Abstract
A major challenge in undergraduate life science curricula is the continual evaluation and development of courses that reflect the constantly shifting face of contemporary biological research. Synthetic biology offers an excellent framework within which students may participate in cutting-edge interdisciplinary research and is therefore an attractive addition to the undergraduate biology curriculum. This new discipline offers the promise of a deeper understanding of gene function, gene order, and chromosome structure through the de novo synthesis of genetic information, much as synthetic approaches informed organic chemistry. While considerable progress has been achieved in the synthesis of entire viral and prokaryotic genomes, fabrication of eukaryotic genomes requires synthesis on a scale that is orders of magnitude higher. These high-throughput but labor-intensive projects serve as an ideal way to introduce undergraduates to hands-on synthetic biology research. We are pursuing synthesis of Saccharomyces cerevisiae chromosomes in an undergraduate laboratory setting, the Build-a-Genome course, thereby exposing students to the engineering of biology on a genomewide scale while focusing on a limited region of the genome. A synthetic chromosome III sequence was designed, ordered from commercial suppliers in the form of oligonucleotides, and subsequently assembled by students into ∼750-bp fragments. Once trained in assembly of such DNA “building blocks” by PCR, the students accomplish high-yield gene synthesis, becoming not only technically proficient but also constructively critical and capable of adapting their protocols as independent researchers. Regular “lab meeting” sessions help prepare them for future roles in laboratory science.
AN ongoing challenge to the design and maintenance of undergraduate biology curricula is the ability to incorporate new conceptual advances and technologies. Few curricula have been updated to reflect recent innovation and most are estimated to be out of date by approximately 2 decades (National Research Council 2003). The advent of recombinant DNA technology >30 years ago enabled research to proceed at unparalleled rates (Cohen et al. 1973), and these concepts and techniques have been almost universally adopted in undergraduate molecular biology courses. More recently, innovations in engineering, computer science, and biotechnology have enabled biological research to enter the genomic era, yet undergraduate programs have been largely unable to keep pace with these developments. These delays become more inevitable and intractable as expansions in the scope of biological research are driven by the rapidity with which new technologies are being introduced; this technology-driven approach is difficult to reconcile with the dogmatic presentation of lecture material and the focus on memorization-based learning that have characterized traditional biology courses. To incorporate a genomic focus, undergraduate courses would be better served by an active learning orientation and an interdisciplinary foundation (National Research Council 2003; Ares 2004; Bialek and Botstein 2004; Gross 2004). Many of these objectives could be achieved by the introduction of synthetic biology courses to the undergraduate curriculum.
Synthetic biology is a new discipline emerging in the scientific, social, and business arenas and is attracting national attention by promising advances in numerous areas, such as biofuel and pharmaceutical development, agriculture, and bioremediation. The field is best understood as the application of engineering principles to biological systems and is therefore inherently interdisciplinary, with a strong foundation in biology, engineering, computer science, and biotechnology. One focus of synthetic biologists is the deconstruction of biological systems into components that can be uncoupled from each other, abstracted into predictable forms, standardized so they are interchangeable, and then reassembled into new functional systems. These rewired components are employed in the creation of devices, modules, and networks that perform novel functions; notable examples include biofilms that take photographs, genetic oscillators that pulse GFP, and yeast cells that produce the antimalarial drug artemisinic acid (for reviews, see Endy 2005; Andrianantoandro et al. 2006; Drubin et al. 2007). The few undergraduate courses in synthetic biology that are offered utilize this approach (Kuldell 2007), offering students an active learning experience through designing and constructing genetic devices. Indeed, undergraduates have shown great interest in synthetic biology, and many universities, both with and without synthetic biology courses, now host their own International Genetically Engineered Machines (iGEM) competition teams (http://www.iGEM.org) that perform student-directed synthetic biology projects and present their work at the iGEM Jamboree (Campbell 2005; Goodman 2008).
Another subset of synthetic biologists does not seek to create novel circuitry per se, but rather seeks to design and fabricate entire genomes. Genome synthesis serves as a highly informative evolutionary short cut, allowing researchers both to push the boundaries of complex systems and to discover new insights into genome structure that would remain undetected with more targeted approaches (Holt 2008). By enabling historically important pathogens such as the 1918 influenza virus or human retroviruses to be resurrected, genome synthesis allows scientists to isolate and manipulate previously inaccessible genomes (Tumpey et al. 2005; Lee and Bieniasz 2007). Genome fabrication further allows scientists to design and create minimal genomes both to define the sets of genes that are essential for life and, theoretically, to create simplified cells that generate more defined products for biotechnology applications (Holt 2008). This unprecedented control over genome content and organization facilitates the discovery of cellular design principles and enables challenge and confirmation of models of genome structure (Chan et al. 2005; Endy 2008).
Genome design relies heavily on knowledge acquired through multiple disciplines, including molecular biology, bioinformatics, genomics, and systems biology, all of which are becoming a more pervasive part of undergraduate and early graduate biology education (Bednarski et al. 2005; Kumar 2005; Kerfeld and Simons 2007). The breadth and scale of genome synthesis projects invites the involvement of a large number of students, yet the parallel nature of the work does not require the same amount of individual mentoring as typical undergraduate independent research; this enables almost unlimited opportunities for students to participate as collaborators and become part of the synthetic biology research community.
We have developed an undergraduate synthetic biology course at Johns Hopkins University, entitled Build-a-Genome, within the context of the Synthetic Yeast Genome Project (http://www.syntheticyeast.org) initiated in our laboratories. Although the goal of the project is to design and synthesize the total genome of Saccharomyces cerevisiae, our immediate goal was the synthesis of the starting materials for chromosome III in an undergraduate laboratory setting. Students are introduced to the field of synthetic biology through a series of lectures and laboratory sessions. After completion of the initial training segment, students work in an open-access lab to assemble designed segments of chromosome III in the form of ∼750-bp DNA building blocks. Assembly of these building blocks into larger 10-kb chunks in future semesters will be an important milestone in the fabrication of a complete synthetic eukaryotic genome. The Build-a-Genome course began as a pilot program in the summer of 2007 with 9 students. In the Fall 2007 and Spring 2008 semesters, 18 and 20 students, respectively, participated in both Build-a-Genome and Build-a-Genome Mentor, a class offered to returning students who have satisfactorily completed the Build-a-Genome course. In this article we focus on the pedagogical and scientific dimensions of the Build-a-Genome course.
IMPLEMENTATION
Lectures:
Lectures offered in the Build-a-Genome course reflect the multifaceted underpinnings of synthetic biology (Table 1). To ensure that students from all academic backgrounds have a strong foundation upon which to appreciate subsequent course materials, the course begins with a short series of lectures introducing students to the fundamentals of genetics, including nucleic acid structure and function, chromosome structure, and genome organization, as well as techniques that will be employed throughout the course, including PCR, molecular cloning, and DNA sequencing. Students are then introduced to concepts central to the field of synthetic biology, including recombinant DNA technology, synthetic circuitry, gene synthesis, and directed evolution. Following this series, the bioinformatic tools used in the course are presented; on principle, we use only software that is free and publicly accessible including the Saccharomyces Genome Database (http://www.yeastgenome.org), which serves as a central repository for genotypic and phenotypic analyses of yeast; GeneDesign (Richardson et al. 2006; http://www.genedesign.org), which facilitates the design of synthetic genes and constituent oligonucleotides; FinchTV and ClustalW (Larkin et al. 2007; Finch TV 1.4.0, Geospiza, http://www.geospiza.com/finchTV), which enable DNA sequence data analysis and alignment, respectively; and Moodle (http://www.moodle.org) course management software. To help students recognize the context in which their work in the course occurs, we present a lecture on the economics of gene synthesis that explores the financial constraints under which research must necessarily occur and a mathematics-based lecture comparing temporal and financial efficiency. New innovations are constantly reducing the cost of gene synthesis, and we encourage students to offer their ideas for shortcuts and new methodologies, some of which are then incorporated into the streamlined and battle-tested protocols that are routinely employed. Ethical dilemmas inherent in the field of synthetic biology are a matter of considerable discussion (Balmer and Martin 2008); students actively participate in a seminar led by a bioethicist, voicing their opinions on which projects should be allowed to proceed unfettered and which should be government regulated, the restrictions that should (or should not) be placed on the field by governing bodies, and how practitioners of synthetic biology ought to respond to societal concerns.
TABLE 1.
Lecture series
| Lecture topic | Field of lecturer/section leader |
|---|---|
| Studying and synthesizing yeast | Molecular biology and genetics |
| Principles of gene synthesis | Molecular biology and genetics |
| Cybersession: Saccharomyces Genome Database and GBROWSE | Molecular biology and genetics |
| Design of synthetic gene chunks, building blocks, and oligos | Molecular biology and genetics |
| Nucleic acid enzymology | Biochemistry |
| Synthetic gene design software: GeneDesign and BioStudio | Bioinformatics |
| DNA sequencing and sequence analysis | Chemical and biological engineering |
| Time vs. money considerations | Biological engineering and systems biology |
| Economics of gene synthesis | Molecular biology and genetics |
| Bioethics of synthetic biology | Bioethics |
Molecular biology boot camp:
After introductory lectures are complete, students move into the laboratory. We begin with eight guided laboratory sessions of molecular biology “boot camp” that serve as a period to review lecture topics necessary for completion of lab work, to demonstrate lab techniques (especially for students that lack previous lab experience), and to simultaneously practice the methods actually used in the project. Students are also introduced to the few crucial lab safety issues, including a review of chemical and biological safety practices, so they are competent to work in the lab unsupervised. Students are initially paired with lab partners so they may help each other solve simple laboratory problems; typically we pair biology and engineering students so they have complementary expertise. “Graduation” from boot camp requires students to submit assignments (“milestones”) that verify proficiency with each step of the gene synthesis protocol, including the use of GeneDesign to design oligonucleotides to synthesize a gene, serial dilution, PCR, agarose gel electrophoresis, molecular cloning, sequence analysis of cloned constructs using FinchTV, and multiple sequence alignment using ClustalW. Reagents used successfully in prior semesters are used during boot camp to minimize reagent costs.
Synthetic gene assembly:
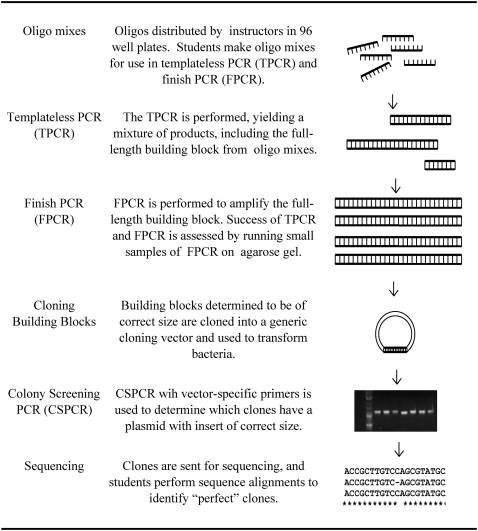
The Synthetic Yeast Genome Project aims to synthesize a designer yeast genome from which repetitive and other sequences likely to be superfluous have been removed or relocated. The synthetic genome is partially constructed by iterative synthesis from oligonucleotides 60–80 bases in length. Oligonucleotides are assembled into building blocks by annealing of partially overlapping single-stranded DNA and extension in two sequential rounds of PCR (Stemmer et al. 1995; Figure 1). For didactic purposes, we refer to the two key steps as “templateless PCR,” which generates a complex pool of reaction intermediates, and “finish PCR,” which uses the outer primer pair to enrich for full-length constructs. This process produces building blocks of DNA averaging 750 bp long and comprises the central experimental focus of the Build-a-Genome course (Figure 1). Following synthesis, students clone their building blocks, and bacterial cultures are sent to a commercial sequencing facility. Students submit 18 clones of each building block for sequencing, as this is the average number of candidates needed in practice for a high probability of achieving one clone of error-free (“perfect”) sequence due to the high error rate of commercial oligonucleotide synthesis. Resulting sequences are analyzed by students, and “perfect” clones are identified. Perfect building blocks are further assembled into multi-kilobase “chunks” that will be subsequently used to functionally replace the native yeast sequence. All protocols used have been extensively tested and are available at http://baderlab.bme.jhu.edu:8888/moodle18/ (click on the “guest access” link).
Figure 1.—
Steps in building block assembly.
Emphasis on independent research:
Following the pilot round of building block synthesis in boot camp, lab work consists of at least one (and often two) additional rounds of gene synthesis to build proficiency. Each team member is initially assigned six to seven building blocks (∼5000 bp) to be synthesized. Frequently only a subset of building blocks will assemble on the first attempt and students must determine if there are any problems in their execution of the protocol and/or devise changes to the standard protocol to successfully synthesize all (or most) of their constructs. This emphasis on troubleshooting and development of problem-solving skills generally supports students' creativity and self-confidence and serves as an important introduction to the process of scientific research. Students develop a sense of ownership of the overall project as their building blocks are contiguous in the yeast sequence, defining a region of the synthetic genome that has their “signature” on it. Further, students are encouraged to develop an individual definition of success; while many students strive to complete only the six to seven building blocks that they have been assigned, several students each semester accomplish synthesis of far more (up to an impressive 10,000 bp in a semester), even creating informal competitions to see who can synthesize the most DNA. We are experimenting with the concept of leader boards and a clickable map showing who synthesized which section(s) to formalize these aspects; however, the amount of DNA made per student is not used in formal evaluation.
While students are encouraged to work independently and have open access to the laboratory (a key ingredient in encouraging student involvement), most choose to retain some aspect of cooperative work and learning, either with their lab partners or in small groups. Interactions between students and with instructors frequently occur during the “open lab” hours when teaching assistants are available in the lab for ∼2 hr, 6 days a week to answer questions, distribute expensive reagents, and help with troubleshooting. Many students choose to work regularly during these hours, whereas others work mostly in periodic binges. The unstructured nature of lab time allows them to develop not only basic time-management skills, but also the ability to multitask, balancing work on several building blocks that may be at different stages of construction.
Students are also encouraged to develop side projects related to the work in the course; this component is central to the Build-a-Genome Mentors course that serves as a successor to the Build-a-Genome course for students who have successfully completed the latter course but want to continue at a more in-depth level. Side projects have included troubleshooting of all building blocks that failed to assemble both by systematic alterations to the protocol and by bioinformatic analysis, development of methods to assemble building blocks from highly complex oligonucleotide pools, creation of software packages/modules, and preparation of posters and write-ups for local presentations. Some students have approached synthetic biology and the Build-a-Genome course with such enthusiasm that they have developed related extracurricular activities; the first Johns Hopkins iGEM team was a spontaneous student-led outgrowth of Build-a-Genome. All of these activities support the development of young scientists who want to continue research both within the Synthetic Yeast Genome Project and in other contexts.
Website and electronic materials:
Computer support is required to fulfill multiple distinct needs: courseware for two-way communication with students; a server for bioinformatics applications; and a laboratory information management system (LIMS) that provides work flow, scheduling, and an electronic notebook essential for large, collaborative projects. We aimed for simplicity by building on a platform provided by freely available, open-source Moodle software.
Designed as a course management system, Moodle provides basic functionality for authenticating users as teachers or students, posting and collecting assignments, and publishing lectures and lab protocols. The course calendar is available online, as are the synthetic yeast DNA sequences assigned to each student team.
Beyond these standard functionalities, the website was enhanced to provide scheduling and workflow capabilities. These require student–student communication, as opposed to the more standard model of teacher–student communication. A student-writable calendar enables 20 students to share limited equipment, especially thermal cyclers, which receive very heavy use. Scheduling ties into workflow for activities such as sequencing, in which students use a web interface to request shared space on plates sent to an external vendor for DNA sequencing.
Workflow and LIMS are essential for front-to-back management of a large-scale synthetic biology project. An initial electronic sequence is designed, broken into smaller chunks, and finally subdivided into oligos ordered from a vendor. At this point the electronic sequences point to physical DNA received from a vendor. The progress of the physical sequences through the lab is tracked as they are distributed to students, joined into larger sequences, cloned, and sent to a facility for sequencing, with the electronic sequencing data archived in an online database. The Moodle platform was further extended to provide an electronic notebook to catalog results at key points through this workflow. For example, each agarose gel image is uploaded directly through the course website into a database, which can subsequently be searched by student name, date, or DNA building block number. Students are also required to submit a biweekly online progress report to a second database, indicating their progress in synthesizing, cloning, and sequencing each building block. This not only is an efficient way to identify problems as they occur, but also allows the instructor to coordinate the overall project; for those building blocks that are not successfully synthesized by the end of a semester, the database allows a succeeding student to see points of difficulty and allows students and instructors to identify building blocks that are persistently difficult to synthesize.
Bioinformatics tasks are simplified by the course website because information can flow seamlessly between applications, relieving bottlenecks and minimizing chances for error. One of the difficult bottlenecks during the first offering of this course was verifying that a clone's sequence was 100% accurate. This required tracking a sequencing plate and well back to a clone and then back to a desired DNA building block, then aligning forward and reverse reads with the desired sequence and scanning for mismatches. While students are still asked to perform these tasks manually to demonstrate knowledge of standard bioinformatics applications and to double-check automated analyses, the overall project requires automation of both the clone-to-desired sequence tracking and the verification analysis. We created a module called CloneQC that automates these tasks, operating on single sequences or in batch mode on entire folders uploaded through the Moodle interface. Similarly, upstream bioinformatics components for designing synthetic DNA generate electronic order sheets that can be transmitted directly to an oligo vendor.
Staffing:
The Build-a-Genome course requires a large team to organize and direct the student-centered research. The course is now directed by two to three faculty mentors (at least one each for molecular/genetic and computational), a course instructor, and two teaching assistants (TAs). Additional lecturers come from diverse backgrounds, including biomedical and chemical engineering, biochemistry, molecular biology and genetics, and bioethics (Table 1). The instructor serves as the coordinator for the course, assigning building blocks to students, grading assignments, managing the course website, delivering lectures, organizing multiwell plate collections of oligonucleotides and bacterial clones, delivering clones for sequencing, providing an introduction to each boot camp lab section, and participating in open lab sessions. Teaching assistants maintain the laboratory, help students to troubleshoot projects both during class time and during open lab hours, and direct individual side projects. Ideally, the instructors and TAs collectively have complementary expertise in molecular biology, genetics, biological engineering, and bioinformatics. For both instructors and TAs the course provides valuable experience in managing a laboratory; both direct student progress in the lab and during lab meetings as well as learn to budget, supply, and equip a laboratory.
Students are recruited from throughout the School of Arts and Sciences and School of Engineering, allowing us to draw highly motivated, research-oriented students with either biology or engineering backgrounds and further advancing the interdisciplinary focus of the courses. To gain admission into the course, students meet with either faculty or instructors in their field for an informal interview, both to stress the level of commitment required for the course and to determine student aptitude for a rigorous lab course. While there are no formal prerequisites for the course, most biology students at the sophomore level have completed or are concurrently enrolled in courses in introductory biology, chemistry, biochemistry, organic chemistry, and cell biology, with approximately one-half having some previous lab experience. Engineering students at the sophomore level typically have completed courses in introductory and organic chemistry, calculus, differential equations, and computer programming. We have recently begun accepting freshmen into the course, and typically these students have significant advanced placement credit, including biology, chemistry, calculus, or computer programming. By selecting the most motivated students, we have thus far been able to run Build-a-Genome course without a single student dropping the course.
Implementation challenges:
Build-a-Genome is a cutting-edge synthetic biology course that encourages students to work at an independent pace. As such, the course requires a dedicated molecular biology laboratory. Minimally, the lab requires incubators for bacterial plates, agarose gel electrophoresis equipment, a set of pipettes for each team, several Internet-connected computer terminals so that students can freely access the course websites and databases and numerous thermal cyclers. Fortunately, much of this equipment can be obtained as excess lab equipment at websites such as eBay; although the thermal cyclers require more maintenance, we find that the low cost of used equipment (∼$800 for a Perkin Elmer 9600 thermal cycler) easily offsets the cost of more frequent repair or replacement. A conference room is needed for lab meetings, and a wireless network is extremely useful in both the lab and conference rooms.
RESULTS AND ASSESSMENT
A core aim of this course is the development of students as skilled, independent researchers. Their competence is assessed foremost in lab meetings held every 2 weeks. In sections of no more than 10 students, each student is asked to present their most recent data and discuss any problems with gene synthesis. The image database in the course website is used with a laptop and video projector so everyone can see every lane of the relevant gel(s); this also provides an opportunity for development of good laboratory organization and note-taking and data presentation practices. Short presentations allow each student to develop fluency with scientific language, and subsequent questioning by the instructors ensures that students understand concepts and that they are able to explain their reasoning when altering protocols. Meetings are informal to encourage participation and group troubleshooting, and the students themselves build significant expertise as they each vary the standard protocols in different ways.
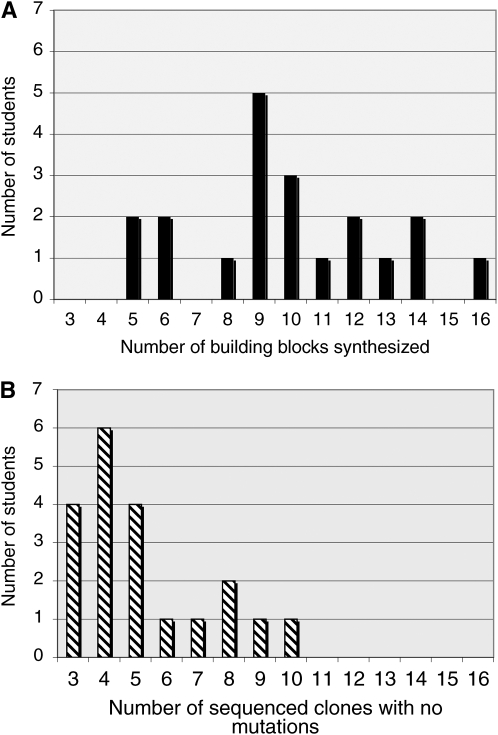
Student success is objectively evaluated according to their ability both to produce building block DNA and to complete the entire gene synthesis protocol by generating clones of perfect sequence. Success was considered to be the synthesis of 7–8 building blocks of the 10–11 assigned during the boot camp and independent phases and the identification of positive (“perfect”) clones from sequencing for at least 3 building blocks. For the spring semester of 2008, the median number of building blocks synthesized was 9.5/student (some students requested more building blocks to synthesize following completion of the initial 10–11), while the median number of perfect clones achieved was 4.5 (Figure 2). Students were remarkably efficient in the synthesis of these building blocks, and the first pass of the entire ∼280-kb synthetic chromosome III sequence was achieved in one academic year (one summer session, two semesters, and one intersession). Additional work will be required to synthesize the missing building blocks and to assemble them into larger chunks.
Figure 2.—
Student progress in the assembly of synthetic building blocks in Spring 2007. (A) Bars represent the number of students completing the synthesis of a given number of synthetic PCR constructs. (B) Bars represent the number of students obtaining a given number of sequenced clones with no mutations.
The success of the course is evident in student evaluations. Most felt that the scope of the course was just right (90.3%), and the course was considered the most popular in the biomedical engineering department last semester; many students listed it as “best lab course I have taken while at JHU” or “best course I have taken at JHU.” Most suggestions for improving the course reflected a desire to shorten the boot camp phase so that students could move more quickly into independent lab work and side projects. Indeed, the commitment and excitement of students toward long-term research after completing this course is evident from the number of students from the Fall 2007 course who continued to work on the project during the Winter 2008 intersession (12 of 18), as well as the number of students from both fall and spring semesters who continued to work on synthetic biology projects during Summer 2008 or on the Johns Hopkins University iGEM Team (18 of 38).
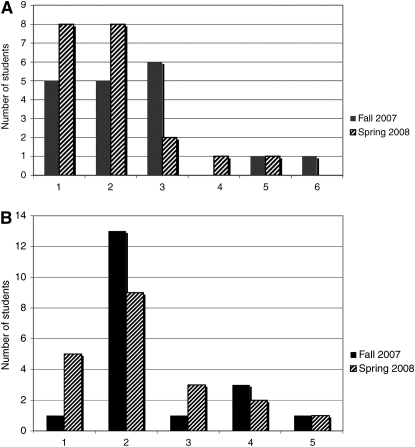
One of our goals was to foster interdepartmental collaboration around synthetic biology. Although the course began with generous support from the biology and biomedical engineering departments of the Krieger School of Arts and Sciences and the Whiting School of Engineering, respectively, the course's popularity has enabled us to expand enrollment to include chemical and biological engineering and computer science majors, as well as others (Figure 3A). Further, we have been encouraged by the ability of a diverse group of students to work effectively together, and we have expanded eligibility for the course to include students who range in experience from freshmen to first-year graduate students (Figure 3B).
Figure 3.—
Students by academic programs (A) and by class years (B) in the Build-a-Genome and Build-a-Genome Mentor courses. (A) 1, Biology; 2, biomedical engineering; 3, chemical and biological engineering; 4, computer science; 5, other science; 6, other humanities. (B) 1, Freshman; 2, sophomore; 3, junior; 4, senior; 5, graduate/postgraduate.
DISCUSSION
Synthetic biology is a developing field founded upon multiple disciplines and as such presents an excellent opportunity to introduce interdisciplinary cooperation and active learning in undergraduate education. Participating in a large, collaborative genome synthesis project allows students a true sense of ownership and the ability to participate in the broader scientific community. The great potential for success with undergraduate synthetic biology courses lies not only in synthetic biology's “cool factor,” but also in the need for new approaches to develop critical thinking skills and technical prowess in undergraduate researchers. In this spirit, we encouraged students to develop their novel, outside-the-box ideas as side projects that stimulate their sense of innovation.
The Build-a-Genome course presents several financial and practical challenges in its implementation elsewhere and is not for the faint of heart when it comes to faculty, TA, and student workload. Foremost among these issues are the cost of gene synthesis and the ability to maintain a high level of organization in an open-access laboratory. To implement a Build-a-Genome course on the scale of the Johns Hopkins course costs ∼$30,000/semester for consumables, oligonucleotides, and sequencing. While the per building block cost of synthesis is high (Table 2), it remains far below the cost of commercial synthesis, which approximates $1/bp for yeast DNA containing considerable stretches of high A/T content. Gene synthesis by undergraduates therefore becomes a more cost-effective method than commercial alternatives. One can imagine this course becoming financially self-sustainable by offering researchers synthetic genes for a fee (something we have not tried).
TABLE 2.
Cost of synthesis per building block
| Cost ($) | |
|---|---|
| Templateless PCR | |
| Oligos (18 70 mers) | 113.40 |
| Reagents | 0.14 |
| Plastics | 0.57 |
| Subtotal | 114.11 |
| Finish PCR: reagents and plastics | 0.97 |
| Ligation and transformation | |
| Ligation kit (Promega pGEM-T) | 2.75 |
| Competent cells (Promega JM109) | 2.75 |
| LB + carbenicillin plate | 0.47 |
| Plastics | 0.18 |
| Subtotal | 6.15 |
| Colony screening PCR | |
| Plastics | 1.17 |
| Reagents | 2.58 |
| Subtotal | 3.75 |
| Sequencing | |
| Plastics | 1.44 |
| Sequencing (18 clones) | 90.00 |
| Subtotal | 91.44 |
| Total | 216.42 |
Although our intent is to eventually synthesize the complete yeast genome, this methodology is widely applicable at institutions that do not have the resources to pursue eukaryotic genome synthesis. While we have assigned 10–11 individual building block sequences per student, it is certainly feasible to have multiple students, or indeed an entire class, working on a single gene, small viral genome, or transposable element (Smith et al. 2003; Han and Boeke 2004; Chan et al. 2005). The synthesis of fewer building blocks reduces the most substantial costs, namely oligonucleotide synthesis and DNA sequencing, as well as the amount of staff support required. An even more cost-effective method would be to create multiple variants in addition to a wild-type synthetic gene; replacement of only one or two oligonucleotides in the initial pool used for synthesis would enable students to incorporate targeted mutations at only a marginally increased cost. Mutants synthesized during a Build-a-Genome course could be used in subsequent molecular biology, biochemistry, cell biology, or biological engineering courses to examine gene function and expression, providing opportunities for cooperation between undergraduate laboratory courses.
Although a full laboratory Build-a-Genome course might be prohibitively expensive for many institutions, synthetic biology could be incorporated into existing molecular biology, molecular genetics, or bioinformatics courses. Class time could be reduced to two lectures: a didactic lecture on the principles of genome synthesis and a second computer-based class that would allow students to design their own synthetic gene and break down the sequence into constituent oligonucleotides using GeneDesign software. These lectures could be coupled to the assembly of building blocks in the laboratory with two or three lab sessions devoted to templateless PCR, finish PCR, and gel electrophoresis. A focus on the PCR steps could retain much of the Build-a-Genome emphasis on independent lab work and troubleshooting.
One area of the Build-a-Genome course where some students had difficulty was the repetitive nature of this project. Repetition of the protocols led to proficiency with each technique, yet beyond two rounds, students understandably became bored with additional gene synthesis (with a few exceptions). In many cases, student attention was refocused by the introduction of side projects. In the future, we plan to expand the scope of the course to include assembly of individual building blocks into larger chunks of 10–30 kb and incorporation of synthetic DNA into yeast as part of the standard curriculum. The assembly of synthetic genes into larger genomes or into synthetic parts and devices offers an unlimited number of directions in which students and faculty alike can expand their involvement in synthetic biology.
Acknowledgments
We thank Marc Ostermeier, Debra Mathews, and our teaching assistants/instructors Eric Cooper, Michalis Hadjithomas, Josh Sims, Jennifer Tullman, and Karthikevan Kandavelou for help in implementation and refinement of the course. We thank the students on the Build-a-Genome team whose enthusiasm for synthetic biology was crucial to the success of the course: Raghav Ramachandran, James DiCarlo, Allison Suarez, Alyson Nickols, Tejasvi Niranjan, Ben Lin, John Chung, Avi Schuldenfrei, Calvin Lau, Jason Feinberg, Matthew Linder, Brian Capaldo, Judy (Rong) Qiu, Nathaniel Sotuyo, Matt Rubashkin, Marina Paul, Viktoriya London, Jaime Liu, Jonathan Ling, David Gladowski, Joy Chang, Richard Carrick, Aaron Moore, Alexandra McMillan, Alex Rhee, Allen Yu, Andy Wong, Denise Lin, Ina Soh, Isabel Emiko Ishizuka, Javaneh Jabbari, Jessi McDade, Jonathan Liu, Kimberly Cirelli, Kristie Charoen, Murat Bilgel, Pasha Hadidi, Ruchi Patel, Wei “Rose” Xie, Yu “Charlie” Ouyang, Viswanath “Visu” Annaluru, Katrina Foelber, Matthew Dumont, Mary Mallaney, Chris van Dollen, Zheyuan Guo, Jingye “Sunny” Chen, Deng “Peter” Pan, Jessica Shiao, and Rishi Trivedi. We thank Karen Beemon, Sally Chiappa, Paula Burger, and Dean David Bell of the Krieger School of Arts and Sciences and Kathy Jancuk, Murray Sachs, and Dean Nick Jones of the Whiting School of Engineering for facilities, infrastructure, and financial support of the course. We thank the Schools of Medicine and Public Health for partial salary support of J.D.B. and S.C., respectively. S.R. was supported by a Department of Energy Computational Science Graduate Fellowship (no. DE-FG02-97ER25308). This project was supported in part by a Microsoft Research Award to J.S.B. and J.D.B. and by grant MCB 0718846 from the National Science Foundation to J.D.B., J.S.B., and S.C.
References
- Andrianantoandro, E., S. Basu, D. K. Karig and R. Weiss, 2006. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2 0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares, M., Jr., 2004. Interdisciplinary research and the undergraduate biology student. Nat. Struct. Mol. Biol. 11 1170–1172. [DOI] [PubMed] [Google Scholar]
- Balmer, A., and P. Martin, 2008. Synthetic Biology: Social and Ethical Challenges. Biotechnology and Biological Sciences Research Council. http://bbsrc.ac.uk/organisation/policies/reviews/scientific_areas/0806_synthetic_biology.html
- Bednarski, A. E., S. C. Elgin and H. B. Pakrasi, 2005. An inquiry into protein structure and genetic disease: introducing undergraduates to bioinformatics in a large introductory course. Cell Biol. Educ. 4 207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek, W., and D. Botstein, 2004. Introductory science and mathematics education for 21st-century biologists. Science 202 788–790. [DOI] [PubMed] [Google Scholar]
- Campbell, A. M., 2005. Meeting report: synthetic biology jamboree for undergraduates. Cell Biol. Educ. 4 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, L. Y., S. Kosuri and D. Endy, 2005. Refactoring bacteriophage T7. Mol. Syst. Biol. 1 0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S. N., A. C. Y. Chang, H. W. Boyer and R. B. Helling, 1973. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. USA 70 3240–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin, D. A., J. C. Way and P. A. Silver, 2007. Designing biological systems. Genes Dev. 21 242–254. [DOI] [PubMed] [Google Scholar]
- Endy, D., 2005. Foundations for engineering biology. Nature 438 449–453. [DOI] [PubMed] [Google Scholar]
- Endy, D., 2008. Genomics: reconstruction of the genomes. Science 319 1196–1197. [DOI] [PubMed] [Google Scholar]
- Goodman, C., 2008. Engineering ingenuity at iGEM. Nat. Chem. Biol. 4 13. [DOI] [PubMed] [Google Scholar]
- Gross, L. J., 2004. Interdisciplinarity and the undergraduate biology curriculum: finding a balance. Cell Biol. Educ. 3 85–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J. S., and J. D. Boeke, 2004. A highly active synthetic mammalian retrotransposon. Nature 429 314–318. [DOI] [PubMed] [Google Scholar]
- Holt, R. A., 2008. Synthetic genomes brought closer to life. Nat. Biotechnol. 26 296–297. [DOI] [PubMed] [Google Scholar]
- Kerfeld, C. A., and R. W. Simons, 2007. The undergraduate genomics research initiative. PLoS Biol. 5 e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuldell, N., 2007. Authentic teaching and learning through synthetic biology. J. Biol. Eng. 1 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., 2005. Teaching systems biology: an active-learning approach. Cell Biol. Educ. 4 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan et al., 2007. Clustal W and clustal X version 2.0. Bioinformatics 23 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee, Y. N., and P. D. Bieniasz, 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council, 2003. Bio2010: Transforming Undergraduate Education for Future Research Biologists. National Academies Press, Washington, DC. http://books.nap.edu/catalog/10497.html. [PubMed]
- Richardson, S. M., S. J. Wheelan, R. M. Yarrington and J. D. Boeke, 2006. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 16 550–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H. O., C. A. Hutchison, III, C. Pfannkoch and J. C. Venter, 2003. Generating a synthetic genome by whole genome assembly: PhiX174 bacteriophage from synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA 100 15440–15445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer, W. P., A. Crameri, K. D. Ha, T. M. Brennan and H. L. Heyneker, 1995. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164 49–53. [DOI] [PubMed] [Google Scholar]
- Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solorzano et al., 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310 77–80. [DOI] [PubMed] [Google Scholar]