Abstract
The process of arginine-dependent extreme acid resistance (XAR) is one of several decarboxylase-antiporter systems that protects Escherichia coli and possibly other enteric bacteria from exposure to the strong acid environment of the stomach. Arginine-dependent acid resistance depends on an intracellular proton-utilizing arginine α-decarboxylase and a membrane transport protein necessary for delivering arginine to and removing agmatine, its decarboxylation product, from the cytoplasm. The arginine system afforded significant protection to wild-type E. coli cells in our acid shock experiments. The gene coding for the transport protein is identified here as a putative membrane protein of unknown function, YjdE, which we now name adiC. Strains from which this gene is deleted fail to mount arginine-dependent XAR, and they cannot perform coupled transport of arginine and agmatine. Homologues of this gene are found in other bacteria in close proximity to homologues of the arginine decarboxylase in a gene arrangement pattern similar to that in E coli. Evidence for a lysine-dependent XAR system in E. coli is also presented. The protection by lysine, however, is milder than that by arginine.
The family Enterobacteriaceae includes some of the most frequently encountered and virulent pathogens, such as Escherichia coli strain O157:H7 and Shigella, Salmonella, and Yersinia spp. (8). Infectiousness in these organisms, as well as colonization by benign ones, relies upon an ability to withstand exposure to strongly acidic pH (pH <3.0) for up to several hours to permit safe passage through the human stomach on the way to the gut (1, 13, 33). The mechanisms by which enteric bacteria achieve extreme acid resistance have been thoroughly investigated during the past few years (1, 9), and for E. coli, the genetic and environmental requirements for the process are understood in broad outline (5, 7, 21, 22, 31). Extreme acid resistance, which we name XAR according to the nomenclature of Hersh et al. (15), refers to the following features: (i) robust survival upon extended exposure to strong acid (>1 h at 37°C in HCl, pH <3), (ii) requirement for fermentative growth in rich medium to stationary phase prior to acid exposure, (iii) no requirement for an adaptation step at a mild acid pH prior to acid shock below pH 3.0, and (iv) dependence on certain amino acids during acid shock (4, 19, 20). We explicitly ascribe to XAR the above-described features to avoid confusion with the previously described E. coli acid tolerance response (9, 26) or with the oxidative acid resistance response (4, 19), the former operating at mild acid shock and the latter not requiring supplementation during extreme acid shock.
Two XAR systems, thus defined, are currently known to exist in E. coli. One depends on the availability of glutamate, and the other depends on arginine (4, 19, 20). The common theme underpinning both systems is amino acid decarboxylation in the cytoplasm, which is the proton-consuming chemical reaction that counteracts lethal intracellular acidification in an HCl-rich environment. The role of amino acid decarboxylases in ameliorating internal acid pH stress was suggested by Gale over 50 years ago (11), and recently, several laboratories have identified the genes involved. The E. coli genome contains genes for two glutamate decarboxylases, gadA and gadB, and an arginine decarboxylase gene, adiA, that carry out this function (4, 19, 20, 28, 29). In addition, each XAR system requires a second component, an inner membrane antiporter used to deliver the amino acid substrate to, and remove the decarboxylated product from, the cytoplasm (4, 27). The glutamate-γ-aminobutyrate (glutamate-GABA) exchanger gene, gadC, serves this function in glutamate-dependent XAR (4, 15, 16). However, the antiporter required to exchange arginine for its decarboxylation product, agmatine, has not previously been found. Here we identify the gene coding for the E. coli arginine-agmatine exchanger used in XAR, a membrane protein spanning 12 amino acids of unknown function, designated yjdE (b4115) and now named adiC by us and, recently, by others (12). The adiC gene is a homologue of a gene, aniC, in Salmonella enterica serovar Typhimurium that is conjectured to be the possible arginine-agmatine exchanger in arginine XAR (25) and was previously identified from a genetic analysis by using lacZ transcriptional fusion proteins activated under conditions of low pH (10). We show that adiC functions specifically in arginine XAR and that the dependence of transport activity on arginine concentration and pH naturally accounts for its role in maintaining cell viability under strong acid stress conditions.
MATERIALS AND METHODS
Genetic techniques, bacterial strains, plasmids, and growth conditions.
The yjdE/adiC deletion (ΔyjdE) strain was constructed with the one-step gene inactivation technique (6). In the Swiss-Prot database, YjdE is listed as a hypothetical transport protein (accession no. P39269). The gene deletion was transferred to the MG1655 background by using standard P1 transduction techniques (23). All experimental results shown here were in the background of the MG1655 E. coli strain, which we designate the wild type. To create rescue plasmid pRI-adiC1, the yjdE coding region was cloned by PCR from genomic DNA and inserted behind a lac promoter between the XbaI and HindIII sites in the plasmid pWSK29 (32). The sequence of the insert was confirmed and is identical to that deposited in the database.
In all acid shock experiments, cells were prepared by growth to stationary phase (18 to 20 h at 37°C) in 50-ml conical tubes in brain heart infusion broth with 0.4% glucose (BHIG). The BHIG contained ∼17 mM disodium phosphate and had a starting pH of 7.4 ± 0.2. After growth to stationary phase, the pH was ∼5.5. For rescue experiments, ΔyjdE cells were transformed with the rescue plasmid and then grown to stationary phase in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside) at the desired concentration.
Acid shock experiments.
Acid shock experiments were carried out in a pH 2.5 minimal shock medium containing 40 mM KCl, 80 mM KH2PO4, 33 mM H3PO4, 1.7 mM Na3 citrate, and 20 mM glucose with or without added amino acids (1 mM in most experiments). For some experiments, pH varied in the range of 2.1 to 4.0 by adjusting the KPi buffers while the solution osmolarity was maintained at 305 ± 5 mosM, a maneuver that necessitated a covariation of 110 to 135 mM in K+ and 140 to 101 mM in Pi. Cell survival was assayed by diluting a 10-μl aliquot of stationary-phase culture into 1 ml of prewarmed acid shock medium and incubating the culture at 37°C for 2 h; after appropriate serial dilution, cells were plated on Luria-Bertani agar plates for counting of well-separated colonies after overnight growth. Survival efficiency is defined as the percentage of survivor colonies relative to the number of colonies plated directly from the stationary-phase culture without the acid shock step.
For measurement of arginine and agmatine transport rates during acid shock, an aliquot of the stationary-phase culture was pelleted and resuspended in 1 ml of prewarmed shock medium (final cell density, 5 × 108 cells/ml). About 15 s after dilution, transport was initiated by adding arginine (10 μM to 1 mM). At each time point, a 55-μl aliquot was removed and added to an Eppendorf tube containing 5.5 μl of 1.1 M KOH, a step that instantly stops transport by neutralizing the medium (16). The suspension was centrifuged for 15 s at 14,000 rpm in an Eppendorf centrifuge, and 20 μl of the supernatant was removed for determination of arginine and agmatine.
Quantitation of arginine and agmatine.
Arginine and agmatine were quantified by reverse-phase high-pressure liquid chromatography after precolumn derivatization with phenyl isothiocyanate (PITC) according to standard methods (14). The acid shock supernatant aliquot containing 1 to 20 nmol of arginine or agmatine was added to 100 μl of a solution of 0.5% (vol/vol) PITC dissolved in freshly mixed acetonitrile-pyridine-triethylamine (10/5/2, by volume). The mixture was incubated for 10 min at room temperature and then evaporated overnight in a vacuum centrifuge. The residue was dissolved in 200 μl of water, and 80 μl of this was injected onto a Vydac C18 column; arginine and agmatine peaks eluting in a gradient (2 to 50% for 20 min) of water-acetonitrile, with 0.1 M ammonium acetate in each solvent, were measured by comparison of either peak heights or integrated areas against appropriate standards.
RESULTS
Amino acid requirement for extreme acid survival.
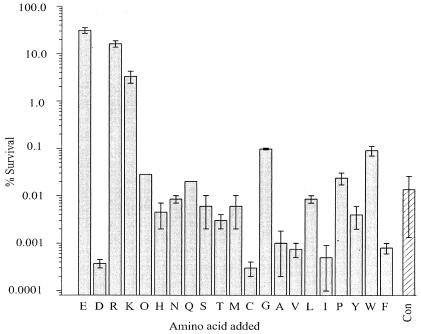
We conducted a brief survey of 21 natural amino acids to compare their relative levels of effectiveness in protecting E. coli cells from strong acid exposure. Cells were challenged with acid shock conditions (37°C for 2 h, pH 2.5) in the presence of a 1 mM concentration of each amino acid, and survival efficiency was assessed (Fig. 1). Control cells with no added amino acid are highly susceptible to acid shock (<0.05% survival), but an addition of 1 mM glutamate or arginine to the shock medium raises the survival efficiency by many orders of magnitude (30 or 15%, respectively), as is well known (4, 19). Figure 1 further shows that lysine also supports XAR, albeit at a lower efficiency (∼3%) than glutamate or arginine. No other amino acid tested produced robust survival.
FIG. 1.
Dependence on amino acids for survival. Wild-type E. coli stationary-phase cells were subjected to acid shock at pH 2.5 in the presence of a 1 mM concentration of the amino acid indicated by the single-letter code (with “O” representing ornithine). Bars and error bars indicate the means ± ranges of two to three measurements. CON, control experiment with no added amino acid.
Identification of the arginine-agmatine transporter underlying arginine-dependent XAR.
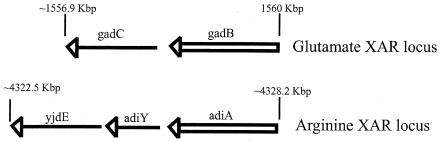
Biological necessity demands that for XAR to operate, E. coli must have a membrane transport protein to feed the arginine decarboxylase gene, adiA, with substrate and to expel the product into the medium. To search for this elusive transporter, we examined the E. coli genome to see whether the arrangement of genes involved in glutamate-dependent XAR is recapitulated near adiA. As shown in Fig. 2, the glutamate XAR locus in E. coli consists of two genes arranged in tandem: gadC, the glutamate-GABA exchanger gene, followed by gadB, the glutamate decarboxylase gene (2). Inspection of the region near adiA reveals a putative 12-amino acid transmembrane protein coding region designated yjdE in unknown-function nomenclature, as well as a region coding for small, AraC-like putative regulatory protein, AdiY. The close physical proximity of yjdE to adiA resembles the arrangement of the glutamate XAR locus and thereby points to YjdE as the arginine-agmatine exchanger.
FIG. 2.
Arrangement of XAR genes in E. coli. Positions of genes are indicated according to the work of Blattner et al. (2). The glutamate XAR system contains additional genes located elsewhere in the chromosome (21, 31).
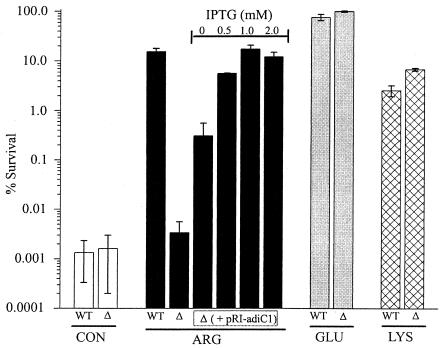
We tested this idea by deleting the yjdE gene from E. coli MG1655. The deletion specifically and completely abolished arginine-dependent XAR (Fig. 3). Survival efficiency for ΔyjdE cells under control conditions (no added amino acids) was very low (<0.01%), but in contrast to what occurred with the wild type, the addition of arginine failed to increase survival. Moreover, the ΔyjdE cells survived normally in the presence of glutamate, a result that further confirms the independence of the two XAR systems (4). Lysine-dependent acid protection was also functional in the ΔyjdE cells. Arginine-dependent XAR can be rescued in the deletion strain by delivering the yjdE coding region under lac promoter control on a low-copy-number plasmid; rescue was partial in the absence of IPTG but rose to wild-type survival efficiency at 1 mM IPTG. These results demonstrate that the absence of yjdE alone was responsible for the acid sensitivity of these mutants, and the results highlight the role of this gene in the arginine-dependent XAR mechanism. We confirmed (data not shown) that arginine decarboxylase activity (16) in ΔyjdE cells is comparable to wild-type values.
FIG. 3.
Effect of yjdE deletion on XAR. E. coli (wild-type [WT] or ΔyjdE [Δ]) cells were assayed for XAR in the absence of any amino acids (control [CON]; open bars) or in the presence of 1 mM arginine (black bars), glutamate (grey bars), or lysine (cross-hatched bars). The rescue experiments were conducted with ΔyjdE cells that had been transformed with pRI-adiC1 and induced with the indicated IPTG concentration. For glutamate experiments, cells were grown in Luria-Bertani agar plus 0.4% glucose medium before acid shock. Each bar represents the mean ± standard error of results from 3 to 14 independent experiments.
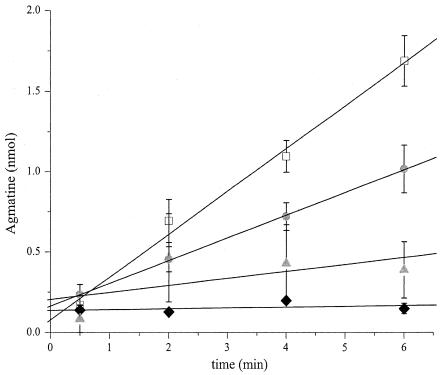
When encountering an acid challenge in the presence of arginine, wild-type E. coli cells take up arginine and extrude agmatine with a 1:1 stoichiometry (16). The immediate release of agmatine in response to acid shock, illustrated in Fig. 4, reflects this exchange process. The initial rate of agmatine release in wild-type cells, 0.26 nmol/min, corresponds to a turnover rate of ∼5,000 molecules/s/cell. Arginine-agmatine exchange in the yjdE cells, however, was completely inhibited. This transport deficiency was partially rescued by transformation of the deletion strain with the yjdE rescue vector, once again in an IPTG-dependent manner. Maximal transport occurred at 1 mM IPTG, at about half the rate seen in the wild type. Inducer concentrations above 1 mM had no additional effect on transport (data not shown).
FIG. 4.
Effect of yjdE deletion on arginine-agmatine exchange during acid shock. Time courses of agmatine release to the extracellular medium were monitored for wild-type cells (open symbols), ΔyjdE cells (black symbols), or ΔyjdE cells transformed with the pRI-adiC1 rescue vector (grey symbols). The transport reactions were initiated by the addition of 1 mM arginine to the shock medium. Initial rates were determined by least-squares fitting (solid lines) and are 0.26 ± 0.02 and 0.005 ± 0.008 nmol/min for wild-type and ΔyjdE cells, respectively. In rescue experiments, cells were either uninduced (triangles) or induced with 1 mM IPTG (circles), giving initial rates of 0.05 ± 0.04 and 0.141 ± 0.002 nmol/min, respectively. Each data point represents the mean ± standard error of results from three to six independent experiments.
pH and substrate dependence of arginine-dependent XAR.
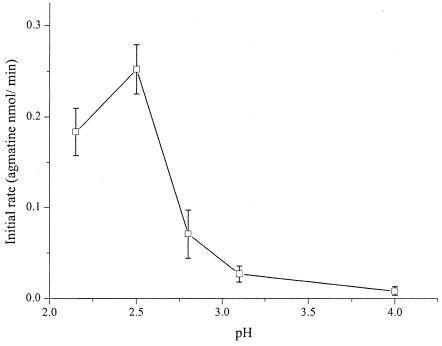
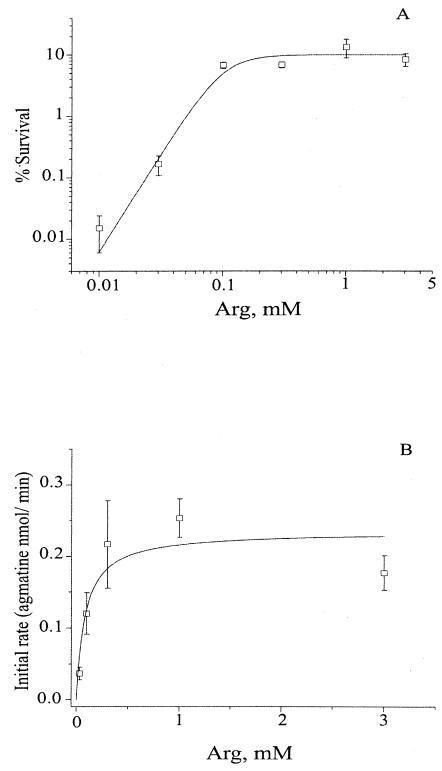
It is known that arginine transport is active under strongly acidic conditions and is silenced at neutral pH (16). We document the pH dependence of the transport phenomenon in more detail in Fig. 5. The agmatine extrusion rate is maximal near pH 2.5, falling sharply above this value and becoming undetectable by pH 4.0. Both survival and exchange rates varied with arginine concentration (Fig. 6). Survival efficiency rose steeply with arginine (Hill coefficient, 3.2) at a half-maximal concentration of ∼100 μM. In parallel, the rate of agmatine release also increased in a saturating fashion with arginine concentration, with a similar value for half-maximal concentration.
FIG. 5.
pH dependence of arginine-agmatine exchange. The initial rate of agmatine release was measured as a function of pH in the acid shock medium, as described for Fig. 4. Points represent means ± standard errors of results from triplicate time courses, except for the pH 2.8 data, which show the range of results from duplicate experiments.
FIG. 6.
Concentration dependence of arginine XAR. Wild-type cells were assayed for survival efficiency or the arginine-agmatine exchange rate. (A) For survival data, the solid curve is calculated with the Hill equation, with a half-maximal concentration of 100 μM and a slope factor of 3.2. (B) Initial transport rate data follow a Michaelis-Menten curve, with a half-maximal concentration of 83 μM. These curves are purely empirical and have no theoretical connotations.
DISCUSSION
For several years, three types of stationary-phase acid resistance phenomena have been described for E. coli: two systems apparent during fermentative metabolism—one dependent on glutamate and the other dependent on arginine—and a third amino acid-independent system expressed during oxidative growth (1, 4). XAR as defined here refers exclusively to the amino acid-dependent fermentative systems. Each of the XAR systems is based on a proton-consuming decarboxylase and an amino acid-amine antiporter protein. We have hypothesized (16) that the antiport of arginine and agmatine at pH 2.5 is electrogenic, resulting in the transient hyperpolarization of the inner membrane. This hyperpolarization may be relieved by the shunting of a negative charge via the ClC-type Cl− channel in E. coli, thus allowing the uninterrupted cycling of the antiporter (16). Thus, deletion of the ClC-type Cl− channel would hinder the cycling of arginine and agmatine by AdiC and lead to acid sensitivity (16). The genes coding for the decarboxylases and for the glutamate-GABA antiporter are known, but the arginine-agmatine exchanger has not been identified in past investigations. We fill this gap here by showing that the unknown-function gene yjdE codes for this exchanger. This gene is located close to the arginine decarboxylase gene adiA, an arrangement reminiscent of the glutamate XAR system genes, gadB and gadC, that are known to cohabit an operon (4). Accordingly, we propose to rename yjdE adiC in a manner analogous to that used to name the gadC glutamate-GABA exchanger. The primary focus of this investigation has been the verification of AdiC as the transporter crucial in arginine XAR, but we have also presented the results of supplementary experiments to fill out some details of these acid resistance systems.
A supplementary finding was that lysine-dependent XAR exists in E. coli. The effectiveness of lysine-dependent acid tolerance response in protecting S. enterica serovar Typhimurium cells in strong acid (pH 3.0) has been thoroughly documented (26). Lin et al. (19) reported lysine to be ineffective in protecting E. coli cells for all of the three strains they tested. We employed E. coli wild-type MG1655 grown in BHIG for these experiments. The differences between the results of Lin et al. (19) and ours may lie in the choice of strain and/or medium for overnight growth. It has been known for some time that E. coli uses a lysine decarboxylase and its accompanying lysine-cadaverine exchanger, the cadBA system, to protect against mild acid shock (24, 34) and that a similar set of genes works analogously in S. enterica serovar Typhimurium, providing protection at pH 3.0 (26). We show here an analogous lysine-mediated protective effect under XAR conditions in E. coli wild-type MG1655 cells, which is an effect not observed for other strains of E. coli tested (19). Park et al. (26) also showed that deleting the lysine decarboxylase gene (cadA) eliminated this lysine-dependent process in S. enterica serovar Typhimurium cells. In this context, the cadBA system is a reasonable guess for the genetic underpinnings of E. coli lysine XAR. Testing the viability of lysine-dependent XAR in a ΔcadB strain is an obvious future experiment. We do know, however, that lysine-dependent XAR does not reflect promiscuity of the arginine system towards lysine, since deletion of adiC, which fully inhibits the arginine system, has no effect on the lysine system.
It is as clear as it is unsurprising that functional transport activity is intimately linked to cell survival during acid shock. The rate of arginine-agmatine exchange and the efficiency of survival vary similarly with arginine concentration, with a half-maximal concentration of ∼100 μM for each. Because of the complexity of the multiple tightly coupled steps in the acid resistance process, this does not mean that transport is the rate-limiting step in survival. Nevertheless, the close correlation of survival and adiC-catalyzed transport buttresses the picture of robust substrate transport as a necessary condition for cell survival in strong acid.
Rates of arginine-agmatine exchange vary sharply with the pH of the medium, with a fivefold level of activation as pH drops from 2.8 to 2.5 and essentially no activity above pH 4. Since the transport assays are done on intact cells rather than an in vitro reconstituted system, we cannot validly assert that it is the transport protein itself that is directly activated by low pH. The difference between the transport pH optimum and the corresponding pH optimum of 5.2 for the purified E. coli-inducible arginine decarboxylase (3) suggests that the pH curve probably reflects a combination of the properties of the enzyme-transporter complex. However, we can say that the transporter functions well at an extracellular pH below 2.5, as is demanded by all standard models of XAR. This finding is precisely as expected if this protein plays an essential role in allowing the bacterium to survive in the stomach during its journey to the intestine. Future biochemical work on the transporter in a reconstituted system (16) would be valuable in establishing the pH dependence more accurately.
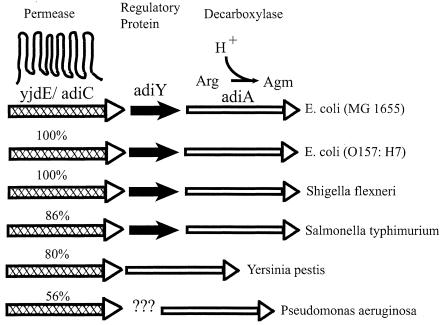
It is instructive to compare the organization of arginine-dependent XAR genes in E. coli with those in other bacteria. As illustrated in Fig. 7, adiC and adiA homologues are present in the enteric pathogens E. coli O157:H7, Shigella flexneri, S. enterica serovar Typhimurium, Yersinia pestis, and a nonenterobacterial pathogen, Pseudomonas aeruginosa. These adiC homologues share convincing sequence identity (50 to 100%), and all lie in proximity to an AdiA-like arginine decarboxylase. Like E. coli, Salmonella carries a homologue of AdiY, an AraC-like putative regulatory protein whose specific function is unknown but which may act as the fulcrum of a proper operon, since it is known to increase the expression of adiA (30). Future experiments are required to test this possibility. The common organization of these genes suggests that the natural substrates of adiC homologues are arginine and agmatine, possibly for acid resistance in enteric bacteria and perhaps for alternative physiological purposes in other organisms. Indeed, Park and colleagues (25), in their work to identify anaerobiosis- and acid-inducible genes in S. enterica serovar Typhimurium, previously named this organism's adiC homologue aniC, (for anaerobiosis inducible) and speculated that it might code for an arginine-agmatine antiporter, citing its sequence similarity to PotE, the well-characterized putrescine-ornithine exchange protein of E. coli (18). Since S. enterica serovar Typhimurium is known to survive pH 2.5 shock in the presence of arginine (17), it is likely that this adiC homologue functions in a manner similar to that described here for E. coli.
FIG. 7.
Genome organization of adiC homologues in several bacterial species. The adiC homologues are marked by cross-hatched arrows; sequence identity to E. coli adiC is as indicated. The putative regulatory protein AdiY is marked by short black arrow, and adiA arginine decarboxylase gene homologues are represented as unfilled arrows. The question marks for P. aeruginosa represent a membrane protein of unknown function. Agm, agmatine.
ADDENDUM
Another article identifying the yjdE (adiC) gene and its involvement in E. coli XAR appeared in print in the Journal of Bacteriology during the time this paper was under process of review for publication. The said paper by Gong et al. (12) has been cited in the reference section of this article.
Acknowledgments
We are grateful to C. Nimigean and A. Accardi for discussions and suggestions during the course of these experiments and to M. Walden for help with a few transport experiments.
REFERENCES
- 1. Audia, J. P., C. C. Webb, and J. W. Foster. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291:97-106. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Boeker, E. A., E. H. Fischer, and E. E. Snell. 1971. Arginine decarboxylase from Escherichia coli. IV. Structure of the pyridoxal phosphate binding site. J. Biol. Chem. 246:6776-6781. [PubMed] [Google Scholar]
- 4.Castanie-Cornet, M.-P., T. A. Penfound, D. Smith, J. F. Elliot, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castanie-Cornet, M.-P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 8.Donenberg, M. S. 2000. Pathogenic strategies of enteric bacteria. Nature 406:768-774. [DOI] [PubMed] [Google Scholar]
- 9.Foster, J. W., and M. Moreno. 1999. Inducible acid tolerance mechanisms in enteric bacteria. Novartis Found. Symp. 221:55-74. [DOI] [PubMed] [Google Scholar]
- 10.Foster, J. W., Y. K. Park, I. S. Bang, K. Karem, H. Betts, H. K. Hall, and E. Shaw. 1994. Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology 140:341-352. [DOI] [PubMed] [Google Scholar]
- 11.Gale, E. F. 1946. The bacterial amino acid decarboxylases. Adv. Enzymol. 6:1-32. [Google Scholar]
- 12.Gong, S., H. Richard, and J. W. Foster. 2003. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 185:4402-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorden, J., and P. L. C. Small. 1993. Acid resistance in enteric bacteria. Infect. Immun. 61:364-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinrikson, R. L., and S. C. Meredith. 1984. Amino acid analysis by reverse-phase high-performance liquid chromatography: precolumn derivatization with phenylisothiocyanate. Anal. Biochem. 136:65-74. [DOI] [PubMed] [Google Scholar]
- 15.Hersh, B. M., F. T. Farooq, D. N. Barstad, D. L. Blankenhorn, and J. L. Slonczewski. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer, R., T. Iverson, A. Accardi, and C. Miller. 2002. A biological role for prokaryotic ClC chloride channels. Nature 419:715-718. [DOI] [PubMed] [Google Scholar]
- 17.Jonge, R., W. S. Ritmeester, and F. M. Leusden. 2003. Adaptive responses of Salmonella enterica serovar typhimurium DT104 and other S. typhimurium strains and Escherichia coli O157 to low pH environments. J. Appl. Microbiol. 94:625-632. [DOI] [PubMed] [Google Scholar]
- 18.Kashiwagi, K., S. Shibuya, H. Tomitori, A. Kuraishi, and K. Igarashi. 1997. Excretion and uptake of putrescine by the PotE protein in Escherichia coli. J. Biol. Chem. 272:6318-6323. [DOI] [PubMed] [Google Scholar]
- 19.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, Z., H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2002. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184:7001-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng, S. Y., and G. N. Bennett. 1992. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J. Bacteriol. 174:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Neely, M. N., C. L. Dell, and E. R. Olson. 1994. Roles of LysP and CadC in mediating the lysine requirement for acid induction of the Escherichia coli cad operon. J. Bacteriol. 176:3278-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, K. R., J.-C. Giard, J. H. Eom, S. Bearson, and J. W. Foster. 1999. Cyclic AMP receptor protein and TyrR are required for acid pH and anaerobic induction of hyaB and aniC in Salmonella typhimurium. J. Bacteriol. 181:689-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, Y. K., B. Bearson, S. H. Bang, I. S. Bang, and J. W. Foster. 1996. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol. Microbiol. 20:605-611. [DOI] [PubMed] [Google Scholar]
- 27.Small, P. L. C., and S. R. Waterman. 1998. Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli. Trends Microbiol. 6:214-216. [DOI] [PubMed] [Google Scholar]
- 28.Smith, D. K., T. Kassam, B. Singh, and J. F. Elliott. 1992. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol. 174:5820-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stim, K. P., and G. N. Bennett. 1993. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J. Bacteriol. 175:1221-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stim-Herndon, K. P., T. M. Flores, and G. N. Bennett. 1996. Molecular characterization of adiY, a regulatory gene which affects expression of the biodegradative acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology 142:1311-1320. [DOI] [PubMed] [Google Scholar]
- 31.Tramonti, A., P. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 33.Waterman, S. R., and P. L. C. Small. 1998. Acid-sensitive enteric pathogens are protected from killing under extremely acidic conditions of pH 2.5 when they are inoculated onto certain solid food sources. Appl. Environ. Microbiol. 64:3882-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson, N., D. S. Dunyak, E. L. Rosey, J. L. Slonczewski, and E. R. Olson. 1992. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J. Bacteriol. 174:530-540. [DOI] [PMC free article] [PubMed] [Google Scholar]