindranath L, Mohamed AS, Zohar O, Chen GH, Chen GH, Etcheberrigaray R, Alkon DL. MAP kinase signaling cascade dysfunction specific to Alzheimer's disease in fibroblasts. Neurobiol Dis. 2002;11:166–183. doi: 10.1006/nbdi.2002.0520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.