Abstract
Cosmids are plasmids that contain the phage λ sequences (cos) required for packaging of the phage DNA into the virion. Induction of a λ prophage in an Escherichia coli strain carrying a cosmid results in lysates containing phage particles that are filled with cosmid DNA. However, the lysates also contain a large excess of infectious phage particles which complicate use of the packaged cosmids. I report that cosmids packaged by induction of a strain carrying a prophage with an altered cos region results in lysates containing very high levels (>1010/ml) of particles that contain cosmid DNA together with very few infectious phage particles. These lysates can be used to transduce cosmid DNA into all of the cells of a growing culture with minimal physiological disturbance. When the cosmid carries a conditionally active origin of replication, transductional introduction of the cosmid under nonreplicative conditions provides a system of transient expression. Transient expression has been used to make a recA strain temporarily recombination proficient and to temporarily introduce a site-specific recombinase. Transductional introduction of a cosmid also allows absolute off-to-on transcriptional control of nonessential genes. Two examples are given showing that when a strain carrying a null mutation in the gene of interest is transduced with a packaged cosmid carrying a functional copy of that gene, the expression of the gene rapidly goes from absolutely off to high-level expression. Additional possible uses of in vivo-packaged cosmids are proposed.
The techniques generally used to introduce plasmid DNA into bacterial cells, chemical transformation (19, 24) and electroporation (13, 19), suffer from two disadvantages in physiological studies. First, these methods either introduce DNA into only a small fraction of the cells, or the treatment required for DNA uptake seriously disturbs cellular metabolism, resulting in death of a large fraction of the cells. In chemical transformation, <10% of the cells take up plasmid (19), whereas in electroporation, although 80% of the surviving cells are transformed (19), only 10 to 50% of the cells survive the electroshock treatment (13). In contrast, infection with bacteriophages is generally highly efficient (1). All of the cells of a culture can be infected, and evolution has optimized the infection process to ensure minimal metabolic disturbance of the host.
To take advantage of these properties, bacterial genes have been introduced into the genomes of several phages, the most popular being phages M13 and λ. These recombinant phages were then used to efficiently introduce bacterial and foreign DNA segments into the cells of growing Escherichia coli cultures (4, 45). However, these phages have advantages and disadvantages as DNA introduction vehicles. A major advantage of phage M13 is that the double-stranded replicative form of the phage genome can be treated as a plasmid in cloning procedures, and vectors have been constructed that contain a multiple cloning site plus lacZ α-complementation screening (53). Moreover, plasmids that carry the M13 fragment containing the phage replication origin and DNA packaging determinant can also be replicated and packaged into M13 virions by use of a helper phage (53). However, a major disadvantage is that infection of E. coli with M13 is slow and incomplete, thereby resulting in mixed populations of uninfected and asynchronously infected cells (37). Moreover, M13 DNA replication is of low fidelity, and deleted versions of the viruses or phagemids often accumulate (53). Phage λ has different advantages and disadvantages. Unlike M13, λ very rapidly and efficiently infects its host. However, λ has a large genome (48.5 kb) and, thus, unique restriction sites are few and difficult to introduce. Moreover, most λ vectors are designed to accept only large (15- to 20-kb) DNA inserts and have lost the ability to form lysogens (33). Bacterial genes can be placed on λ phages by homologous recombination, but this requires construction of suitable acceptor phages and donor plasmids plus a screen or selection to identify recombinant phages (e.g., see reference 4). Moreover, the emergence of high-capacity plasmid vectors such as bacterial artificial chromosomes has resulted in decreased familiarity with the techniques of lambda cloning. In contrast, plasmid cloning is a dominant technique of molecular genetics and, thus, a plasmid that could be packaged into λ phage particles would provide both ease of cloning and efficient introduction of plasmid DNA without major physiological disturbance. Such plasmids, called cosmids, which carry the cos region of λ were described many years ago (9, 23). The 223-bp cos region contains the determinants for processive packaging of the oligomeric phage DNA and includes the site (cosN) at which the cohesive ends of λ virion DNA are generated (16, 51).
When the cos sequence is present in a plasmid and the plasmid DNA is of sufficient length (about 37 to 52 kb), it can be packaged into phage particles either in vitro (22) or in vivo (15, 31). In vitro packaging is often used for preparation of clone banks, but the titers (<108 cosmid-containing particles/ml) of packaged cosmids are too low for facile physiological experiments. In vivo packaging of cosmids upon induction of a λ lysogen was discovered over 20 years ago (47, 48), but has only occasionally been utilized in genetic analysis (e.g., [50]). In vivo packaging is known to result in particles that contain either cosmid DNA or phage DNA (31, 48). The major drawbacks of the prior cosmid packaging systems were low yields of packaged cosmids and the large excess (10- to 1,000-fold) of infectious phage particles over the particles containing cosmid DNA. I report the construction and properties of a modified λ lysogen in which the cosmid DNA is much more efficiently packaged than the phage DNA. Examples of the use of this system to give transient expression and absolute transcriptional control of genes in all of the cells of a growing culture are reported.
MATERIALS AND METHODS
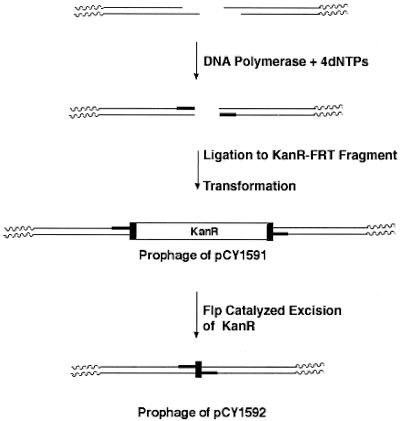
Construction of λ helper phages (Fig. 1).
FIG. 1.
Construction of the prophages of strains pCY1591 and pCY1592. The thin lines denote the cohesive ends of λ DNA, whereas the wavy lines denote the rest of λ (the circular molecule). The thick lines denote the nucleotides added by the DNA polymerase, and the upright solid rectangles at each end of the Kanr determinant represent the FRT sites. Construction of the prophage of strain pCY1590 followed the same scheme as that for pCY1591 except that the Kanr determinant lacked FRT sites, and so a strain analogous to pCY1592 could not be made. The drawing is not to scale.
λ phage DNA (Promega) carrying cI857 and Sam7 was heated to melt the cohesive ends which were filled-in with E. coli DNA polymerase I (Klenow fragment) and the four deoxynucleotide triphosphates. Another sample of λ phage DNA was similarly treated with phage T4 DNA polymerase. The resulting blunt-ended λ DNAs were then ligated to blunt-ended kanamycin resistance cassettes derived by digestion of plasmid pKD4 (11) with either XbaI or HindIII, followed by treatment with Klenow polymerase as above. The ligation mixtures were used to transform strain DH10B (19) by electrophoresis, and transformants resistant to 25 μg of kanamycin/ml were selected at 30°C and screened for an inability to grow at 42°C. The plasmid pKD4 (11) kanamycin cassette is flanked by directly repeated Flp recombinase target (FRT) sites allowing excision of the cassette upon expression of Flp recombinase. Strain CY1591 was cured of the kanamycin resistance determinant (Fig. 1) by transforming the strain with plasmid pCP20 (7) and growing the transformed strain in Luria-Bertani (LB) medium lacking antibiotic for many generations at 33.5°C followed by screening single colonies for sensitivity to both kanamycin and chloramphenicol. The resulting lysogen (strain CY1592) had cosmid packaging properties very similar to those of the parental strain.
Cosmid and plasmid constructions.
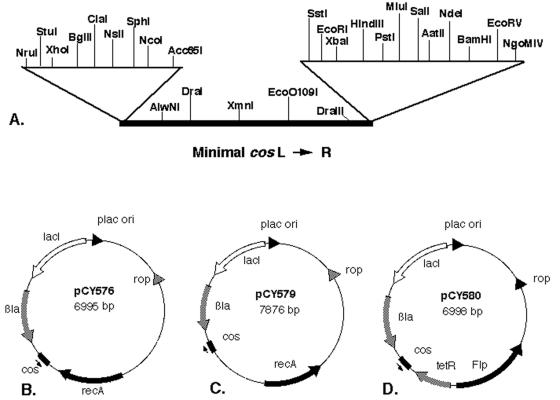
The 223-bp HaeIII cos fragment of pCY128 (21), which is the minimal cos sequence that retains full function (32), was inserted into the SmaI site of pMTL23P (6). Both orientations of cos were obtained; the cos of pCY566 has the same orientation as the truncated lacZ gene, whereas pCY567 has the opposite orientation (Fig. 2A). Derivatives of both plasmids (called pCY572 and pCY573, respectively) were made in which cos was tagged with a chloramphenicol resistance determinant by insertion of the SstI-derived chloramphenicol resistance cassette of pGC1 into the SstI site adjacent to cos. Plasmid pGC1 is a ΔBspHI ampicillin-sensitive derivative of p34S-Cm2 (12) constructed by G. Cronan of this laboratory. Plasmid pCY574 was constructed by digestion of pCY566 with NsiI and HindIII and ligation of the cos-Cm2 fragment of pCY572 to pTrueBlue-rop (Genomics One) digested with the same enzymes, followed by selection for transformants resistant to both ampicillin and chloramphenicol. Plasmid pCY575 was constructed by parallel manipulations, except that the enzymes used were AatII and ClaI and the recipient plasmid was pHC123 (8). Plasmid pCY580 was constructed in two steps. First, plasmids pAM34 (18) and pFT-K (36) were digested with SalI and BbuI and ligated, and transformants resistant to ampicillin and kanamycin were selected at 30°C on plates lacking isopropyl-β-d-thiogalactopyranoside (IPTG) and then screened for an IPTG requirement at 42°C. The resulting fusion plasmid was then digested with ClaI and SstI and ligated to the ClaI-SstI cos-containing fragment of pCY573, resulting in pCY580 (Fig. 2D). Plasmid pCY579 was constructed by ligation of the BamHI recA fragment of pBEU14 (46) to BglII-digested pCY566. The resulting plasmid was digested with XhoI and SstI, and the recA plus cos fragment was ligated to pAM34 digested with SalI and SstI followed by transformation in the presence of 0.5 mM IPTG to give pCY579 (Fig. 2C). A PCR product containing the recA gene of Salmonella enterica serovar Typhimurium strain LT-2 was obtained using chromosomal DNA and the primers STM RecAFor (5′-CCACAGCACGCATTATGC) and STM RecARev (5′-GTTAAGCAACCCGCGT) by N. De Lay of this laboratory. The PCR product was inserted into pCR2.1 TOPO (Invitrogen) and transformed into a recA strain. Two recombinants were shown to have functional recA genes by their ability to act as recipients in P1 transduction. One of these plasmids was digested with SnaBI, which cuts downstream of the recA coding sequence, and ligated to a cos-containing fragment of pCY567 obtained by digestion with BbuI plus PstI, followed by treatment with T4 DNA polymerase as described above. A recombinant plasmid having cos in the orientation opposite that of recA was digested with SpeI and HindIII and ligated to pAM34 digested with XbaI and HindIII followed by transformation in the presence of IPTG to give plasmid pCY576 (Fig. 2B). The phage T7 RNA polymerase expression plasmid pCY598 was constructed by ligation of the NsiI-NgoMIV AraC plus T7 gene 1 fragment of pTara (52) to the PstI-NgoMIV fragment of plasmid pDLK29 (35), which carries the RSF1030 replication origin plus kanamycin resistance.
FIG. 2.
Plasmids constructed. (A) The polylinker and cos region of plasmid pCY566 (the backbone of the plasmid is very similar to that of pUC18). Plasmid pCY567 has cos in the opposite orientation. (B to D) Structures of cosmids pCY576 (B), -579 (C), and -580 (D), which contain the IPTG-dependent replication origin of pAM34 plus the recA of S. enterica serovar Typhimurium, the recA of E. coli, or the FLP gene of Saccharomyces cerevisiae, respectively. The recA genes are expressed from their native promoters, whereas FLP is expressed from the Tn10 tetA promoter.
Media and bacterial methods.
The media used were LB (30) and RB (10 g of tryptone, 1 g of yeast extract, 5 g of NaCl per liter). Maltose, arabinose, glucose, and IPTG were added to final concentrations of 0.2, 0.2, and 0.4% and 0.5 to 1 mM, respectively. Final antibiotic concentrations were sodium ampicillin (100 μg/ml), kanamycin sulfate (25 to 50 μg/ml), chloramphenicol (10 μg/ml), and tetracycline HCl (10 μg/ml). The FLP gene of pCY575, which is expressed from the tetA tetracycline promoter, was induced by addition of anhydrochlorotetracycline (synthesized by dehydration of chlorotetracycline [44]) to a final concentration of 250 ng/ml. All bacterial growth was at 37°C unless otherwise stated. Cosmids were introduced into lysogenic strains by chemical transformation followed by plating for antibiotic-resistant colonies at 30°C (19). Cultures of antibiotic-resistant transformants were grown at 30°C to mid-exponential phase in glucose-supplemented LB medium supplemented with glucose and 50 mM Tris-HCl buffer (pH 7.5), then induced by temperature shift to 43°C for 15 to 25 min, and then shifted to 37°C (3). Induction was then allowed to proceed for 3 to 5 h. The cells were then harvested, resuspended in a 1/10 volume of λ dilution buffer (20 mM Tris-HCl [pH 7.5], 0.1 M NaCl, 10 mM MgSO4, 0.01% gelatin), and lysed by addition of chloroform and shaking at 37°C for 10 to 15 min. The lysates were then treated with a few crystals of DNase for 5 min, and the cellular debris was removed by centrifugation. The titer of packaged cosmids was expressed in cosmid transduction units (ctu) of lysates that were determined by transduction of a sensitive host strain to resistance to the antibiotic determinant carried by the cosmid (15, 31). Titers of infectious phages were determined as PFU on the supF strain LE392. In the case of cosmids derived from pAM34, glucose was omitted from the growth medium and IPTG was added to 1 mM in order to give full induction of the lac promoter that drives plasmid replication (18). Cultures to be transduced with cosmids with high efficiency were grown in the presence of maltose (43). Generally, a Mg2+ concentration that approximated that normally used for λ adsorption (1 to 2 mM) was obtained upon adding the lysate to the cultures. If not, additional Mg2+ was added. Transductions with phage P1vir were done as previously described (28).
RESULTS
Construction of the cosmid packaging strains.
The λ lysogenic strains used in preparation of cell extracts for in vitro packaging of λ or cosmid DNAs (22, 40) are not suitable for in vivo packaging of small cosmids because the prophages of these strains lack functional gam and red genes, both of which are required for efficient packaging of small cosmids (15, 31, 47). The gam gene product is required to inactivate the host recBC nuclease to allow rolling circle replication (15, 47). Induction of λ DNA replication in a host strain carrying a cosmid is known to shift cosmid molecules into the rolling circle mode of DNA replication, although the mechanism is not well understood (15, 31, 47). Rolling circle replication is required in order to synthesize concatemers of small cosmids that are sufficiently large (37 to 52 kb) to be packaged by λ. Function of the red genes gives a marked increase in the level of packageable cosmid DNA, presumably due to the potent homologous recombination activity of the Red proteins. Recombination generates packageable DNA molecules by combining DNA molecules that are not otherwise substrates for packaging (e.g., the circles resulting from conventional plasmid theta replication) (15, 31, 47).
In prior work, it was found that the in vivo packaging of cosmid DNA was 10- to 1,000-fold less efficient than that of the λ helper phage responsible for packaging (15, 31, 48, 50). The low titers of packaged cosmids are a problem, but more important is the presence of a large excess of λ phage in the lysates, because under conditions where all of the cells in a culture receive a cosmid each cell will also become infected with λ phage. If a λ lysogenic recipient strain were used, a 10-fold excess of λ phage over packaged cosmid particles, the prophage-encoded immunity would block phage infection. However, appreciably higher phage/cosmid ratios would overwhelm immunity, resulting in phage infection. To avoid these problems, I constructed λ helper phages that have a cis-acting defect in packaging. This was expected to prevent phage infection of the recipient cells and perhaps also to increase the yields of packaged cosmids, since there would be no λ helper DNA to compete with the cosmids for packaging. λcI857 Sam7 phages defective in packaging were constructed (Fig. 1) by inserting a kanamycin resistance gene between the filled-in cohesive ends followed by electroporation and selection for kanamycin-resistant lysogens. Two temperature-sensitive strains were tested, one constructed with T4 DNA polymerase (CY1590) and the other constructed with Klenow polymerase (CY1591). These strains had identical packaging efficiencies and were used interchangeably in the early experiments. However, CY1591 became the favored strain, since the FRT sites of the λ prophage allowed removal of the kanamycin resistance marker. The cI mutation allows thermal induction of lysogens, and the amber mutation in the S gene (Sam7) blocks lysis of the host, thereby allowing increased time for packaging plus the means to readily obtain concentrated preparation of phage particles in a defined buffer (3). The filling of the cohesive ends together with insertion of the kanamycin resistance cassette prevents packaging by disrupting the strict spacing of the three DNA sequence elements (cosQ, cosN, and cosB) required for λ DNA packaging (10, 51). When the host was made recombination proficient such that high-titer phage P1vir stocks could be made, the cos::kan prophages were easily moved to other host strains by transduction.
Properties of the cosmid packaging strains.
In the first experiments, the lysogens were induced and the lysate titers for infectious phage were determined. Both lysogens gave very low phage yields (about 5 orders of magnitude less than those of lysogens carrying the parental phage). The λ phage produced are attributed to λred-mediated recombination with the cos sequences of defective prophages resident in the E. coli K-12 chromosome (5, 16). The packaging defect of the phage DNAs having a disrupted cos was confirmed by assays done on plasmids derived from the mutant phages. DNA from induced cultures was digested with BspLU11I and ligated to NcoI-digested plasmid pMTL23P, and transformants resistant to both kanamycin and ampicillin were selected. Restriction mapping of the resulting plasmids showed that the two cos::kan λ phages each contained a single kanamycin resistance fragment oriented in the direction opposite to that of the λ genes. These cos::kan plasmids were then shown to be resistant to in vitro cutting by λ terminase (Epicentre) and to be inactive as packaging substrates in vivo (data not shown).
Although the cos::kan prophages were unable to package the viral DNA, induction of these lysogens resulted in very efficient cosmid packaging. Note than when the packaging strain contained a cosmid, the resulting lysates showed about a 10- to 100-fold increase in the infectious phage titer over that seen in the absence of a cosmid. Since the host strain is recA, this increase is presumably due to λred-mediated recombination with the cos sequence of the multicopy cosmid. However, depending on the cosmid tested, the titer of packaged cosmids (ctu) exceeded the number of infectious particles (PFU) by 100- to 10,000-fold. The infectious phage particles present in the lysates very occasionally gave rise to a detectable infection of transduced cultures. These infections presumably resulted from a helper phage that had acquired a functional S gene by recombination or by reversion of the amber mutation. To eliminate this problem, transduced cultures were plated in the presence of 1.25 mM sodium pyrophosphate to disrupt any phage particles released (49). Use of cosmids carrying a minimal cos fragment (223 bp) did not significantly decrease cosmid-prophage recombination relative to that with a cosmid with 1.6 kb of λ sequence, consistent with the high activity of the λred recombination pathway on short DNA sequences (11).
It should be noted that the packaging strains were derived from a Δ(hsdSMR) strain and lack E. coli K-12 modification. Since many of the standard E. coli K-12 strains (including all those used in the present work) carry hsdR mutations, this is not generally a problem. However, derivatives of strains CY1590, CY1591, and CY1592 carrying a wild-type hsd region have been constructed by introduction of F101, an F′ that carries the wild-type hsd genes (27). F101 maintenance was selected by complementation of the leu-ara deletion of these strains or by tetracycline selection of an F101 derivative marked by a Tn10 insertion into mdoB, a gene closely linked to hsd. λ phage released by these strains plated equally well on wild-type and isogenic hsdR strains. The cos::kan λ prophage of strain CY1591 has also been transduced into several strains having wild-type E. coli K-12 modification.
Packaging of cosmids.
Several cosmids were constructed by use of a 223-bp cosmid-containing fragment bracketed by two polylinker sequences that was used as a portable cassette to convert plasmids to cosmids (Fig. 1A). Cosmids with differing replication origins (ColE1, p15A, and F) have previously been shown to be packaged into λ phage particles (31, 47, 48). However, it was unclear whether the orientation of cos relative to the replication origin was important in the efficiency of packaging. I tested the effects of cos orientation in plasmids with the unidirectional ColE1 origin and found that reversal of the cos orientation had no effect on packaging efficiency. Cosmids constructed from plasmids (29) carrying the π protein-dependent origin of plasmid R6K were also tested and were found to be packaged with extremely low efficiencies regardless of cos orientation. I therefore switched to another conditional replication plasmid, pAM34, which has a ColE1 origin controlled by a lac promoter in place of the native P2 promoter (18). In the presence of IPTG, pAM34-derived cosmids were packaged as efficiently as cosmids having the native replication origin, and lysates of >1010 ctu/ml (assayed in the presence of IPTG) were readily obtained.
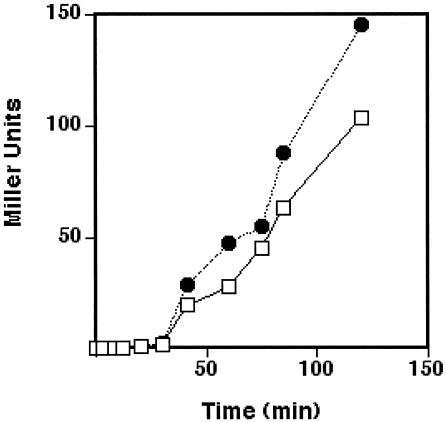
Cultures to be infected with cosmid-containing λ particles were grown in the presence of 0.2% maltose, resulting in a homogeneous distribution of 6,000 to 9,000 λ receptors/cell (41, 43). Schwartz (43) has measured a rate constant of λ adsorption to such cells of 2.1 × 10−10 ml/bacterium per s, and from his data it can be calculated that at a cell density of 5 × 108/ml, 80% of added λ particles adsorb to cells within 19 s. Therefore, at a ratio of 5 packaged cosmids (ctu) per cell, 98% of the cells will adsorb a cosmid-containing phage particle in 19 s and virtually all of the cells will adsorb a particle by 35 s. Hence, the introduction of cosmids into the cells of a culture was expected to be both rapid and quantitative. To test the efficiency and rate of cosmid entry into the intracellular gene pool, a cosmid was constructed from the lacZ α-complementation vector pTrueBlue-rop. This cosmid (called pCY574) was packaged in strain CY1591 and used to transduce strain DH5αZ, which expresses the LacZ ΔM15 protein (an α-complementation recipient) at a multiplicity of 5 ctu/cell. IPTG was added together with the λ particles in order to derepress both the plasmid and chromosomal lac promoters, and samples were then taken for β-galactosidase assays. IPTG was also added to a parallel control culture of strain DH5αZ previously transformed with the same cosmid. The two cultures had very similar induction profiles, indicating that entry of cosmid DNA was essentially immediate within the time scale required to begin gene expression (Fig. 3). Note that β-galactosidase activity was first detected at 7 to 8 min after cosmid and/or IPTG addition in the two cultures, which is about twice the time reported for the first β-galactosidase molecules to be observed upon IPTG induction of cells containing the intact lacZ gene (2). The greater lag shown in Fig. 3 can be attributed to assembly time of the α-complementation complex and the relative insensitivity of the assay due to the low β-galactosidase activities (which are maximally only about 10 to 15% of those given by a wild-type cell). It should be noted that cosmid transduction had no effect on cell viability and that >99% of the colonies resulting from plating dilutions of cultures transduced with cosmid pCY574 gave sectored blue colonies when plated in the absence of ampicillin, indicating that virtually all of the cells had received a cosmid.
FIG. 3.
Time course of β-galactosidase synthesis following cosmid transduction. Strain DH5αZ was grown in LB medium to about 6 × 108 cells/ml and then infected with 3 × 109 CFU of a lysate of pCY574 packaged in pCY1591 (see Materials and Methods). IPTG was added to 1 mM at the time of phage particle addition. Samples were taken at the times indicated, treated with chloroform-sodium dodecyl sulfate, stored on ice, and then assayed as described by Miller (30). A culture of strain CY1617, which is DH5αZ transformed with pCY574, was grown in LB plus ampicillin medium to about 6 × 108 cells/ml and then induced with IPTG (1 mM), and samples were taken in parallel with the cosmid-transduced culture. After all samples had been taken, they were assayed for β-galactosidase activity (30). Symbols: •, strain DH5αZ transduced with cosmid pCY574 in the presence of IPTG; □, strain CY1617 induced with IPTG. The recipient strain DH5αZ contained no detectable β-galactosidase activity.
Absolute off-to-on control of gene expression.
The β-galactosidase experiment (Fig. 3) is an example of absolute off-to-on control of gene expression. The lacZ ΔM15 allele is a null mutation and, thus, prior to introduction of the cosmid the recipient cells lacked functional β-galactosidase. A second example took advantage of the properties of the phage T7 RNA polymerase, which is insensitive to rifampin, an antibiotic that completely blocks activity of E. coli RNA polymerase (14). In prior work my laboratory showed that mutational trapping of the normally periplasmic enzyme E. coli thioesterase I (TesA) in the cytosol resulted in massive accumulation of newly synthesized free fatty acids, indicating a great increase in the rate of fatty acid synthesis (8). Two models were proposed to explain this phenomenon: one was independent of mRNA synthesis, whereas the second model required mRNA synthesis. Our prior experiment had an experimental deficiency that weakened interpretation in that tesA mRNA synthesis was not totally blocked, and thus detectable amounts of trapped TesA were present at the beginning of the experiment. With the in vivo cosmid packaging system, a more rigorous test of the two models could be done. T7 RNA polymerase was expressed in a strain that lacked the mutant tesA gene, and then rifampin was added. λ particles containing pCY575, a cosmid that expressed the mutant tesA gene under control of a T7 promoter, were added and free fatty acid (FFA) production was assayed (Fig. 4). FFA production was observed in the absence of chromosomal gene transcription, demonstrating that thioesterase production alters lipid synthesis independently of chromosomal transcription. In the control culture, the strain was transduced with the α-complementation cosmid pCY574 (Fig. 3), thus providing both a null control for thioesterase expression plus assay of rifampin action (which completely blocked β-galactosidase synthesis).
FIG. 4.

Production of free FFA in rifampin-treated cells. Strain DH5αZ carrying the T7 RNA polymerase production plasmid pCY598 was grown on RB medium supplemented with kanamycin and maltose. When the culture reached about 3 × 108 cells/ml, the cells were harvested by centrifugation and resuspended in the same medium except that 0.2% arabinose replaced the maltose. Following growth for 1 h, the cells were again harvested by centrifugation and resuspended in RB medium supplemented with kanamycin. Following 30 min of additional growth, rifampin was added to a final concentration of 0.2 mg/ml. After 30 min of additional incubation, the culture was split in half. One half was transduced with a lysate of the ′tesA cosmid pCY575 packaged in pCY1591, whereas the second half was transduced with the same lysate of pCY574 packaged in pCY1591 used in the experiment shown in Fig. 3. After 2 h of further incubation, both cultures were labeled for 30 min with [1-14C]acetate and then extracted with chloroform-methanol for assay of FFA production (8). The figure shown is an autoradiogram of a thin-layer chromatographic separation of the chloroform-methanol-soluble lipids of the cultures. Lane 1 is the lipids of the pCY574-transduced control culture, whereas lane 2 is the lipids of the ′tesA pCY575-transduced culture. Maltose was removed to avoid inhibition of arabinose induction of T7 RNA polymerasesynthesis, and arabinose was subsequently removed to prevent accumulation of acetate and dilution of the radioactive label.
Transient expression of recombinases.
Cosmids encoding either the Flp or recA recombinases were constructed from the conditional replication plasmid pAM34. These cosmids were then packaged, and the lysates were used to transiently express the recombinase in growing cultures. The assay for Flp recombinase activity was excision of a chloramphenicol resistance cassette (by catalysis of site-specific recombination between the FRT sites that flank the cassette) from strains carrying marked deletions constructed by the λred expression system (11). In the simplest protocol used, cells of a culture grown in the presence of maltose were transduced with a lysate of the packaged FRT cosmid pCY580 (5 ctu/cell) and then plated on a medium that contained anhydrochlorotetracycline and pyrophosphate and lacked selective antibiotics and IPTG. After overnight incubation of the plates at 37°C, 20 to 50 of the resulting colonies were patched to three anhydrochlorotetracycline (to induce FLP expression) plates lacking IPTG. One plate contained ampicillin (the cosmid marker), another contained chloramphenicol (the marker to be cured), and the third lacked chloramphenicol and ampicillin. After growth at 37°C for 5 to 7 h, colony patches that had been cured of resistance to both chloramphenicol and ampicillin were occasionally seen, but in general the patches failed to grow on the ampicillin plate and grew slowly on the chloramphenicol plate. The slow growth in the presence of chloramphenicol indicated growth of a minority population of uncured cells within the patches. Cells were picked from the patches on the plate lacking chloramphenicol and ampicillin that corresponded to the patches that grew slowly in the presence of chloramphenicol, and then they were streaked for single colonies on an anhydrochlorotetracycline plate lacking chloramphenicol and ampicillin. After overnight incubation, virtually all of the resulting colonies were found to be sensitive to both chloramphenicol and ampicillin.
In the case of recA, we tested the ability of a recA strain to be transduced by a phage P1 lysate grown on a strain with a chromosomal antibiotic maker. Protocols similar to those used with FLP were followed. A maltose-grown culture of a recA strain was transduced first with a lysate of the packaged recA cosmid pCY579 and, after 10 min, 5 mM CaCl2 and the phage P1 transducing lysate were added (Ca2+ inhibits λ adsorption [3]). Following P1 adsorption, the cells were then plated on LB medium lacking IPTG and containing the appropriate antibiotic and pyrophosphate. The transduction frequencies obtained were comparable to those of wild-type strains, whereas no transductants were observed in the absence of cosmid. Virtually all of the colonies formed were ampicillin sensitive, indicating loss of the cosmid. In a few cases, cosmid-free colonies that had the wild-type recA allele (as assayed by resistance to UV light) were observed. These colonies were attributed to homologous recombination between the chromosome and the wild-type recA gene on the cosmid, thereby transferring a wild-type recA allele to the chromosome. Although these colonies could be detected by their faster growth rates, their presence complicated the protocol. Therefore, pCY576, a cosmid carrying the recA gene of S. enterica serovar Typhimurium, was constructed. Although the recA genes of the two bacteria are fully interchangeable in regards to expression and protein function, they differ by 10% at the DNA level. The DNA sequence heterogeneity (due largely to mismatches at the third positions of codons) is scattered throughout the sequence and therefore is expected to result in inhibition of recombination by the mismatch repair system (39).
DISCUSSION
Efficient in vivo packaging of cosmids allows several unique physiological and genetic manipulations as well as facilitation of other operations that can be done by more laborious procedures. The main attribute of the use of cosmids packaged in vivo is that a given gene can be synchronously introduced into every cell of a growing bacterial culture. This quantitative aspect of cosmid transduction is particularly important in experiments that require chemical analyses to be done on an unfractionated cell population where the presence of cells lacking the gene would increase the background and thus could obscure the experimental results. Although recombinant λ phages have been similarly used to introduce bacterial genes (4, 45), in vivo-packaged cosmids have several advantages. The main advantage is that construction of cosmids is much simpler and more straightforward than cloning into phage λ, and existing plasmids can be readily converted to cosmids. Another advantage is that greatly elevated gene dosages are obtained with in vivo-packaged cosmids, whereas λ is limited to one or a few gene copies. However, most advantages of in vivo-packaged cosmids over plasmids depend on λ biology. For example, cells can simultaneously receive several different cosmids, as in the mixed infections with different λ mutants that form the basis of λ genetics. Moreover, the kinetics and distribution of cosmid introduction events can be directly calculated from those known for λ, since the cosmid transducing particle is a λ particle. Therefore, it can be calculated that 99.3% of the cells will receive cosmid DNA at a ctu/cell ratio of 5 and that, of these cells, only 3% will receive a single cosmid. Therefore, immediately after transduction the number of cosmid copies per cell will be variable, although with growth the control mechanisms controlling plasmid replication should ameliorate these differences. In a recA strain, the transduced cosmid will remain as circular oligomers, but in recombination-proficient strains the extent of oligomerization will drift towards lower states of oligomerization (31), as also seen with multimeric plasmids (26).
The facile and efficient adsorption of λ particles allows plasmid molecules (as cosmids) to be readily moved into a large number of individual colonies. In vivo-packaged cosmids should also be useful in detecting gene expression and assigning proteins to genes. An approach discovered by Ptashne (38) that was very useful in aligning the genes and proteins of phage λ (20) was infection of a heavily UV-irradiated host cell followed by labeling with radioactive amino acids. DNA damage caused by UV irradiation effectively blocked transcription of chromosomal genes and, thus, radioactive label was incorporated only into phage-encoded proteins. By use of λ specialized transducing phages, this method was used to identify the products of many important bacterial genes such as RecA (28) and DnaK (17). However, due to the difficulties of making recombinant λ phages, this method fell into disuse and was replaced by the maxicell procedure (42), which used plasmids in place of phage. However, the maxicell procedure gave much less robust labeling of cellular proteins, due to the lengthy incubation under nongrowing conditions required for chromosomal DNA degradation. It seems obvious that in vivo-packaged cosmids could be used in place of λ phage in the Ptashne method and, thus, the proposed method should give both the convenience of plasmid cloning as in the maxicell procedure plus the robust expression of the Ptashne method. Although the need for assigning proteins to E. coli genes is passé, the cosmid-UV irradiation method could be useful in assigning genes from other organisms. Such manipulations are not limited to E. coli, since expression of the E. coli lamB gene in other enterobacteria confers the ability to adsorb and express packaged cosmids (34).
Possible improvements to the present cosmid packaging strains are readily envisioned. The main improvement would be production of in vivo-packaged cosmid preparations that contain no λ DNA. This would require blocking homologous recombination between the helper phage and the cos sequences present in both the cosmid and the defective prophages of the E. coli chromosome. Recombination with the cosmid could be avoided by constructing a deletion within the helper phage cos region that removes all of the λ sequences that are present in the cosmid. Blocking recombination with the defective prophages would prevent any remaining acquisition of cos by the helper phage, but it is more difficult to achieve. An ideal host strain would be the E. coli K-12 strain of Kolisnychenko and coworkers (25), in which all cryptic prophages have been deleted. Note that a strain that in vivo packages only cosmid DNA together with the well-developed methods for production of pure preparations of λ phage particles (3) would provide a means to make large preparations of cosmid DNA free of contaminants such as other DNA molecules, RNA, and endotoxin.
Acknowledgments
I thank A. Kuzminov for strains and valuable discussion.
This work was supported by National Institutes of Health grant AI15650 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Adams, M. H. 1959. Bacteriophages. Interscience, New York, N.Y.
- 2.Adamson, L., C. Gross, and A. Novick. 1970. Is subunit assembly a rate-limiting factor on induction?, p. 317-323. In J. Beckwith and D. Zipser (ed.), The lactose operon. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 3.Arber, W., L. Enquist, B. Hohn, N. E. Murray, and K. Murray. 1983. Experimental methods for use with lambda, p. 433-466. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 4.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, A. M. 1996. Cryptic prophages, p. 2041-2046. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C.
- 6.Chambers, S. P., S. E. Prior, D. A. Barstow, and N. P. Minton. 1988. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139-149. [DOI] [PubMed] [Google Scholar]
- 7.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Cho, H., and J. E. Cronan, Jr. 1995. Defective export of a periplasmic enzyme disrupts regulation of fatty acid synthesis. J. Biol. Chem. 270:4216-4219. [DOI] [PubMed] [Google Scholar]
- 9.Collins, J., and B. Hohn. 1992. Cosmids: a type of plasmid gene-cloning vector that is packageable in vitro in bacteriophage lambda heads. Bio/Technology 24:193-197. [PubMed] [Google Scholar]
- 10.Cue, D., and M. Feiss. 2001. Bacteriophage lambda DNA packaging: DNA site requirements for termination and processivity. J. Mol. Biol. 311:233-240. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubendorff, J. W., and F. W. Studier. 1991. Creation of a T7 autogene. Cloning and expression of the gene for bacteriophage T7 RNA polymerase under control of its cognate promoter. J. Mol. Biol. 219:61-68. [DOI] [PubMed] [Google Scholar]
- 15.Feiss, M., D. A. Siegele, C. F. Rudolph, and S. Frackman. 1982. Cosmid DNA packaging in vivo. Gene 17:123-130. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, R., and M. Feiss. 1980. Reversion of a cohesive end site mutant of bacteriophage lambda by recombination with a defective prophage. Virology 107:160-173. [DOI] [PubMed] [Google Scholar]
- 17.Georgopoulos, C. P., B. Lam, A. Lundquist-Heil, C. F. Rudolph, J. Yochem, and M. Feiss. 1979. Identification of the E. coli dnaK (groPC756) gene product. Mol. Gen. Genet. 172:143-149. [DOI] [PubMed] [Google Scholar]
- 18.Gil, D., and J. P. Bouche. 1991. ColE1-type vectors with fully repressible replication. Gene 105:17-22. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63-113. [DOI] [PubMed] [Google Scholar]
- 20.Hendrix, R. W. 1971. Identification of proteins coded in phage lambda, p. 355-370. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Henry, M. F., and J. E. Cronan, Jr. 1991. Direct and general selection for lysogens of Escherichia coli by phage lambda recombinant clones. J. Bacteriol. 173:3724-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hohn, B. 1979. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 68:299-309. [DOI] [PubMed] [Google Scholar]
- 23.Hohn, B., and J. Collins. 1980. A small cosmid for efficient cloning of large DNA fragments. Gene 11:291-298. [DOI] [PubMed] [Google Scholar]
- 24.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 25.Kolisnychenko, V., G. Plunkett III, C. D. Herring, T. Feher, J. Posfai, F. R. Blattner, and G. Posfai. 2002. Engineering a reduced Escherichia coli genome. Genome Res. 12:640-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolodner, R. 1980. Genetic recombination of bacterial plasmid DNA: electron microscopic analysis of in vitro intramolecular recombination. Proc. Natl. Acad. Sci. USA 77:4847-4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makovets, S., A. J. Titheradge, and N. E. Murray. 1998. ClpX and ClpP are essential for the efficient acquisition of genes specifying type IA and IB restriction systems. Mol. Microbiol. 28:25-35. [DOI] [PubMed] [Google Scholar]
- 28.McEntee, K., J. E. Hesse, and W. Epstein. 1976. Identification and radiochemical purification of the recA protein of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 73:3979-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Miwa, T., and K. Matsubara. 1983. Formation of oligomeric structures from plasmid DNA carrying cos lambda that is packaged into bacteriophage lambda heads. J. Bacteriol. 153:100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miwa, T., and K. Matsubara. 1982. Identification of sequences necessary for packaging DNA into lambda phage heads. Gene 20:267-279. [DOI] [PubMed] [Google Scholar]
- 33.Murray, N. E. 1983. Phage lambda and molecular cloning, p. 395-432. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Palva, E. T., A. Harkki, H. Karkku, H. Lang, and M. Pirhonen. 1987. Lambda vehicles provide new tools for genetic analysis of gram-negative bacteria. Microb. Pathog. 3:227-230. [DOI] [PubMed] [Google Scholar]
- 35.Phillips, G. J., S. K. Park, and D. Huber. 2000. High copy number plasmids compatible with commonly used cloning vectors. BioTechniques 28:400-402, 404, 406. [DOI] [PubMed] [Google Scholar]
- 36.Posfai, G., M. D. Koob, H. A. Kirkpatrick, and F. R. Blattner. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratt, D., and W. S. Erdahl. 1968. Genetic control of bacteriophage M13 DNA synthesis. J. Mol. Biol. 37:181-200. [DOI] [PubMed] [Google Scholar]
- 38.Ptashne, M. 1967. Isolation of the lambda phage repressor. Proc. Natl. Acad. Sci. USA 57:306-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayssiguier, C., D. S. Thaler, and M. Radman. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396-401. [DOI] [PubMed] [Google Scholar]
- 40.Rosenberg, S. M., M. M. Stahl, I. Kobayashi, and F. W. Stahl. 1985. Improved in vitro packaging of coliphage lambda DNA: a one-strain system free from endogenous phage. Gene 38:165-175. [DOI] [PubMed] [Google Scholar]
- 41.Ryter, A., H. Shuman, and M. Schwartz. 1975. Integration of the receptor for bacteriophage lambda in the outer membrane of Escherichia coli: coupling with cell division. J. Bacteriol. 122:295-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sancar, A., A. M. Hack, and W. D. Rupp. 1979. Simple method for identification of plasmid-coded proteins. J. Bacteriol. 137:692-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz, M. 1976. The adsorption of coliphage lambda to its host: effect of variations in the surface density of receptor and in phage-receptor affinity. J. Mol. Biol. 103:521-536. [DOI] [PubMed] [Google Scholar]
- 44.Stephens, C. R., L. H. Conover, R. Pasternak, F. A. Hochstein, W. T. Moleland, P. P. Regna, F. J. Pilgrim, K. J. Brunings, and R. B. Woodward. 1954. The structure of aureomycin. J. Am. Chem. Soc. 76:3568-3575. [Google Scholar]
- 45.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 46.Uhlin, B. E., and A. J. Clark. 1981. Overproduction of the Escherichia coli recA protein without stimulation of its proteolytic activity. J. Bacteriol. 148:386-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umene, K., K. Shimada, and Y. Takagi. 1978. Packaging of ColE1 DNA having a lambda phage cohesive end site. Mol. Gen. Genet. 159:39-45. [DOI] [PubMed] [Google Scholar]
- 48.Umene, K., K. Shimada, T. Tsuzuki, R. Mori, and Y. Takagi. 1979. Lambda bacteriophage-mediated transduction of ColE1 deoxyribonucleic acid having a lambda bacteriophage-cohesive end site: selection of packageable-length deoxyribonucleic acid. J. Bacteriol. 139:738-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 50.White, F. F., H. J. Klee, and E. W. Nester. 1983. In vivo packaging of cosmids in transposon-mediated mutagenesis. J. Bacteriol. 153:1075-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wieczorek, D. J., and M. Feiss. 2001. Defining cosQ, the site required for termination of bacteriophage lambda DNA packaging. Genetics 158:495-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wycuff, D. R., and K. S. Matthews. 2000. Generation of an AraC-araBAD promoter-regulated T7 expression system. Anal. Biochem. 277:67-73. [DOI] [PubMed] [Google Scholar]
- 53.Zinder, N. D., and J. D. Boeke. 1982. The filamentous phage (Ff) as vectors for recombinant DNA—a review. Gene 19:1-10. [DOI] [PubMed] [Google Scholar]