Abstract
Streptomyces linear plasmids and linear chromosomes can replicate also in a circular form when their telomeres are deleted. The 17-kb linear plasmid pSLA2 has been a useful model in studies of such replicons. Here we report that the minimal origin initiating replication of pSLA2-derived plasmids as circular molecules cannot propagate these plasmids in a linear mode unless they also contain a novel plasmid-encoded locus, here named rlrA (required for linear replication). In contrast with the need for rlrA to accomplish replication of telomere-containing linear plasmids, expression of rlrA, which encodes two LuxR family regulatory domains, interferes with the establishment of pSLA2 in circular form in Streptomyces lividans transformants. The additional presence of an adjacent divergently transcribed locus, rorA (rlrA override), which strongly resembles the kor (kil override) transcription control genes identified previously on Streptomyces plasmids, reversed the detrimental effects of rlrA on plasmid establishment and additionally stabilized circular plasmid inheritance by spores during the S. lividans life cycle. While the effects of the rlrA/rorA locus of pSLA2 were seen also on linear plasmids derived from the unrelated SLP2 replicon, they did not extend to plasmids whose replication was initiated at a cloned chromosomal origin. Our results establish the existence of, and provide the initial description of, a novel plasmid-borne regulatory system that differentially affects the propagation of linear and circular plasmids in Streptomyces.
Streptomyces species are among the few eubacteria known to include both linear chromosomes and linear plasmids (10, 17, 20). The telomeres of Streptomyces linear replicons contain a series of short inverted repeat DNA sequences (7, 9, 12, 18, 22) and are capped by terminal proteins linked covalently to 5′ ends of double-stranded DNA (1, 9, 20, 33). Unlike adenovirus and bacteriophage φ29, which also have terminal protein linked covalently to 5′ DNA ends but which replicate by a strand displacement mechanism (25), Streptomyces linear plasmids contain a centrally located origin and replicate bidirectionally (4). This process has been shown to leave 280 nucleotides of single-stranded DNA at the 3′ ends of pSLA2 replication intermediates; these are then filled in (4, 5), possibly by a fold-back mechanism involving the inverted repeats of telomeres (22). As Streptomyces chromosomes also appear to duplicate their genes bidirectionally (21) and Streptomyces coelicolor and Streptomyces lividans chromosomal telomeres are highly similar to those of pSLA2 (12, 22), the filling in of recessed 5′ ends of linear chromosomes is presumed to occur by a similar mechanism.
Streptomyces linear replicons can replicate in both linear and circular form when their telomeres are deleted (4, 19, 20, 27, 30). The site of initiation of DNA replication for Streptomyces linear plasmid pSLA2 has been identified experimentally (4) and found to include a region containing short direct repeats (iterons) as well as a DNA helicase gene (5). A related region has been shown to encode replication functions of linear plasmid SCP1 (24), and an analogous locus identified in linear plasmid SLP2 by sequence analysis is inferred to contain the replication origin of that plasmid (11). Although pSLA2 linear plasmids normally exist stably at high copy number in Streptomyces cell populations (10), pSLA2 circular plasmids containing the minimal origin replicate at low copy number and are not stably inherited (5), suggesting that genes and/or sites involved in plasmid copy number control and maintenance are located outside of the pSLA2 minimal origin. Similarly, plasmids containing a cloned chromosomal DNA region capable of replication extrachromosomally are maintained at a very low copy number and show extremely unstable inheritance (34).
During investigations of linear plasmid DNA replication in Streptomyces, we discovered that the minimal replication origin region, which accomplishes propagation of the linear plasmid pSLA2 in a circular mode (4, 5), is not sufficient for plasmid replication in a linear form when the plasmids contain functional telomeres. Here we report the identification and characterization of an additional pSLA2 locus, named rlrA, required for linear plasmid DNA replication. We show that the actions of this locus, which can affect plasmid copy number during replication in a circular mode, are regulated by an adjacent divergently transcribed rlrA-override (rorA) gene that strongly resembles the kor loci that control expression of conditionally lethal (kil) conjugation-mediating genes identified previously on certain Streptomyces replicons.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general methods.
Escherichia coli strain DH5 (Life Technologies) and plasmids pSP72 (Promega) and pBluescript II SK (Stratagene) were used as cloning host and vectors. Streptomyces rochei 7434-AN4, kindly provided by K. Sakaguchi, was the source of linear plasmid pSLA2 (10). S. lividans ZX7 (35) was host for propagating pSLA2-derived linear and circular plasmids. Culture and transformation of Streptomyces followed the methods of Kieser et al. (16). To obtain good sporulation, strain ZX7 grew on complete medium (23). Plasmid pLUS450, kindly provided by Carton Chen, contained 2.6-kb chromosomal telomere sequences. Plasmid pXQ25, kindly provided by Mingxuan Xu, contained the SLP2 minimal origin. High-quality and concentrated plasmid DNAs were obtained by CsCl-ethidium bromide gradient centrifugation (26). For ligation reactions, DNA fragments from agarose gels were recovered by using the QiaQuick gel extraction kit (Qiagen). Linear plasmid DNAs were isolated from yeast extract-malt extract (YEME) (16) liquid medium or from R2/yeast extract (R2YE) (16) transformant plates following the methods of Qin and Cohen (22). NaOH treatment to remove the terminal protein linked covalently at 5′ DNA ends followed the procedures of Hirochika et al. (9) and Shiffman and Cohen (27). Sequencing of pSLA2 rlrA/rorA and other DNA fragments used an Applied Biosystems Genetic Analyser 310 or the Stanford PAN laboratory facility. The intensities of chromosomal and plasmid DNA bands were measured and analyzed by using Kodak ID Image software. The plasmid copy number per cell was calculated as follows: [(linear chromosomal DNA size)/(linear plasmid DNA size)] × (intensity ratio of chromosomal versus plasmid DNA bands).
RESULTS
Requirements for function of the minimal replication origin of pSLA2 in a linear mode.
Intact telomere-containing Streptomyces linear plasmids previously have been cloned in E. coli, and it was shown that linearization of these replication-competent constructs by restriction endonuclease cleavage external to the telomeres yielded linear plasmids when the DNA was introduced into Streptomyces cells by transformation (4, 5, 22, 27). However, during investigations of the replication functions of Streptomyces plasmid pSLA2, we found that a construct (pQC36; Fig. 1, right) containing two functional 365-bp plasmid telomeres and a pSLA2 region sufficient to accomplish DNA replication in S. lividans strain ZX7 in a circular mode (i.e., the rep1 and rep2 loci; see reference 5) (Fig. 1), did not generate the expected linear plasmids in vivo after treatment with DraI and introduction into S. lividans protoplasts by transformation (Fig. 2; cf. reference 22). To identify possible additional pSLA2 loci necessary for the minimal replication origin to propagate DNA in a linear mode, we digested full-length pSLA2 DNA with various endonucleases and, using E. coli as host, cloned the resulting fragments into the BclI or MluI site of pQC36 (Fig. 1 and 2). The resulting pQC36-derived constructs (Fig. 2) were analyzed by electrophoresis on agarose gels after endonuclease treatment. The plasmids were linearized with DraI and introduced by transformation into ZX7 cells. Transformants expressing the thiostrepton resistance gene (tsr) were isolated, and transformant clones containing linear plasmids were identified as previously described (22); as linear plasmids contain 5′ DNA termini protected by covalent linkage to a terminal protein, their DNA is resistant to attack by bacteriophage λ exonuclease (1, 4, 10, 22, 27) despite its sensitivity to E. coli exonuclease III (Fig. 3).
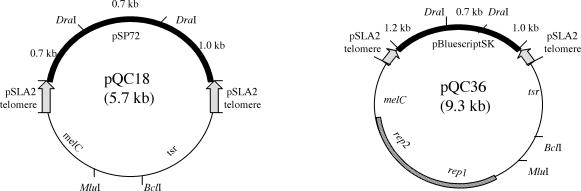
FIG. 1.
Maps of plasmids used to identify pSLA2 linear DNA replication genes. pSLA2 telomeres (indicated as arrowheads), the thiostrepton resistance gene (tsr), and a melanin gene (melC) were cloned into E. coli plasmid pSP72 (filled arc) to obtain pQC18 (also see reference 22). Using similar methods, two telomeres, tsr, melC, and the 3.2-kb pSLA2 minimal origin (rep1 and rep2 genes [striped box]) were cloned into plasmid pBluescript II SK (filled arc) to obtain pQC36. Cloning sites used for insertion of pSLA2 library fragments were MluI and BclI. DraI treatment of the plasmids removed a 0.7-kb E. coli fragment.
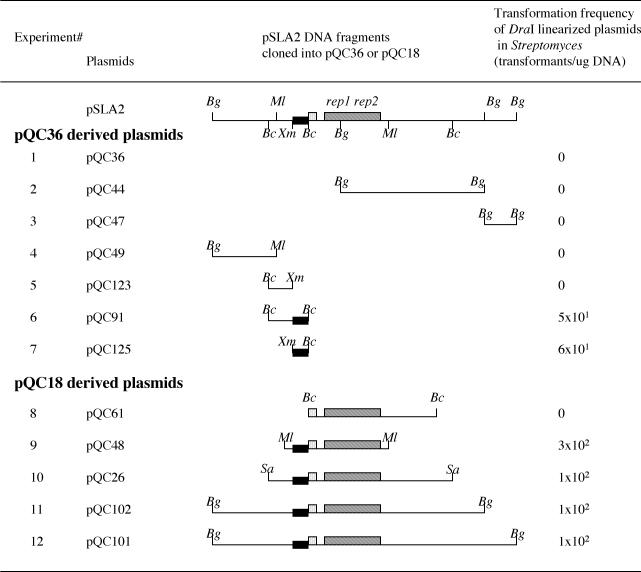
FIG. 2.
Identification of the rlrA gene. pSLA2 fragments were cloned into pQC18 or pQC36 in E. coli, and the resulting circular plasmid DNAs were linearized with DraI and introduced into S. lividans ZX7 cells by transformation. To clone pSLA2 fragments, full-length pSLA2 DNA was digested by various restriction enzymes. Resulting DNA fragments were cloned into pQC36 or pQC18. Fragments cloned into the BclI site of pQC36 included an 8.5-kb BglII fragment (in pQC44), a 1.3-kb BglII fragment (in pQC47), and a 2.3-kb BclI fragment (in pQC91). To obtain pQC125, a 1.1-kb XmaI-BclI fragment was first cloned into the polylinker of E. coli plasmid pSP72 and then a 1.1-kb BamHI-BglII fragment was cloned into pQC36. The same technique was used to clone a 1.2-kb BclI-XmaI fragment isolated in pQC123. A 3.4-kb BglII-MluI fragment was cloned into the BclI-MluI sites of pQC36 to obtain pQC49. Fragments cloned into the BclI site of pQC18 were 16.5- and 15-kb BglII fragments from partially digested pSLA2 DNA, a 12-kb Sau3A fragment from partially digested DNA, and a 7.5-kb BclI fragment, to obtain pQC101, pQC102, pQC26, and pQC61, respectively. A 6-kb MluI fragment cloned into the MluI site of pQC18 gave rise to pQC48. The resulting DraI-linearized plasmid DNAs were introduced into S. lividans ZX7 cells by transformation. Transformation frequencies are shown. The pSLA2 origin containing rep1 and rep2 genes is indicated as a striped box. Sequences comprising rlrA (filled box) and rorA (dotted box) are indicated. Abbreviations: Bg, BglII; Bs, BstI; Ml, MluI; Bc, BclI; Xm, XmaI; Sa, Sau3A.
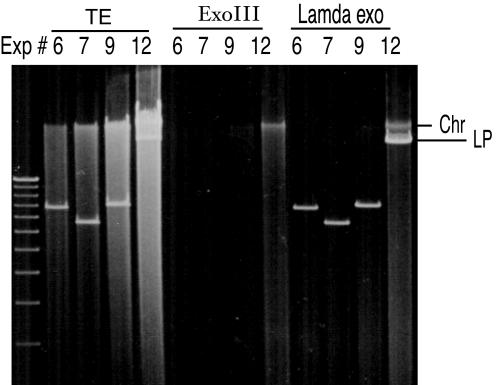
FIG. 3.
Confirmation of linearity of S. lividans replicons. Native plasmid DNAs were isolated from the pool of transformants grown in the R2YE medium by the previously reported method (22). Aliquots of the DNAs were treated with 100 U of E. coli exonuclease III or 10 U of λ exonuclease at 37°C for 1 h and then electrophoresed in a 0.5% agarose gel at 55 V for 6 h. Gels were then stained by ethidium bromide. Experiment numbers were same as in Fig. 2. A 1-kb DNA ladder was used as a size marker. The position of residual chromosomal DNA (Chr) detected in lane 12 after treatment with either λ exonuclease or exonuclease III is indicated.
Identification of rlrA as the gene necessary for pSLA2 replication in a linear mode.
The smallest pSLA2 fragment that allowed the minimal origin to initiate replication of a pQC36-derived plasmid construct in a linear mode was the 1.1-kb XmaI-BclI fragment cloned in pQC125 (Fig. 2). Sequencing of this fragment suggested that it contains an open translational reading frame truncated distally by XmaI cleavage. The sequence of a larger BclI-BclI pSLA2 fragment that overlaps the XmaI/BclI fragment and that was cloned in the pQC91 plasmid showed a putative full-length open reading frame (ORF) encoding a 308-amino acid (aa) protein (Fig. 4A), as analyzed by the MacFrame program (3; K. Kendall, unpublished data). Based on the phenotypic properties associated with expression of this locus, the gene it contains was designated rlrA (required for linear replication A).
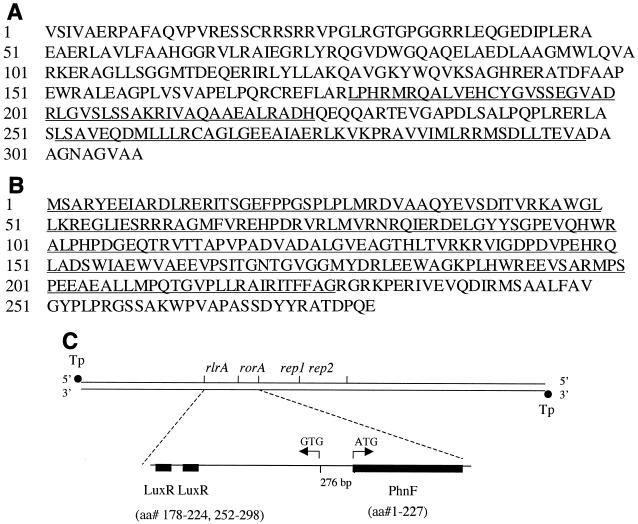
FIG. 4.
Sequences and locations of the RlrA and RorA proteins on linear plasmid pSLA2. (A) The 308-aa sequence of the RlrA protein. LuxR motifs are underlined. (B) The 279-aa sequence of the RorA protein. The PhnF motif is underlined. (C) The protein domains of RlrA and RorA are shown by the filled boxes. The translational starting codons and directions of translation are indicated by arrowheads. Terminal proteins (Tp) are indicated by filled circles.
Motif analysis using the eMatrix search program (31, 32) revealed the presence of two sequences at aa 178 to 224 and aa 252 to 298 that showed similarity (expectation values of 4.35 × 10−3 and 3.33 × 10−4, respectively) to members of the LuxR family of bacterial regulatory proteins (Fig. 4A). The helix-turn-helix DNA-binding motif of these proteins is located, as are the LuxR motifs in RlrA, in the C-terminal region of the protein sequence (29).
Identification of a regulator of the rlrA gene.
In other experiments, the MluI or BclI site of pQC18 (Fig. 1), which lacks any replication origin capable of functioning in Streptomyces, was used to clone fragments containing both the pSLA2 minimal origin of replication and nearby sequences. All plasmid derivatives able to replicate in a linear mode were found to include the rlrA sequence described above in addition to a segment containing the minimal replication origin, supporting the conclusion that rlrA is required for plasmid DNA replication in a linear form. During these experiments, we also observed that transformants from uncleaved pQC36 or pQC18 derivatives containing rlrA occurred at an efficiency 1,000-fold lower than that of transformants lacking this gene (Table 1) and that this reduction of transformation frequency was reversed by the presence of a DNA segment located between rlrA and the minimal origin of replication (Fig. 2, lower panel). Sequencing of this segment showed a putative ORF encoding a 279-aa sequence (Fig. 4B) and transcribed divergently from rlrA (Fig. 4C). As this gene, which is adjacent to rlrA, was able to override a detrimental effect of rlrA on transformation by nonlinear DNA, it was designated rorA (for rlr override). A BLAST search identified a conserved PhnF transcriptional regulatory domain in RorA, and a comparison with the NCBI nonredundant database using BLAST showed similarity (expectation value = 4 × 10−7) between RorA and the transcriptional regulatory KorA protein of circular plasmid pIJ101 (15, 28), as well as to other putative KorA-like proteins found by sequencing on various other Streptomyces plasmids (8, 14). The divergently transcribed rlrA and rorA genes are separated by a 276-bp spacer region (Fig. 4C), a distance approximately the same as the 269-bp spacer separating the korA gene of pIJ101 from its divergently transcribed target, kilA (15, 28).
TABLE 1.
Effects of rlrA and rorA on the transformation frequency in Streptomyces of circular pSLA2 plasmid constructs
| Expt no. | Plasmid | rlrA | rorA | Transformation frequency in Streptomyces (transformants/μg of DNA)a |
|---|---|---|---|---|
| 1 | pQC36 | − | − | 3 × 104 |
| 6 | pQC91 | + | − | 3 × 101 |
| 7 | pQC125 | + | − | 4 × 101 |
| 8 | pQC61 | − | + | 1 × 104 |
| 9 | pQC48 | + | + | 4 × 104 |
| 12 | pQC101 | + | + | 3 × 104 |
A 0.1-μg aliquot of pSLA2 circular plasmid DNA isolated from E. coli was introduced into S. lividans ZX7 by transformation (16). Transformant colonies were counted after thiostrepton selection.
rlrA and rorA affect the inheritance of pSLA2 circular plasmids by spores.
We observed that the rlrA-rorA gene combination on the pSLA2-derived circular plasmids pQC48 and pQC101 stabilized the inheritance of these plasmids in ZX7 (28 and 78%, respectively) during an S. lividans life cycle, as indicated by the incidence of plasmids in spores isolated during a cycle of growth in the absence of selection for thiostrepton resistance (Table 2). While the presence of rlrA alone on circular plasmids (pQC125 and pQC91) reduced plasmid inheritance to 5 and 1%, respectively, of the level observed in the absence of rlrA (pQC36), the addition of rorA reversed the effect of rlrA and also resulted in a 300-fold increase in inheritance over what was seen in the absence of either gene. Telomere-free plasmids that we constructed showed similar stabilization of inheritance by the rlrA-rorA gene combination (Table 3).
TABLE 2.
Effects of rlrA and rorA on the inheritance by spores of pSLA2 plasmids replicating in a circular mode
| Expt no. | Plasmid | rlrA | rorA | Frequency of plasmid inheritance (%)a |
|---|---|---|---|---|
| 1 | pQC36 | − | − | 0.1 |
| 2 | pQC125 | + | − | 0.005 |
| 3 | pQC91 | + | − | 0.001 |
| 4 | pQC61 | − | + | 0.7 |
| 5 | pQC48 | + | + | 28 |
| 6 | pQC101 | + | + | 78 |
pSLA2 circular plasmids were introduced into ZX7, and thiostrepton-resistant colonies were inoculated into CM medium containing thiostrepton (23). After 7 days at 30°C, spores were streaked on CM plates without selection for 7 days at the same temperature. Harvested spores were diluted in water and plated equally on LB and on LB medium containing thiostrepton. After 3 days of incubation, we counted the number of colonies on plates. The frequency of plasmid inheritance = 100 × ratio of colonies on LB containing thiostrepton-resistant colonies on LB.
TABLE 3.
Effects of rlrA and rorA on circular plasmid inheritancea
| Expt no. | Plasmid | Telomeres | rlrA | rorA | Frequency of plasmid inheritance (%) |
|---|---|---|---|---|---|
| 1+ | pQC36 | + | − | − | 0.1 |
| 1− | pCIR1051 | − | − | − | 0.1 |
| 2+ | pQC48 | + | + | + | 28 |
| 2− | pQC578 | − | + | + | 57 |
| 3+ | pQC125 | + | + | − | 0.005 |
| 3− | pQC711 | − | + | − | 0.004 |
| 4+ | pQC101 | + | + | + | 78 |
| 4− | pQC354 | − | + | + | 71 |
Methods were as described for Table 2. For each experiment, telomere-free (−) plasmids were constructed. The 2.6-kb BclI fragment containing tsr and melC genes (see reference 22) was cloned into the BglII site of plasmid pSP72 to obtain pQC156. A 6-kb pSLA2 MluI fragment (Fig. 2) was cloned into the MluI site of pQC156 to obtain pQC578. A 16.5-kb BglII fragment was cloned into the BamHI site of pQC156 to obtain pQC354. A 6-kb NdeI-SphI fragment from pQC125 (Fig. 2) was cloned into the NdeI-SphI sites of pSP72 to obtain pQC711.
Effect of rlrA-rorA on copy number of circular pSLA2-derived replicons.
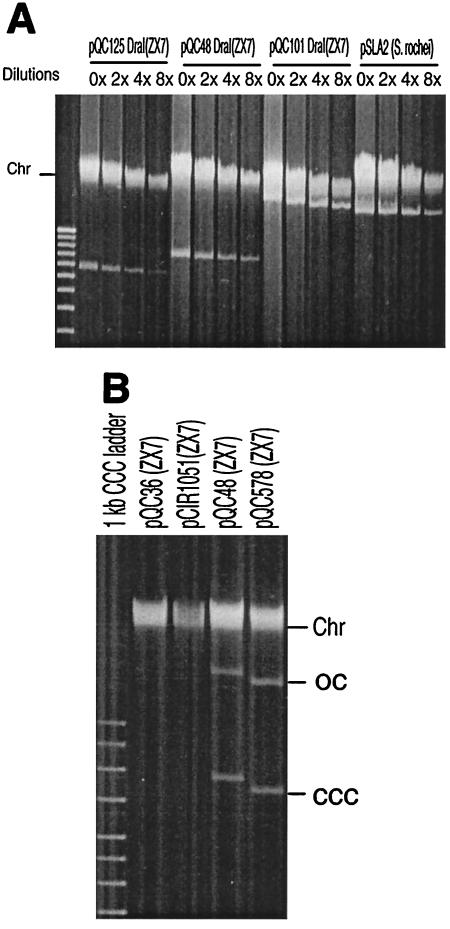
As seen in Fig. 5A, plasmid copy number estimates from the relative intensity of chromosomal and plasmid DNA bands (see Materials and Methods) showed that native pSLA2 linear plasmids replicated at high copy number (∼113 per cell; cf. estimate of 60 per cell using radioactive labeling in the original host, S. rochei [see reference 10]). Our findings indicate that the pSLA2-derived linear plasmids replicate in S. lividans at a similarly high copy number (estimated at 145, 179, and 130 per cell for pQC125, pQC48, and pQC101, respectively). In contrast, a circular pSLA2-derived plasmid, pCIR1051, containing the minimal origin but lacking the region now known to contain the rlrA and rorA genes replicated in S. lividans strain TK23 as a very-low-copy-number plasmid whose DNA was detectable by Southern blotting but not by ethidium bromide staining (5). Consistent with this finding, no circular DNA band was detectable by agarose gel electrophoresis in pCIR1051-transformed hygromycin-resistant ZX7 cells (Fig. 5B; a faint band was seen by Southern blotting [data not shown]), suggesting that plasmid copy number in this Streptomyces species is also very low. Similar results were obtained using a pQC36 circular plasmid containing the pSLA2 minimal origin and telomeres (Fig. 2). However, we observed that the addition of rlrA/rorA genes to these circular replicons resulted in a prominent increase in plasmid DNA for these circular plasmids (∼156 and 198 per cell for circular pQC48 and for pQC578 [telomere-free], respectively) (Fig. 5B).
FIG. 5.
Effect of the rlrA/rorA genes on copy number of pSLA2-derived plasmids. (A) Agarose gel electrophoresis analysis of pSLA2-derived linear plasmids. Approximately the same volumes of Streptomyces mycelium from YEME liquid culture were used for DNA isolation (22). DNAs were diluted 0, 2, 4, and 8× in TE (10 mM Tris Hcl, 1 mM EDTA) and loaded in equal volumes in a 0.5% agarose gel at 65 V for 7 h. Gels were then stained with ethidium bromide. A 1-kb DNA ladder was used as a size marker. (B) Agarose gel analysis of pSLA2-derived circular plasmids. Transformants from colonies were picked from R2YE medium and added to a lysis solution (16). After proteinase K-sodium dodecyl sulfate treatment and phenol-chloroform extraction (22), supernatants were loaded in a 0.5% agarose gel at 65 V for 7 h. Gels were then stained with ethidium bromide. A 1-kb supercoiled DNA ladder was used as a size marker.
The rlrA gene also enables plasmids containing the SLP2 origin, but not a chromosomal origin, to replicate in a linear mode.
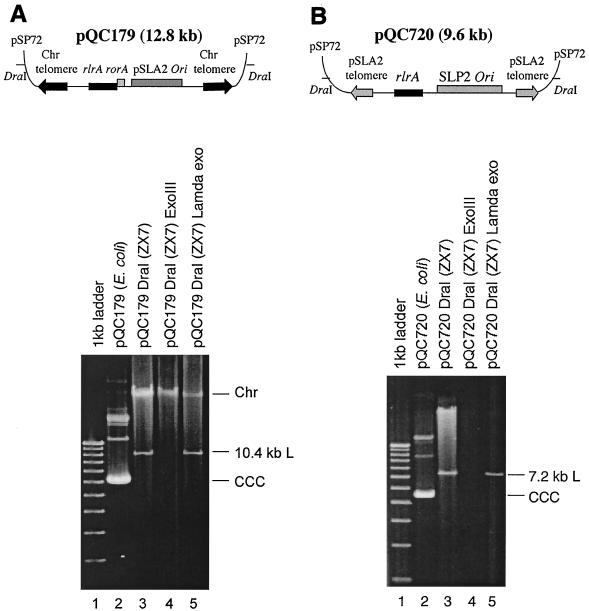
Replacing pSLA2 telomeres by chromosomal telomeres did not prevent pSLA2-derived DraI-treated plasmids from replicating in a linear mode (Fig. 6A; cf. pQC179 with pQC48 in Fig. 3), indicating that the ability of the rlrA/rorA locus combination to promote linear plasmid DNA replication does not rely on the presence of plasmid telomeres. To further investigate the target of the observed rlrA effects on linear DNA replication, we replaced the pSLA2 replication origin with either a chromosomal origin or with the replication origin of another plasmid native to Streptomyces, the SLP2 plasmid of S. lividans. Plasmids carrying the chromosomal origin, which contains two clusters of 19 DnaA boxes separated by a 134-bp spacer (13), were unable to replicate extrachromosomally in a linear mode, even in the presence of rlrA. However, rlrA enabled the replication origin of plasmid SLP2 which, like the pSLA2 origin contains two direct repeat iterons and a helicase gene (see references 5 and 11; M. Xu et al., unpublished results), to propagate plasmid DNA in a linear mode (Fig. 6B).
FIG. 6.
Effect of the rlrA gene on replication of plasmids in a linear mode. (A) Propagation of linear plasmids containing the pSLA2 origin and chromosomal telomeres. Construction of pQC179 was as follows: 0.8-kb chromosomal telomeres from plasmid pLUS450 (kindly provided by Carton Chen) were subcloned into pSP72, to obtain pQC154. They were then used to construct pSLA2-derived plasmid pQC177, using the strategy described above for the formation of pQC18 (Fig. 1). A 6-kb pSLA2 MluI fragment containing the minimal origin and rlrA/rorA genes was cloned into the pQC177 MluI site to obtain pQC179. (Lower panel) Native chromosomal and plasmid pQC179 DNAs were isolated from ZX7 transformants and analyzed by agarose gel electrophoresis (lane 3). Aliquots of the DNAs were treated with 100 U of E. coli exonuclease III (lane 4) and with 10 U of λ exonuclease (lane 5) at 37°C for 1 h and electrophoresed in a 0.5% agarose gel at 37 V for 12 h. A 1-kb DNA ladder (lane 1) and pQC179 circular plasmid DNA isolated from E. coli (lane 2) were used as size markers. (B) Propagation of linear plasmids containing the SLP2 origin and pSLA2 telomeres. The BamHI-BglIII fragment containing rlrA (Fig. 2, pQC125) was cloned at the BclI site of pQC18 to obtain pQC707 in E. coli. The 3.2-kb SLP2 fragment containing the SLP2 minimal origin (kindly provided by Xu Mingxuan) was cloned into the MluI site of pQC707 to obtain pQC720. (Lower panel) pQC720 DraI-linearized DNA was used to transform ZX7 cells. Transformant DNAs from thiostrepton-resistant clones were analyzed by agarose gel electrophoresis (lane 3). DNA aliquots were treated with 100 U of E. coli exonuclease III (lane 4) or 10 U of λ exonuclease (lane 5) at 37°C for 1 h and then electrophoresed in a 0.5% agarose gel at 37 V for 12 h. A 1-kb DNA ladder (lane 1) and pQC720 circular plasmid DNA isolated from E. coli (lane 2) were used as size markers.
DISCUSSION
The results reported here reveal a striking distinction in the ability of the pSLA2 replication origin to accomplish propagation of linear versus circular DNA molecules. rlrA, a novel gene identified in pSLA2, is required for the pSLA2 minimal origin to replicate telomere-containing plasmids in a linear mode but is dispensable for the same origin to accomplish circular plasmid DNA replication. Presence of the rlrA gene of pSLA2 enabled the origin regions of plasmids pSLA2 and SLP2 to propagate linear replicons using either plasmid telomeres or chromosomal telomeres.
Notwithstanding the dispensability of rlrA for the replication of Streptomyces chromosomes as linear DNA molecules (i.e., S. lividans cells that contain a linear chromosome but lack rlrA are viable), plasmids containing the cloned chromosomal origin are maintained poorly as extrachromosomal circular replicons (34). We found that the addition of rlrA did not enable replication of these molecules in a linear mode, whether the chromosomal origin was joined to plasmid telomeres or chromosomal telomeres. These findings suggest that the effects of rlrA are mediated through the replication origins of linear plasmids rather than through their telomeres. This notion is supported by evidence that the presence of rlrA on circular plasmids propagated by the minimal origin increased plasmid copy number. Our results also imply that the linear chromosome of S. lividans, which can replicate in cells lacking the plasmid-encoded rlrA gene, is likely to include an origin-specific locus that is functionally analogous to rlrA and which acts only in cis.
Our analysis of the published DNA sequence of the SLP2 plasmid (11) indicates that it contains a putative ORF showing 40% identity (expectation value, 9 × 10−6) to pSLA2 RorA. Like pSLA2 RorA, this ORF, which was designated korSLP2 by Huang et al. (12), includes a PhnF motif. Adjacent to the SLP2 ORF, which we now suggest be renamed as rorA-SLP2, we observed an ORF that is transcribed in the opposite direction and which is separated from rorA-SLP2 by 211 bp. While the divergently transcribed SLP2 ORF adjacent to rorA-SLP2 lacks LuxR family motifs and shows no detectable homology to rlrA of pSLA2, we have found that its presence on plasmids containing a pSLA2 minimal origin and pSLA2 telomeres enables these plasmids to replicate in a linear mode (data not shown).
The replication origin of pSLA2 is located within a gene encoding an essential DNA binding protein (rep1; see reference 5). The pSLA2 minimal origin includes another trans-acting locus, which encodes a DNA helicase gene, as well as two cis-acting loci (5). One of these cis-acting loci—the replication origin itself—contains two 21-mer repeat sequences (i.e., iterons), which are located at the site of replication initiation. While the inferred SLP2 origin contains an iteron structure similar to that of pSLA2 (i.e., 23-mer repeat sequences), we found no DNA sequence homology between the iterons of the two plasmids. The DnaA box of the S. lividans chromosomal replication origin (oriC), which differs from the replication origins of pSLA2 and SLP2 in both sequence and overall structure, contains two clusters of 19 DnaA boxes separated by 134 bp (13, 34). The actions of the rlrA locus, which our data argue are mediated through the replication origin, may be directed at the plasmid iteron structure or they may affect other cis-acting loci within the minimal origins of these Streptomyces plasmids (see reference 5). Alternatively, they may affect the helicase genes adjacent to the plasmid DNA origins. While the molecular mechanism that underlies the requirement for rlrA for linear—but not circular—DNA replication has at this time not been elucidated, evidence suggesting that the central origin of bidirectional replication is the target of rlrA action leads us to speculate that rlrA may be involved in coordinating the initiation of plasmid DNA replication from the origin (see reference 4) with telomere-patching DNA synthesis at the termini of linear plasmids.
Whereas the detrimental effect of rlrA on transformation by circular plasmid DNA and its reversal by an override locus are reminiscent of kill (kil) and kill-override (kor) systems identified previously on other Streptomyces plasmids (4, 8, 14, 15), the negative effects of the rlrA gene when unaccompanied by its override gene did not result in either cell lethality or the inability of circular plasmids to replicate. Rather, once circular plasmids containing the minimal pSLA2 replication origin and rlrA were established in bacterial cells, they were maintained in the absence of rorA at higher copy number than circular replicons lacking rlrA. The effects of the rlrA locus on circular plasmids were dependent on its position relative to the plasmid replication origin (unpublished data), suggesting that they may be mediated in part through localized topological alterations. Earlier work has shown the effects of such localized alterations in DNA supercoiling on both the inheritance (i.e., partitioning) and copy number of the circular pSC101 plasmid of E. coli (2, 6).
Acknowledgments
Z.Q. is grateful to Annie C. Y. Chang and Christine Miller and other members of our laboratory for technical help and discussions and to Ruisheng Jiao, Yunliu Yang, Guoping Zhao, and Weihong Jiang for their encouragement. We thank Simone Manteuil-Brutlag for assistance with preparation of the manuscript, Douglas Brutlag for motif analysis of our sequence data, David Hopwood for S. lividans strain ZX7, Carton Chen for the plasmid pLUS450, and Mingxuan Xu for the plasmid pXQ25.
These investigations were supported by National Institutes of Health grant AI08619 to S.N.C. and by grants from the Chinese National Foundation of Science (30170019 and 30270030) and the Chinese National “863” project (2002AA227021) to Z.Q.
REFERENCES
- 1.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaucage, S. L., C. A. Miller, and S. N. Cohen. 1991. Gyrase-dependent stabilization of pSC101 plasmid inheritance by transcriptionally active promoters. EMBO J. 10:2583-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibb, M. J., P. R. Findlay, and M. W. Johnson. 1984. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30:157-166. [DOI] [PubMed] [Google Scholar]
- 4.Chang, P. C., and S. N. Cohen. 1994. Bidirectional replication from an internal origin in a linear Streptomyces plasmid. Science 265:952-954. [DOI] [PubMed] [Google Scholar]
- 5.Chang, P. C., E. S. Kim, and S. N. Cohen. 1996. Streptomyces linear plasmids that contain a phage-like, centrally located, replication origin. Mol. Microbiol. 22:789-800. [DOI] [PubMed] [Google Scholar]
- 6.Conley, D. L., and S. N. Cohen. 1995. Effects of the pSC101 partition (par) locus on in vivo DNA supercoiling near the plasmid replication origin. Nucleic Acids Res. 23:701-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goshi, K., T. Uchida, A. Lezhava, M. Yamasaki, K. Hiratsu, H. Shinkawa, and H. Kinashi. 2002. Cloning and analysis of the telomere and terminal inverted repeat of the linear chromosome of Streptomyces griseus. J. Bacteriol. 184:3411-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagège, J., J. L. Pernodet, G. Sezonov, C. Gerbaud, A. Friedmann, and M. Guerineau. 1993. Transfer functions of the conjugative integrating element pSAM2 from Streptomyces ambofaciens: characterization of a kil-kor system associated with transfer. J. Bacteriol. 175:5529-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirochika, H., K. Nakamura, and K. Sakaguchi. 1984. A linear DNA plasmid from Streptomyces rochei with an inverted terminal repetition of 614 base pairs. EMBO J. 3:761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirochika, H., and K. Sakaguchi. 1982. Analysis of linear plasmids isolated from Streptomyces: association of protein with the ends of the plasmid DNA. Plasmid 7:59-65. [DOI] [PubMed] [Google Scholar]
- 11.Huang, C. H., C. Y. Chen, H. H. Tsai, C. Chen, Y. S. Lin, and C. W. Chen. 2003. Linear plasmid SLP2 of Streptomyces lividans is a composite replicon. Mol. Microbiol. 47:1563-1576. [DOI] [PubMed] [Google Scholar]
- 12.Huang, C. H., Y. S. Lin, Y. L. Yang, S. W. Huang, and C. W. Chen. 1998. The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol. Microbiol. 28:905-916. [DOI] [PubMed] [Google Scholar]
- 13.Jakimowicz, D., J. Majkadagger, G. Konopa, G. Wegrzyn, W. Messer, H. Schrempf, and J. Zakrzewska-Czerwinska. 2000. Architecture of the Streptomyces lividans DnaA protein-replication origin complexes. J. Mol. Biol. 298:351-364. [DOI] [PubMed] [Google Scholar]
- 14.Kataoka, M., T. Seki, and T. Yoshida. 1991. Regulation and function of the Streptomyces plasmid pSN22 genes involved in pock formation and inviability. J. Bacteriol. 173:7975-7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kendall, K. J., and S. N. Cohen. 1987. Plasmid transfer in Streptomyces lividans: identification of a kil-kor system associated with the transfer region of pIJ101. J. Bacteriol. 169:4177-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 17.Kinashi, H., E. Mori, A. Hatani, and O. Nimi. 1994. Isolation and characterization of linear plasmids from lankacidin-producing Streptomyces species. J. Antibiot. (Tokyo) 47:1447-1455. [DOI] [PubMed] [Google Scholar]
- 18.Kinashi, H., and M. Shimaji-Murayama. 1991. Physical characterization of SCP1, a giant linear plasmid from Streptomyces coelicolor. J. Bacteriol. 173:1523-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, Y. S., and C. W. Chen. 1997. Instability of artificially circularized chromosomes of Streptomyces lividans. Mol. Microbiol. 26:709-719. [DOI] [PubMed] [Google Scholar]
- 20.Lin, Y. S., H. M. Kieser, D. A. Hopwood, and C. W. Chen. 1993. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 10:923-933. [DOI] [PubMed] [Google Scholar]
- 21.Musialowski, M. S., F. Flett, G. B. Scott, G. Hobbs, C. P. Smith, and S. G. Oliver. 1994. Functional evidence that the principal DNA replication origin of the Streptomyces coelicolor chromosome is close to the dnaA-gyrB region. J. Bacteriol. 176:5123-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin, Z., and S. N. Cohen. 1998. Replication at the telomeres of the Streptomyces linear plasmid pSLA2. Mol. Microbiol. 28:893-903. [DOI] [PubMed] [Google Scholar]
- 23.Qin, Z., K. Peng, X. Zhou, R. Liang, Q. Zhou, H. Chen, D. A. Hopwood, T. Kieser, and Z. Deng. 1994. Development of a gene cloning system for Streptomyces hygroscopicus subsp. yingchengensis, a producer of three useful antifungal compounds, by elimination of three barriers to DNA transfer. J. Bacteriol. 176:2090-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redenbach, M., M. Bibb, B. Gust, B. Seitz, and A. Spychaj. 1999. The linear plasmid SCP1 of Streptomyces coelicolor A3(2) possesses a centrally located replication origin and shows significant homology to the transposon Tn4811. Plasmid 42:174-185. [DOI] [PubMed] [Google Scholar]
- 25.Salas, M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:39-71. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Shiffman, D., and S. N. Cohen. 1992. Reconstruction of a Streptomyces linear replicon from separately cloned DNA fragments: existence of a cryptic origin of circular replication within the linear plasmid. Proc. Natl. Acad. Sci. USA 89:6129-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein, D. S., K. J. Kendall, and S. N. Cohen. 1989. Identification and analysis of transcriptional regulatory signals for the kil and kor loci of Streptomyces plasmid pIJ101. J. Bacteriol. 171:5768-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vannini, A., C. Volpari, C. Gargioli, E. Muraglia, R. Cortese, R. De Francesco, P. Neddermann, and S. D. Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volff, J. N., P. Viell, and J. Altenbuchner. 1997. Artificial circularization of the chromosome with concomitant deletion of its terminal inverted repeats enhances genetic instability and genome rearrangement in Streptomyces lividans. Mol. Gen. Genet. 253:753-760. [DOI] [PubMed] [Google Scholar]
- 31.Wu, T. D., C. G. Nevill-Manning, and D. L. Brutlag. 2000. Fast probabilistic analysis of sequence function using scoring matrices. Bioinformatics 16:233-244. [DOI] [PubMed] [Google Scholar]
- 32.Wu, T. D., C. G. Nevill-Manning, and D. L. Brutlag. 1999. Minimal-risk scoring matrices for sequence analysis. J. Comput. Biol. 6:219-235. [DOI] [PubMed] [Google Scholar]
- 33.Yang, C. C., C. H. Huang, C. Y. Li, Y. G. Tsay, S. C. Lee, and C. W. Chen. 2002. The terminal proteins of linear Streptomyces chromosomes and plasmids: a novel class of replication priming proteins. Mol. Microbiol. 43:297-305. [PubMed] [Google Scholar]
- 34.Zakrzewska-Czerwinska, J., and H. Schrempf. 1992. Characterization of an autonomously replicating region from the Streptomyces lividans chromosome. J. Bacteriol. 174:2688-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, X., Z. Deng, J. L. Firmin, D. A. Hopwood, and T. Kieser. 1988. Site-specific degradation of Streptomyces lividans DNA during electrophoresis in buffers contaminated with ferrous iron. Nucleic Acids Res. 16:4341-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]