Abstract
Synthesis of the small regulatory RNA DsrA is under temperature control. The minimal dsrA promoter of 36 bp contains sufficient information to ensure such regulation. In vivo, we have analyzed the critical elements responsible for the temperature control of dsrA by using a collection of chimeric promoters combining various elements of the dsrA promoter and the lacUV5 promoter, which does not respond to temperature. Our results favor an RNA polymerase-DNA interaction model instead of a trans-acting factor for temperature regulation. While all of the elements of the dsrA promoter contribute to temperature-sensitive expression, the sequence of the −10 box and the spacer region are the essential elements for the thermal response of the dsrA promoter. The proper context for these promoter elements, including at least one of the flanking elements, the −35 region or the start site region, is also required. Point mutations demonstrate that the sequence of the −10 box imposes constraints on the length and the sequence of the spacer and/or its AT richness, even at low temperature. These results show a complex interdependence of different regions in the promoter for temperature regulation.
Temperature is one environmental variable that all organisms must deal with. Escherichia coli, an enteric bacterium, is able to grow at temperatures of around 37°C in birds or mammalian hosts but also at significantly lower temperatures in the outside environment (11). To survive, Escherichia coli has to respond to sudden up or down temperature shifts and to maintain its physiology after adaptation to a new constant temperature. In general, temperature shifts trigger transient cellular processes involving specific systems, such as cold or heat shock proteins. During this transient period, changes in gene expression and other processes help adapt cell physiology to the new temperature condition. Besides the direct effect of temperature on enzymatic reactions, temperature response involves remodeling of the expression pattern of genes, affecting transcription, RNA stability, translation efficiency, and/or proteolysis (7, 8, 13, 20, 27).
RpoS is a stationary-phase and stress response sigma factor in E. coli and many other bacteria. In addition to other mechanisms, expression of RpoS-dependent genes is controlled by the level of RpoS. Thus, stresses that presumably need RpoS-dependent gene expression cause an increase in RpoS levels by blocking degradation and/or stimulating synthesis (reviewed in reference 10). One environmental cue that increases RpoS synthesis is low temperature (below 37°C). This increase is completely dependent upon DsrA, a small noncoding RNA (25). DsrA was shown to stimulate RpoS translation by pairing with a portion of the mRNA upstream of the RpoS translation start that can pair with and occlude the ribosome-binding site of the transcript (16). DsrA is more abundant in E. coli at low growth temperatures than at higher temperatures, resulting in the increased RpoS expression at low temperatures (22). Temperature affects both the synthesis and the stability of DsrA, leading to thermocontrol of RpoS translation.
In a previous study, we mapped several regulatory sites in the promoter of the dsrA gene (dsrAp) and demonstrated that thermocontrol of DsrA synthesis is primarily due to the minimal promoter of the gene, namely, the 36 bp upstream of the transcription start site. In this promoter, sequences at −10 (TAAGGT) and −35 (TTGTCA) appear to be relatively close to the consensus recognized by the σ70 RNA polymerase (respectively, TATAAT and TTGACA). The distance between the −10 and −35 boxes in dsrA is also optimal (17 bp). Indeed, transcriptional fusions revealed an activity at 25°C equivalent to that seen with the lacUV5 promoter (lacUV5p), which is generally considered a fairly active promoter. However, while lacUV5 was equally as active or more active at 37 and 42°C, the activity of dsrAp was much lower at these elevated temperatures (22). In the present study, we performed an in vivo analysis of the critical sequences within dsrAp that are responsible for this temperature-sensitive behavior. We used a collection of chimeric promoters and point mutations, and our data show that the sequence of the −10 box is the major determinant of the thermocontrol of DsrA synthesis; this unusual −10 sequence can only function in some contexts, even at low temperature.
MATERIALS AND METHODS
Genetic procedures, bacterial strains, plasmids, and phages.
Standard procedures were used for growth of bacteria and bacteriophages (17, 24). The parental bacterial strain used in this study was MC4100 (4). All of the other strains were lysogens of MC4100 with phage λRS468 (18) recombined with appropriate plasmids. The plasmids used were derivatives of pFRΔ (22). Isolation of plasmid DNA, digestion with restriction enzymes, and ligation with T4 DNA ligase were carried out in accordance with the manufacturers' (New England Biolabs and Promega) protocols or as described by Sambrook et al. (23). Transformations were performed as described by Chung et al. (5), with strain DH5α (New England Biolabs) or MC4100 (4). The sequences of all of the constructs described in this report (plasmids as well as fusions inserted into the chromosome) were confirmed by sequencing (National Cancer Institute-National Institutes of Health DNA Core facility, Bethesda, Md.).
Constructions of plasmids, phages, and strains carrying the transcriptional fusions for the in vivo study of dsrAp.
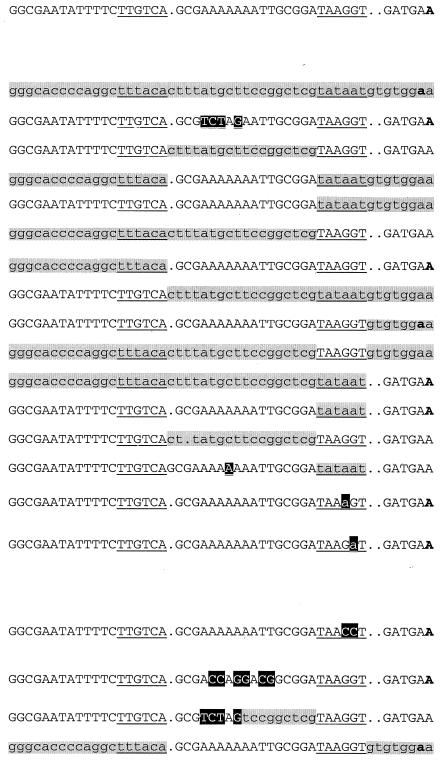
The strain carrying 46-bp wild-type dsrAp was constructed as previously described by Repoila and Gottesman (22), and the corresponding strain was named FRΔ4 (Table 1). All of the other constructs carrying the desired promoter fused to the frΔ reporter system were generated by two complementary primers used in a PCR with no template. The complementary portion of the pair of primers corresponds to the spacer sequence of the desired promoter. Each 5′ primer carried an EcoRI site at the 5′ extremity, and 3′ primers carried a BamHI site at the 5′ extremity. The same PCR conditions were used to generate the fragments for cloning, for sequencing, and for the screening of recombinant clones, and they have been previously described by Repoila and Gottesman (22). The sequence of each promoter used in this study is shown in Table 1. After the PCR, each product was cleaved with EcoRI and BamHI and then ligated to the pFRΔ vector cut with the same enzymes. The ligation mixture was then transformed into DH5α. Ampicillin-resistant colonies were screened by PCR for the presence of a fragment of approximately 250 bp with oligonucleotides FR.EcoRI-415 (5′-GCCATAAACTGCCAGGAATTGG-3′) and Deeplac (5′-CGGGCCTCTTCGCTA-3′). The sequence of each construct was confirmed by using the primer Deeplac, annealing about 150 bp within the α peptide of lacZ downstream of the fusion point. Each recombinant plasmid was introduced by transformation into MC4100 and recombined with λRS468 as described previously (22). Recombinant phages were selected by their ability to give blue plaques on Luria broth plates with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside-isopropyl-β-d-thiogalactopyranoside (IPTG) on the indicator strain MC4100. After purification, the recombinant phage were used to lysogenize MC4100. Single-copy Lac+ lysogens were selected for each fusion as described by Powell et al. (21). One isolate of each fusion was confirmed by sequencing and saved (Table 1). Each promoter fused to the frΔ reporter system is mentioned in the text as a construct; i.e., construct Δ4 designates the 46 bp of wild-type dsrAp driving the expression of lacZ.
TABLE 1.
Activities of chimeric promoters used in this study
For each promoter construct, the portion of the sequence identical to dsrA is shown in uppercase; lowercase sequences are identical to lacUV5 and are shaded light grey. A dot in the sequence indicates the absence of a base; changes from the original dsrA sequence are in black boxes. The −35 and −10 boxes are aligned vertically and underlined. The transcription start site generated from the frΔ fusion is indicated by the bold letter only for those constructs tested by primer extension.
The 25 and 42°C columns contain the average β-galactosidase activities measured at those temperatures. The values are machine units determined as described in Materials and Methods.
Ratio of the value at 25°C to that at 42°C. β-Galactosidase values of less than or equal to 17 are considered not significant since such a level corresponds to the maximal value measured with the empty frΔ reporter system (22).
Growth conditions used for β-galactosidase.
The growth conditions used were identical to those previously described by Repoila and Gottesman (22). Promoter activities were monitored during growth by measuring the optical density at 600 nm (OD600) of the cultures and assaying the β-galactosidase activity as described by Zhou and Gottesman (28), with a SpectraMax microtiter machine (Molecular Devices). The specific activities provided (Table 1) correspond to the slope of the line (Vmax) divided by (OD600 × vol), where vol is the volume of the culture used for the assay expressed in milliliters. These “machine units” have been estimated to be about 2.5-fold lower than Miller units. The activities in Table 1 correspond to an average of at least two series of independent measurements performed during exponential-phase growth over an OD600 range of 0.35 to 0.8 as previously reported by Repoila and Gottesman (22). For each strain, the observed variation between two series of measurements was less than 20% and less than 15% for β-galactosidase activities of 700 U or below.
RNA preparation and primer extension.
Total RNA was prepared from cells growing under the same conditions as described for β-galactosidase assays, with TRIZOL (Gibco BRL) in accordance with the manufacturer's protocol. Briefly, primer extensions were performed with an avian myeloblastosis virus reverse transcriptase kit (Promega) on either 12 μg of total RNA to detect transcripts expressed from fusions inserted as single-copy lysogens in the MC4100 genome or 6 μg of total RNA when expressed from the pFRΔ-derived multicopy plasmids carried by MC4100 (22). Total RNA was extracted from cultures at an OD600 of 0.45 to 0.55. Oligonucleotide FR.rtl (5′-CAGTGAATCCGTAATCATGGTCATAGCTGTTTCC-3′) was labeled with T4 polynucleotide kinase (Promega) and [γ-32P]ATP (Amersham). FR.rtl anneals within the 5′ coding sequence of the lacZ open reading frame, 10 bp downstream of the translation initiation site, and was used as the primer for avian myeloblastosis virus reverse transcriptase at 42°C. Products of the reverse transcription reaction were run on 8% sequencing gels. Signals were quantified with the National Institutes of Health Image program.
RESULTS
In vivo mapping of the elements responsible for the thermocontrol of dsrAp.
To identify the elements of dsrAp responsible for thermoregulation, we constructed a set of chimeric promoters carrying elements from dsrAp and lacUV5p (Table 1). lacUV5p is not sensitive to temperature and has an activity similar to that of dsrAp at 25°C (22). In a previous study, we had shown that the minimal dsrAp sequence (36 bp) carries sufficient information to ensure thermoregulated transcription initiation. However, the activity of this promoter is very low, basically no higher than the background at high temperatures (37 or 42°C). Extension of the promoter region to −46 bp, which includes an apparent UP sequence, increases its activity about 10-fold but does not significantly change the temperature regulation (22). In the present study, we used promoter fusions that included this upstream region. Chimeras were made by exchanging each of four different portions of the promoter: (i) the −35 box, including the upstream 10 bp; (ii) the spacer region; (iii) the −10 box; and (iv) the region between −10 and the start of transcription (start site region). Each fusion was first constructed on a plasmid in front of the lac α fragment and then transferred to a lambda vector and lysogenized at the lambda att site. We had difficulty isolating the lambda versions of a few of these promoter fusions (see below).
Features of dsrAp.
Isogenic strains carrying the different chimeric promoters were assayed during growth at 25 and 42°C. The results are summarized in Table 1. The values shown are averages of at least two independent measurements. Wild-type dsrAp (Δ4) gives about a fourfold difference in expression between 25 and 42°C; wild-type lacUV5p (Δ31) gives a ratio of 0.8 (Table 1). Δ13, which differs from wild-type dsrAp by only four nucleotide changes in the spacer, also shows a ratio of about 4. Only two other chimeras, Δ24 and Δ26 (Table 1), are significantly more active at 25°C than at 42°C. Both constructs are based on the dsrAp structure; Δ24 carries the −35 region from lacUV5, and Δ26 carries the start site region of lacUV5 (Table 1). These data indicate that neither the −35 region nor the start site is essential for the temperature response. However, the magnitude of the response for Δ24 and Δ26 is lower than that for the wild type (about 2-fold compared to 4.5-fold). Thus, the −35 region and the start site both amplify the temperature-sensitive activity associated with the rest of the promoter. In construct Δ41, both the −35 region and the start site region of dsrAp were replaced with sequences from lacUV5; this chimeric promoter is no longer temperature sensitive (Table 1). Therefore, although we conclude from results discussed below that the essential elements for the temperature response of dsrAp are the −10 box and the spacer sequence, to display such behavior, these require at least one of the surrounding elements, namely, the −35 region or the start site region.
Constraints on promoter activity: the −10 box and spacer incompatibilities.
Although dsrAp and lacUV5p are similarly active at 25°C, certain combinations of elements are very defective for activity. In general, any construct that contained the −10 region from lacUV5 (consensus, TATAAT) with any other combination of elements was at least as active as the original promoters (Table 1, constructs Δ21, Δ22, Δ25, Δ29, and Δ34) and was temperature independent. Δ28, which contains the consensus −10 region, as well as the spacer and −35 region, from lacUV5 but the start site from dsrA, had slightly decreased but temperature-independent activity (Table 1).
In contrast, many constructs containing the −10 box from dsrA (TAAGGT) were defective for promoter activity at all temperatures, suggesting that this −10 region has special requirements for function. In particular, any construct that contained the spacer region from lacUV5, which is 18 rather than 17 bases long, is not tolerated by the dsrA −10 region at any of the temperatures tested (Δ20, Δ23, and Δ27; Table 1). In experiments done earlier on somewhat different constructs, addition or deletion of one A · T base pair in natural dsrAp also drastically decreased expression to the level of the vector alone (<20 U at any temperature) (data not shown and reference 22). Therefore, we concluded that the −10 region of dsrA requires a 17-bp spacer to activate the promoter.
The sequence of the spacer matters as well. Removal of one base from the 18-bp spacer of lacUV5 improves expression to a measurable range, but values are still 10-fold lower at 25°C than for the promoter carrying the dsrA spacer (Δ32 compared to Δ20 and to Δ4; Table 1). Other single-base deletions in the lacUV5 spacer with an architecture similar to that of Δ32 have been tested; comparable results (not shown) were obtained. The dsrA spacer is AT rich, including a stretch of seven A's and two T's, a sequence known, in other contexts, to curve DNA (reviewed in references 6 and 19). Two sets of mutations that conserve the 17-bp spacing were introduced into the spacer of dsrAp (Δ13 and Δ40; Table 1). Δ40, in which the left side of the spacer contains the Δ13 sequence but the right side contains the lacUV5 sequence, is quite defective for activity and does not show temperature regulation (20-fold down at 25°C compared to Δ13; Table 1). These results suggest that sequences critical for activity with the dsrA −10 region are those in the right-hand portion of the spacer and not necessarily the AT-rich stretch as a whole.
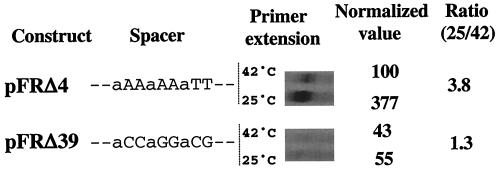
In a separate series of experiments, we replaced the AT-rich region of the spacer with ACCAGGACG, in which the AT content is profoundly modified (only three out of nine bases were conserved; construct Δ39; Table 1). For unexplained reasons, the transcriptional fusion bearing the Δ39 construct could not be inserted as a single copy into the chromosome. Therefore, its expression was tested by primer extension directly on the lacZ transcript expressed from plasmid pFRΔ carrying the fusion (see Materials and Methods). In parallel experiments, the same vector containing the construct Δ4 (wild-type dsrAp) was used as a control (Fig. 1). The lacZ transcript detected for Δ4 is about 3.8-fold more abundant at 25°C than at 42°C, which is quite consistent with the levels of β-galactosidase activity detected when the fusion was inserted as a single copy into the chromosome (4.5-fold higher at 25°C than at 42°C; Table 1). In contrast, Δ39 exhibited extremely low activity; about sevenfold less RNA was detected compared to that of Δ4 at 25°C. In addition, the amounts of transcript were similar at 25 and 42°C, indicating a loss of temperature control (Fig. 1). A comparison of Δ39 and Δ40, both very weakly active and not thermoregulated, with Δ13 and the wild-type sequence, both active and responding to temperature, suggests an important role for either the AATT sequence in the middle of the spacer or the AT content for promoter activity at 25°C and the thermoresponse.
FIG. 1.
Primer extension analysis of the transcripts generated by dsrAp (Δ4) and spacer mutant construct Δ39. Both fusions are carried by the plasmid vector pFRΔ (see Materials and Methods). Spacer sequence variations between the two constructs are shown with uppercase letters. For each construct, quantitation of the transcripts at 25 and 42°C was done, and the values were normalized to the activity of wild-type dsrAp at 42°C, which was set arbitrarily as equal to 100 (normalized-value column). The ratio of the abundances of the transcripts detected at 25 and 42°C for a given construct is provided in the rightmost column.
The −10 region is critical for temperature regulation.
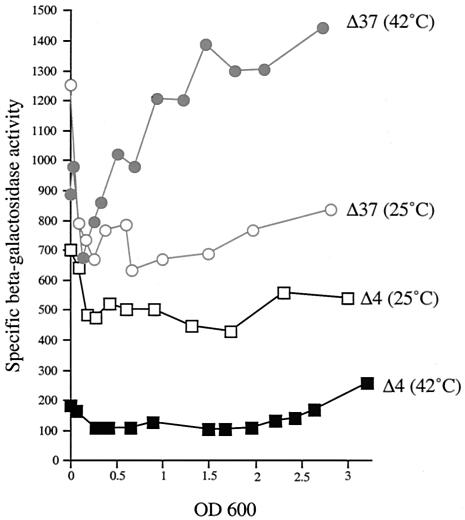
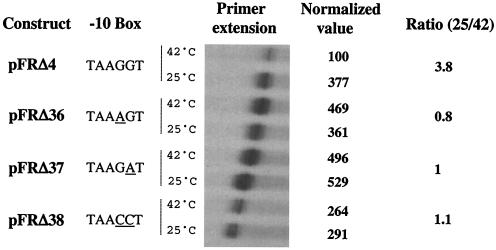
The data presented above suggest that the dsrA −10 region cannot function at all with many spacer sequences. Further analysis suggests that it is this “impaired” −10 region that is critical for temperature regulation. Constructs based on lacUV5p in which the −35 box (Δ25), the spacer sequence (Δ21), or the start site region (Δ28) was replaced with the corresponding sequences from dsrAp were still active but did not provide temperature regulation to these chimeric promoters (Table 1). Even constructs in which everything but the −10 region, or the −10 region and the start site region, was from dsrA (Δ29 and Δ22, respectively) were not temperature regulated and were very active (Table 1). These results all suggested a key role for the dsrA −10 region in temperature regulation, i.e., limiting of high-temperature expression. The sequence of the −10 box of dsrA (taAGGt; see Table 1) differs by three bases from the consensus one (taTAAt) found in lacUV5p. In vitro, dsrAp is effectively used by the σ70 transcription factor from a supercoiled template (22). Replacement of the three bases of dsrAp with the consensus sequence (Δ29) abolishes temperature regulation (Table 1). In the dsrA −10 box, the two G's in the fourth and fifth positions are rather unusual in E. coli promoters (15). In order to determine the importance of these two bases for the thermocontrol of dsrAp, we changed GG to AG (construct Δ36), to GA (construct Δ37), or to CC (construct Δ38) in an otherwise dsrAp context (Table 1). Figure 2 shows the result of a representative experiment with the Δ37 construct, present as a single-copy lacZ fusion, compared to the activity of wild-type dsrAp (Δ4); average activities are also shown in Table 1. At 25°C, both fusions have constant and similar activities at all growth points, although Δ37 has somewhat higher activity. At 42°C, the fusion carrying wild-type dsrAp displayed its characteristic lower activity compared to that at 25°C (about 4.5-fold lower). Remarkably, the fusion carrying the mutant promoter (construct Δ37) did not show a decrease in its activity; in fact, its activity was significantly higher than at 25°C, particularly at higher ODs (Fig. 2). Thus, a single base pair change in the −10 region abolishes the temperature response of dsrAp.
FIG. 2.
Expression during growth of dsrAp (Δ4) and point mutant construct Δ37 at 25 and 42°C. The curves provided correspond to a representative experiment. Each construct is carried as a single-copy fusion at the lambda att site of MC4100. Constructs vary from each other by the sequence of the −10 box: TAAGGT for Δ4 and TAAGAT for Δ37.
We could not isolate Δ36 and Δ38 as single-copy fusions in the chromosome and therefore assayed them in primer extension experiments with Δ37 and wild-type dsrAp as controls (Fig. 3). For all three point mutant constructs, Δ36, Δ37, and Δ38, transcripts expressed were not down-regulated by temperature. In fact, both Δ36 and Δ37 were higher at 42°C than at 25°C, as seen for β-galactosidase activity for Δ37 (Fig. 2). The level of expression for Δ36 was similar to that for Δ37. Therefore, changing either G within the −10 box to A is sufficient to make dsrAp temperature independent, increasing expression modestly at 25°C and drastically at 42°C. Thus, dsrAp acts as a strong promoter that cannot function at high temperature because of these G's in the −10 sequence. Δ38 had comparable activities at 25 and 42°C but had reduced activity compared to that of Δ37 or Δ36 at both temperatures (about twofold down; Fig. 3). Therefore, the replacement of the two G's in the fourth and fifth positions with two C's not only abolished the temperature control but also reduced the activity of the promoter modestly.
FIG. 3.
Primer extension analysis of the transcripts generated by dsrAp (Δ4) and −10 box point mutant constructs Δ36, Δ37, and Δ38 carried by plasmid vector pFRΔ. Sequences of the −10 boxes are provided for each construct. Results of quantitation are presented as described in the legend to Fig. 1.
DISCUSSION
dsrAp is regulated by temperature, operating efficiently at 25°C but poorly at 42°C. The chimeras tested here strongly suggest that the sequence of the promoter itself, and particularly the sequence of the −10 region, places constraints on promoter function that result in temperature-sensitive activity. We found that single nucleotide changes in the −10 region lead to high, temperature-independent activity, consistent with an inability of the dsrA −10 region to function at high temperature. Furthermore, the dsrA −10 region has a stringent requirement for a 17-bp spacer region, and, more surprisingly, for particular sequences within the spacer region. In the presence of other spacers, the promoter is inactive even at low temperature. Other components of the promoter (the −35 region and sequences upstream of it and sequences between the −10 region and the start site) also affect the activity of this promoter but not as drastically. Taken together, they all impinge on its activity at 42°C.
This picture is most consistent with a promoter poised at the edge of inactivity. Might this be due to a trans-acting protein? While we cannot fully rule this out, we find it difficult to imagine how such a protein would act, given the effects of various mutant promoters. If a protein stimulates at 25°C, it is unclear how a single base change in the −10 sequence (G to A) would render the promoter active at 42°C. Again, if a protein inhibits at 42°C, we would have to propose that the G-to-A change prevents protein binding and would need another explanation for why the activity of dsrAp is dependent on the sequence of the spacer at 25°C. In addition, the effects of flanking sequences are difficult to explain by a model requiring a protein mediating temperature control, other than RNA polymerase, which is known to have multiple contacts with the promoter region. Attempts to identify multicopy plasmids that perturbed temperature regulation (by overexpression of this hypothetical protein) were unsuccessful. One protein previously implicated in thermoregulation, Hns, usually is found to repress expression at low temperature, in contrast to what was found here (reviewed in reference 8). Mutations in hns did not show any significant effects on DsrA amounts at high temperature in Northern blots (data not shown).
In previous in vitro experiments (22), dsrAp was transcribed without additional factors, from a supercoiled DNA template, but we were not able to duplicate the temperature sensitivity seen in vivo. Varying of the supercoiling of the template plasmid and/or other properties of the in vitro reaction would have to be carried out to fully understand whether some additional factor is necessary for temperature regulation. The availability of single base changes that obliterate temperature sensitivity should provide excellent control substrates for in vitro studies to define the aspect of polymerase-DNA interaction that is perturbed at high temperature.
What are the characteristics of this promoter that limit high-temperature activity? We have noted already the importance of the two G's in the −10 region. In a compilation of 472 E. coli promoter sequences (12), only 6 were found with apparent −10 sequences of either TATGGT or TAAGGT. While parallel experiments have not been done with these, we might predict that they would all be relatively intolerant of changes in the sequence and length of their spacer regions, dependent on positive activators, or otherwise impaired. In terms of what these constraints are for dsrA, while retention of all of the AT-rich spacer is not necessary (see, for instance, Δ13), any of the three constructs in which the central, conserved portion of the spacer was changed from AATT gave a significant decrease in expression (sevenfold or greater; Table 1, Δ32, Δ39, and Δ40) at 25°C. While further analysis of the spacer sequence might clarify the situation somewhat, we note some previous studies on spacer sequences. The galP1 promoter has a −10 region not unlike that for dsrA (TATGGT) but also contains an extended −10 sequence (TGN). Any changes in this extended −10 region significantly decreased the activity of the promoter; even when the extended −10 region was present, other spacer sequence changes affected promoter activity when the −35 region was not optimal (3). DsrA does not carry an extended −10 region, and the effect of such a sequence was not tested here, but when the favorable G at −14 was changed (Δ40), activity at 25°C dropped significantly (Table 1). In studies of the consequences of changes in the spacer sequence for the PRM promoter (−10 sequence, TAGATT) (1, 2), the more modest effect on activity seen there was ascribed to changes in the orientation of the −10 and −35 regions; such changes could well be responsible for the strong effects of changes in the spacer seen here. Recent studies on the in vitro bend provided by an A tract as a function of temperature show a decrease in the bend at elevated temperatures (14, 26). If such changes in orientation occur in vivo for dsrAp, we suggest that it is in the context of suboptimal −10 sequences (and a unique start site sequence, see below) that this becomes most important. For lacUV5 with a consensus −10 region, both 18- and 17-bp spacers were well tolerated, although use of the 17-bp dsrA spacer in place of the 18-bp lacUV5 spacer did increase activity almost twofold (compare Δ31 and Δ21).
The start site region also contributes to the temperature sensitivity of dsrAp. There are four active pairs of promoters in our work that differ by whether they carry the start site region from dsrA or that from lacUV5. Once again, the nature of the −10 region changes the sensitivity of the promoter to the start site sequence. Thus, replacement of the dsrA start site sequence in either wild-type lacUV5p (compare Δ28 to Δ31) or a promoter with the dsrA spacer and −35 region but the lacUV5 −10 box (compare Δ29 to Δ22) reduces activity modestly (1.5-fold in one case and 1.1-fold in another, at both 25 and 42°C). By contrast, in the two pairs in which the −10 box is from dsrA, the lacUV5 start site sequence allows 5- to 6-fold higher activity at 42°C (compare Δ26 to Δ4 and Δ41 to Δ24) and a more modest (1.8- to 1.9-fold) increase in activity at 25°C. Primer extension experiments with a number of these constructs confirmed that the start site was dictated by the sequence between the −10 box and the expected start site (data not shown). These results suggest that however the dsrA start site sequence participates in transcription initiation, at high temperature this works poorly, helping to restrict the activity of dsrAp at high temperatures. We found two other promoters in the E. coli promoter database with a similar sequence between the −10 region and the start site; both had a start site somewhat closer than expected to the −10 region, as does dsrA, confirming that this unusual sequence leads to an unusually spaced start (12). One, uvrCP3, also has an unusual −10 sequence (TATGCT) and has AATT at the same position in the spacer region; possibly this promoter is controlled in a manner similar to that of dsrA.
The sequence of the −35 box, TTGTCA, is fairly close to the consensus for RNA polymerase associated with σ70 (TTGACA) (9, 12). Just upstream of the −35 box, we previously defined an activator sequence, AATATTT, recognized by Eσ70 in vitro. While we found temperature regulation in the absence of this A-rich region (22), we noted a modest effect of the −35 region on both temperature regulation and dsrAp activity. Thus, as for the start site region, in the context of the lacUV5 −10 region, changing the −35 region only slightly changes promoter activity at either 25 or 42°C (compare Δ31 to Δ25 [1.4-fold increase at both temperatures with the dsrA −35 region] and Δ21 to Δ22 [no change in expression with the dsrA −35 region]). However, for constructs with the −10 region from dsrA, the dsrA −35 region increases activity at 25°C about 1.5-fold and decreases it at 42°C (compare Δ24 to Δ4 and Δ41 to Δ26).
In summary, dsrAp appears to achieve temperature regulation by a complex combination of promoter elements, most strikingly, an unusual −10 region. While we cannot rule out trans-acting protein effectors, our results are most consistent with the notion that the geometry of the promoter, coupled with intrinsic changes in DNA bending and possible changes in supercoiling with temperature, dictates temperature regulation via changing interactions with RNA polymerase. Detailed in vitro studies are necessary to demonstrate this unequivocally.
Acknowledgments
We thank Nadim Majdalani, Eric Massé, Yan-Ning Zhou, and Sankar Adhya for comments on the manuscript and discussions.
REFERENCES
- 1.Auble, D. T., T. L. Allen, and P. L. deHaseth. 1986. Promoter recognition by Escherichia coli RNA polymerase: effects of substitutions in the spacer DNA separating the −10 and −35 regions. J. Biol. Chem. 261:11202-11206. [PubMed] [Google Scholar]
- 2.Auble, D. T., and P. L. deHaseth. 1988. Promoter recognition by Escherichia coli RNA polymerase: influence of DNA structure in the spacer separating the −10 and −35 regions. J. Mol. Biol. 202:471-482. [DOI] [PubMed] [Google Scholar]
- 3.Burr, T., J. Mitchell, A. Kolb, S. Minchin, and S. Busby. 2000. DNA sequence elements located immediately upstream of the −10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res. 28:1864-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crothers, D. M., T. E. Haran, and J. G. Nadeau. 1990. Intrinsically bent DNA. J. Biol. Chem. 265:7093-7096. [PubMed] [Google Scholar]
- 7.Drlica, K., and N. R. Perl-Rosenthal. 1999. DNA switches for thermal control of gene expression. Trends Microbiol. 7:425-426. [DOI] [PubMed] [Google Scholar]
- 8.Eriksson, S., R. Hurme, and M. Rhen. 2002. Low-temperature sensors in bacteria. Philos. Trans. R. Soc. Lond. B 357:887-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herendeen, S. L., R. A. VanBogelen, and F. C. Neidhardt. 1979. Levels of major proteins of Escherichia coli during growth at different temperatures. J. Bacteriol. 139:185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hershberg, R., G. Bejerano, A. Santos-Zavaleta, and H. Margalit. 2001. PromEC: an updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 29:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurme, R., and M. Rhen. 1998. Temperature sensing in bacterial gene regulation—what it all boils down to. Mol. Microbiol. 30:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Jerkovic, B., and P. H. Bolton. 2000. The curvature of dA tracts is temperature dependent. Biochemistry 39:12121-12127. [DOI] [PubMed] [Google Scholar]
- 15.Lisser, S., and H. Margalit. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Pepe, C. M., C. Suzuki, C. Laurie, and R. W. Simons. 1997. Regulation of the “tetCD” genes of transposon Tn10. J. Mol. Biol. 270:14-25. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Martin, J., F. Rojo, and V. Lorenzo. 1994. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 58:268-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phadtare, S., J. Alsina, and M. Inouye. 1999. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2:175-180. [DOI] [PubMed] [Google Scholar]
- 21.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, M. P. Rivas, and C. L. J. Turnbough. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Repoila, F., and S. Gottesman. 2001. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 183:4012-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 26.Tchernaenko, V., M. Radlinska, C. Drabik, J. Bujnicki, H. R. Halvorson, and L. C. Lutter. 2003. Topological measurement of an A-tract bend angle: comparison of the bent and straightened states. J. Mol. Biol. 326:737-749. [DOI] [PubMed] [Google Scholar]
- 27.Yura, T., M. Kanemori, and M. T. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 28.Zhou, Y.-N., and S. Gottesman. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 180:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]