Abstract
The ability of Mycobacterium tuberculosis auxotrophs to survive long-term starvation was measured. Tryptophan and histidine auxotrophs did not survive single-amino-acid starvation, whereas a proline auxotroph did. All three auxotrophs survived complete starvation. THP-1 cells were also able to restrict the growth of the tryptophan and histidine auxotrophs.
Mycobacterium tuberculosis is exposed to low or restricted nutrient concentrations in the host and is able to survive these conditions for long periods of time (2). Several studies suggest that certain amino acids are not available within the compartment of the macrophage in which the bacteria reside, since leucine, tryptophan, and arginine auxotrophs do not survive or multiply in macrophages (1, 5, 6, 13). I investigated the ability of three auxotrophs and one wild-type strain to survive different starvation conditions (Table 1). The proline auxotroph (Tame 2) has been described previously (9). The unmarked tryptophan and histidine auxotrophs were constructed by homologous recombination. Suicide (nonreplicating) delivery vectors were constructed with a rapid cloning system (11); the central 0.3-kbp NotI fragment internal to trpD was deleted, and the central 0.7-kbp PstI fragment of hisDC was deleted. A two-step procedure to replace the wild type with the unmarked, deleted gene was carried out (11). The expected genotype of the mutants was confirmed by Southern blotting, and both these strains showed the expected auxotrophy (data not shown).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Genotype | Source or reference |
|---|---|---|---|
| M. tuberculosis strain | |||
| H37Rv | Wild type | trpD+proC+hisDC+ | ATCC 25618 |
| Tame 2 | Proline auxotroph | trpD+proCΔ::hyg hisDC+ | 9 |
| Tame 5 | Tryptophan auxotroph | trpDΔ proC+hisDC+ | This study |
| Tame 24 | Histidine auxotroph | trpD+proC+hisDCΔ | This study |
| Plasmid | |||
| pSM128 | Promoter probe vector, lacZ reporter gene, Sm | 4 | |
| pTRIP5 | 0.2 kbp upstream of trpECBA operon in pSM128 | This study | |
| pTRIP7 | 0.2 kbp upstream of trpD gene in pSM128 | This study | |
| pTRIP10 | 0.8 kbp upstream of trpE2 gene in pSM128 | This study | |
| pHIP5 | 0.52 kbp SmaI upstream of hisD in pSM128 | This study |
We had previously shown that a tryptophan auxotroph is severely attenuated in both macrophage and mouse models of infection (13), suggesting that the organisms are sequestered in an intracellular compartment from which they cannot obtain tryptophan. We do not know if clearance of the bacteria is caused by death due to tryptophan starvation or by active killing of the bacteria by the macrophage. In order to address this question in vitro, I examined the survival ability of the tryptophan auxotroph on withdrawal of tryptophan in axenic culture.
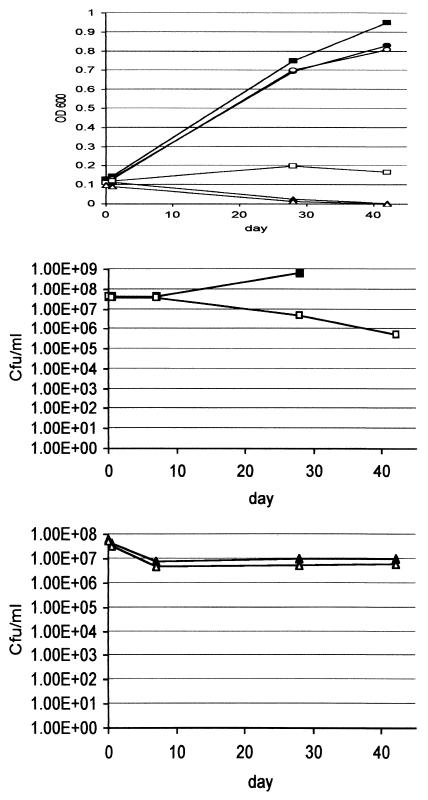
Bacteria were subjected to single-amino-acid starvation (Middlebrook 7H9 liquid plus 10% oleic acid-albumin-dextrose-catalase [OADC; Becton Dickinson] and 0.05% [wt/vol] Tween 80 without amino acid supplementation) and complete starvation (no nutrients, i.e., sterile distilled water). Lysis was measured by monitoring the optical density at 600 nm (OD600), and viability was measured by determining the numbers of CFU per ml on Middlebrook 7H10 agar containing 10% (vol/vol) OADC supplemented with 40 μg of amino acid per ml as required (Fig. 1). Both the wild type and tryptophan auxotroph grew normally in supplemented medium (Middlebrook 7H9 liquid containing 10% OADC, 0.05% [wt/vol] Tween 80, and 40 μg of amino acid per ml). There was no difference in their ability to survive complete starvation. The cultures in water showed an immediate drop in OD600 to below the limit of detection and an accompanying log reduction in CFU. There was no further death over 7 weeks. The fact that these bacteria have a remarkable ability to survive complete starvation for long periods of time suggests that there must be a coordinated response to the lack of nutrients which changes their physiology from actively growing cells to persistent (nongrowing) cells. In the nonsupplemented medium, the wild-type strain grew normally; however, the tryptophan auxotroph showed an unusual phenotype. Although the OD600 remained stable, indicating that the cells did not lyse, there was a 2-log reduction in cell viability over the course of the 7-week experiment. Whatever response the bacteria made to complete starvation was not induced solely in response to tryptophan depletion.
FIG. 1.
Short-term starvation survival of the tryptophan auxotroph. The tryptophan auxotroph and wild-type strains were inoculated into the indicated media to an OD600 of 0.1 and incubated at 37°C. OD600 readings (A) and CFU (B and C) were measured over 42 days. Filled shapes, wild type; open shapes, tryptophan auxotroph; squares, nonsupplemented 7H9; circles, tryptophan-supplemented 7H9; triangles, water. Cultures were assayed in duplicate with a variation of less than 10%. Note that no wild-type CFU were measured at day 42.
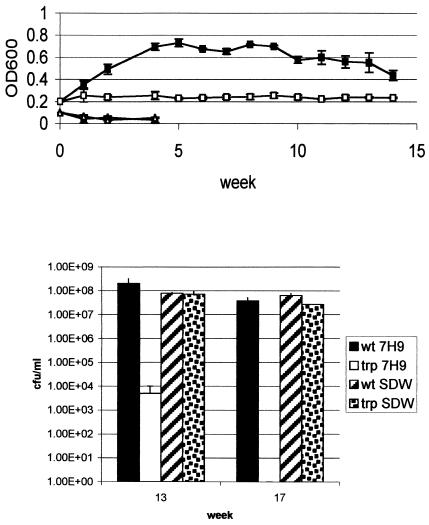
I repeated the survival experiment over a longer time scale to determine if this pattern of survival would be maintained (Fig. 2). Over 15 weeks, the OD600 of the tryptophan auxotroph remained constant in nonsupplemented medium, while the OD600 in water rapidly declined in the first week. Again, this did not mirror the viability counts. The wild type showed a small (<1 log) reduction in viability after extended stationary-phase growth (17 weeks) in 7H9 medium, while the tryptophan auxotroph lost viability over this time scale with a 4-log reduction at 13 weeks and a complete loss of viability (approximately 8 log) by 17 weeks. In contrast, there was no difference in the two strains' ability to survive in water, with both strains losing less than 0.5-log viability over 17 weeks. I also determined whether the tryptophan auxotroph could be recovered from the nonsupplemented medium by subculturing into supplemented liquid medium, but no growth occurred after repeated subculture, thus indicating that the cells were nonviable and nonculturable (data not shown).
FIG. 2.
Long-term starvation survival of the tryptophan auxotroph. The tryptophan auxotroph and wild-type strains were inoculated into the indicated media to an OD600 of 0.2 and incubated at 37°C. OD600 readings (A) and CFU (B) were measured over 17 weeks. Filled shapes, wild type; open shapes, tryptophan auxotroph; squares, nonsupplemented 7H9; triangle, water.
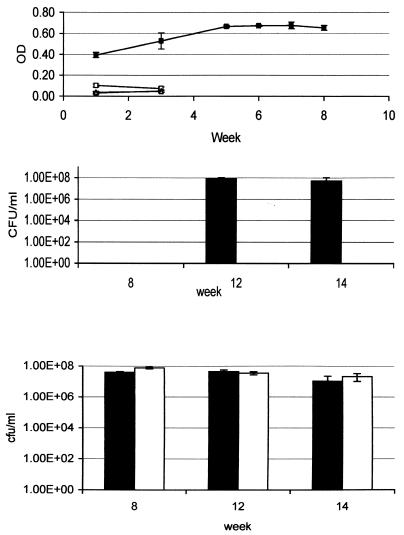
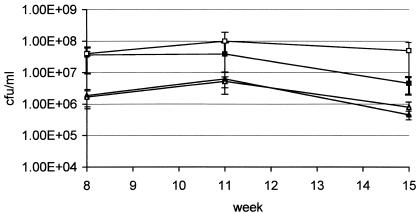
In order to determine if the inability to survive amino acid starvation was a general feature of auxotrophs, I studied the histidine and proline auxotrophs in the same system. The histidine auxotroph behaved in a manner similar to that of the tryptophan auxotroph (Fig. 3). Prolonged survival of complete starvation (i.e., over 14 weeks) was seen after an initial log reduction in viability, whereas deprivation of histidine led to an approximate 8-log reduction in viability (no viable bacteria left) after only 8 weeks; this was a much more rapid loss of viability than that exhibited by the tryptophan auxotroph. In marked contrast, the proline auxotroph did not lose any viability in nonsupplemented media over 15 weeks (Fig. 4). Survival of the proline auxotroph in water was the same as that for the other strains. Thus, the ability of auxotrophs to survive starvation is dependent on the particular amino acid that they require. Histidine and tryptophan auxotrophs cannot withstand individual amino acid deprivation, whereas a proline auxotroph can.
FIG. 3.
Long-term starvation survival of the histidine auxotroph. The histidine auxotroph and wild-type strains were inoculated into the indicated media to an OD600 of 0.1 and incubated at 37°C. OD600 readings (A) and CFU (B and C) were measured over 14 weeks. Filled shapes, wild type; open shapes, histidine auxotroph; squares, nonsupplemented 7H9; triangles, water. Panel B depicts results for which nonsupplemented 7H9 was used; panel C depicts results achieved with water. Note that no wild-type CFU were measured at week 8.
FIG. 4.
Long-term starvation survival of the proline auxotroph. The proline auxotroph and wild-type strains were inoculated into the indicated media to an OD600 of 0.1 and incubated at 37°C. CFU were measured over 15 weeks. Filled shapes, wild type; open shapes, proline auxotroph; squares, nonsupplemented 7H9; triangles, water.
The possibility that amino acids were biologically available from the bovine serum albumin in the OADC supplement was considered unlikely, as none of the auxotrophs were able to grow without individual amino acid supplementation. I also repeated the experiment in a chemically defined medium (1.5 g of K2HPO4 per liter, 0.5 g of KH2PO4 per liter, 0.5 g of MgSO4 per liter, 0.5 mg of CaCl per liter, 0.1 mg of ZnSO4 per liter, 0.1 mg of CuSO4 per liter, 50 mg of ferric chloride per liter, and 30 mM NH4SO4) with and without OADC. Survival was assessed after 12 weeks. As was the case for Middlebrook media, the histidine and tryptophan auxotrophs showed a significant decrease in viability, whereas the proline auxotroph and wild-type strains remained viable, thereby confirming that the bovine serum albumin does not directly affect the viability of these strains.
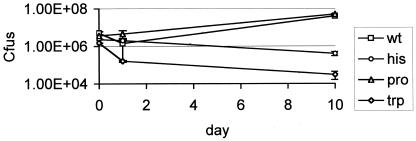
Previously, both the proline auxotroph and a different (marked) tryptophan auxotroph had been shown to be attenuated to different degrees in murine macrophages and mice (13). The histidine auxotroph had not previously been assayed in any model of virulence. I used the human THP-1 cell line in order to quickly determine whether the two new mutants were attenuated. THP-1 cells were infected at a multiplicity of infection of 1 as described previously (8) (Fig. 5). As before, the tryptophan auxotroph showed attenuation in this model, and a reduction in bacterial numbers over 10 days was observed. The histidine auxotroph was attenuated to a similar degree. The proline auxotroph showed the same growth profile as the wild type. These results mirror the starvation results in that the tryptophan and histidine auxotrophs could not survive amino acid starvation, whereas the proline auxotroph could. These findings led to the hypothesis that the intracellular environment does not contain certain amino acids but does contain other nutrients, as the bacteria did not enter the same state of persistence that they did in the complete-starvation model.
FIG. 5.
Intracellular survival. THP-1 cells were infected at a multiplicity of infection of 1, and the number of intracellular viable bacteria was measured over 10 days. Results are given as the means of triplicate wells. wt, wild type.
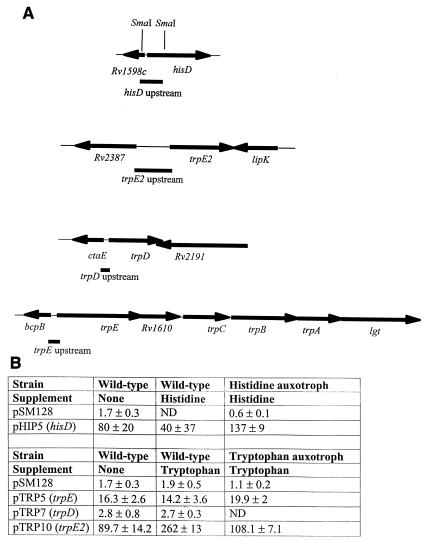
In many bacteria, including M. tuberculosis, the stringent response is activated in response to nutrient starvation, including amino acid deprivation (12). This response is mediated by intracellular levels of (p)ppGpp and the RelA protein, resulting in the downregulation of most genes and the upregulation of a specific subset of genes. This subset includes amino acid biosynthesis genes and proteases, including the histidine operon (3). If this response occurred in the histidine auxotroph, then all of the amino acids recycled from other proteins could be directed into making histidine biosynthetic enzymes, possibly resulting in uncontrolled expression. This could also account for the same phenotype in the tryptophan auxotroph. In order to address this possibility, I looked at the promoter activity of the three trp loci and the his operon. If runaway transcription were occurring, then we would expect to see the transcription of these operons being controlled by the appropriate amino acid. The upstream regions of the three trp loci were PCR amplified and subcloned into the promoter probe vector pSM128. The 0.5-kbp SmaI fragment upstream of the his operon was subcloned from pHIS1 into pSM128. Plasmids were electroporated into wild-type and auxotrophic strains, transformants were selected on 20 μg of streptomycin (and amino acid where required) per ml, and promoter activity was assayed (Fig. 6). None of the promoters were repressed by the relevant amino acid and, counterintuitively, the trpE2 promoter was upregulated twofold by the addition of tryptophan. The plasmid pSM128 is an integrating vector which occurs in one copy per cell, so the lack of transcriptional control is not due to a copy number effect. In Streptomyces spp., the main trp operon is regulated by growth phase rather than by tryptophan availability (7). The apparent induction of trpE2 may therefore result from an increased growth rate upon the addition of tryptophan rather than from a direct effect of the amino acid. The histidine operon was not controlled by histidine, although there was a slightly higher level of promoter activity in the hisDC mutant strain. Thus, the enzymes of these two biosynthetic pathways are not controlled transcriptionally in response to amino acid availability. This may reflect a general pattern in M. tuberculosis in which amino acid biosynthesis genes are constitutively expressed, and the finding meshes with the previous observation that the impA gene resides within the his operon because of its constitutive nature (10).
FIG. 6.
Promoter activity analysis. (A) Constructs to assay promoter activity. The indicated regions upstream of each gene were cloned into the promoter probe vector pSM128. (B) Promoter activity. The plasmids depicted in panel A were transformed into the wild-type and auxotrophic strains. Transformants were grown in 10 ml of medium with the indicated supplement and assayed for beta-galactosidase activity. Each value represents the mean ± standard deviation for three independent transformants, each assayed in duplicate. ND, not determined.
In conclusion, both auxotrophic and wild-type strains of M. tuberculosis are able to survive long periods of complete starvation. However, histidine and tryptophan auxotrophs are unable to survive single-amino-acid starvation. These results have implications for live, attenuated vaccine development in situations in which the persistence of auxotrophic strains is assumed to be required for generation of protective immunity and for drug development scenarios for which drugs that target bacteria in nongrowing states are required. The results presented here demonstrate that M. tuberculosis has a different response to different types of starvation and that models of persistence involving starvation may need to be refined.
Acknowledgments
I thank Paul Wheeler and Todd Primm for useful discussions.
REFERENCES
- 1.Bange, F. C., A. M. Brown, and W. R. Jacobs. 1996. Leucine auxotrophy restricts growth of Mycobacterium bovis BCG in macrophages. Infect. Immun. 64:1794-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 3.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. The stringent response, p. 1497-1512. In F. C. Neidheart (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 4.Dussurget, O., J. Timm, M. Gomez, B. Gold, S. W. Yu, S. Z. Sabol, R. K. Holmes, W. R. Jacobs, and I. Smith. 1999. Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. J. Bacteriol. 181:3402-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordhan, B. G., D. A. Smith, H. Alderton, R. A. McAdam, G. J. Bancroft, and V. Mizrahi. 2002. Construction and phenotypic characterization of an auxotrophic mutant of Mycobacterium tuberculosis defective in l-arginine biosynthesis. Infect. Immun. 70:3080-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hondalus, M. K., S. Bardarov, R. Russell, J. Chan, W. R. Jacobs, and B. R. Bloom. 2000. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infect. Immun. 68:2888-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu, D. S. J., D. W. Hood, R. Heidstra, and D. A. Hodgson. 1999. The expression of the trpD, trpC, and trpBA genes of Streptomyces coelicolor A3(2) is regulated by growth rate and growth phase but not by feedback repression. Mol. Microbiol. 32:869-880. [DOI] [PubMed] [Google Scholar]
- 8.Lukey, P. T. 2001. Macrophage virulence assays, p. 271-280. In T. Parish and N. G. Stoker (ed.), Mycobacterium tuberculosis protocols. Humana Press, Totowa, N.J.
- 9.Parish, T., B. G. Gordhan, R. A. McAdam, K. Duncan, V. Mizrahi, and N. G. Stoker. 1999. Production of mutants in amino acid biosynthesis genes of Mycobacterium tuberculosis by homologous recombination. Microbiology 145:3497-3503. [DOI] [PubMed] [Google Scholar]
- 10.Parish, T., J. Liu, H. Nikaido, and N. G. Stoker. 1997. A Mycobacterium smegmatis mutant with a defective inositol monophosphate phosphatase gene homolog has altered cell envelope permeability. J. Bacteriol. 179:7827-7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 12.Primm, T. P., S. J. Andersen, V. Mizrahi, D. Avarbock, H. Rubin, and C. E. Barry. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182:4889-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith, D. A., T. Parish, N. G. Stoker, and G. J. Bancroft. 2001. Characterization of auxotrophic mutants of Mycobacterium tuberculosis and their potential as vaccine candidates. Infect. Immun. 69:1142-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]