Abstract
Mutagenic DNA repair (MDR) employs low-fidelity DNA polymerases capable of replicating past DNA lesions resulting from exposure to high-energy ultraviolet radiation (UVR). MDR confers UVR tolerance and activation initiates a transient mutator phenotype that may provide opportunities for adaptation. To investigate the potential role of MDR in adaptation, we have propagated parallel lineages of the highly mutable epiphytic plant pathogen Pseudomonas cichorii 302959 with daily UVR activation (UVR lineages) for ∼500 generations. Here we examine those lineages through the measurement of relative fitness and observation of distinct colony morphotypes that emerged. Isolates and population samples from UVR lineages displayed gains in fitness relative to the ancestor despite increased rates of inducible mutation to rifampicin resistance. Regular activation of MDR resulted in the maintenance of genetic diversity within UVR lineages, including the reproducible diversification and coexistence of “round” and “fuzzy” colony morphotypes. These results suggest that inducible mutability may present a reasonable strategy for adaptive evolution in stressful environments by contributing to gains in relative fitness and diversification.
SEQUENCE variation in the form of mutation supplies the fuel for natural selection. However, boosting variation by raising the mutation supply rate does not automatically accelerate adaptation, as only a small minority of mutations are expected to be advantageous. The propensity for deleterious mutations maintains continual selection pressure in favor of lower genomic mutation rates (Sniegowski et al. 2000; De Visser 2002; De Visser and Rozen 2005). This notion is reinforced by the conservation of multiple DNA error-avoidance and error-repair processes. Therefore, bacteria must balance a mutation supply rate adequate to permit adaptation with the maintenance of genome stability, as both are necessary for long-term population survival (Rainey 1999).
There are an increasing number of examples of bacterial strains that do not maintain low genomic mutation rates and still manage to survive. General mutators (or hypermutators) exhibit increased mutation rates, as much as 100–1000 times greater than wild type, due to defects in DNA proofreading and repair functions. Mutator alleles themselves have not been shown to confer any direct benefit to organismal fitness. Their advantage arises from an increased probability of producing rare, beneficial mutations compared to a wild-type population of the same size (De Visser et al. 1999; Tenaillon et al. 1999; Giraud et al. 2001). Mutator abundance within a population is likely the result of hitchhiking when the benefit from secondary adaptive mutations counterbalances the cost of accumulating deleterious mutations (Sniegowski et al. 2000; Tenaillon et al. 2001; De Visser 2002). The widespread existence of mutator genotypes in both environmental and clinical populations suggests that the evolutionary strategies of bacteria include mechanisms for increasing cellular mutability (Hall and Henderson-Begg 2006; Sundin and Weigand 2007).
Similarly, an inducible mutator phenotype can transiently increase mutability in response to specific stress conditions. One such transient mutator strategy is mutagenic DNA repair (MDR), executed by specialized low-fidelity DNA polymerases in response to ultraviolet radiation (UVR) exposure. DNA lesions in the form of cyclobutane pyrimidine dimers and 6-4 photoproducts result from direct UVR exposure and distort the helical structure of DNA. These lesions disrupt replication fork progression and induce expression of the SOS response. The SOS regulon is a genotoxic stress response that coordinates the control of ∼40 unlinked genes involved in DNA repair, recombination, and cell cycle control (Sutton et al. 2000; Michel 2005; Erill et al. 2007; Schlacher and Goodman 2007).
Among those genes induced in the SOS regulon are those encoding low-fidelity DNA polymerases polIV (Escherichia coli dinB) and polV (E. coli umuDC) from the Y family of DNA polymerases (Jarosz et al. 2007). The genetic and biochemical nature of the Y-family class of DNA polymerases has been the subject of intense study (Goodman 2002; Rattray and Strathern 2003). These repair polymerases exhibit high processivity and low fidelity by nature of their more relaxed active sites, permitting them to perform translesion DNA synthesis (TLS) across damaged regions of DNA in a template-independent manner where normal, replicative DNA polymerases cannot (Goodman 2002; Rattray and Strathern 2003). TLS activity confers UVR tolerance by rescuing stalled replication forks and allowing complete replication of the genome but carries the cost of an increased mutation rate.
Members of the Y family of DNA polymerases can be found across all three domains of life (Ohmori et al. 2001), yet their function in such divergent backgrounds is not well understood. According to the second-order selection hypothesis, the genetic diversity associated with an inducible increased mutation rate facilitates adaptation in stressful environments (Radman 2001; Tenaillon et al. 2004). Alternatively, the pleiotropic hypothesis suggests that inducible mutability is simply the unfortunate by-product of a last-ditch effort to enhance survival in the face of DNA damage (Tenaillon et al. 2004). Inducible mutagenesis either provides positive opportunities for adaptive evolution under stressful conditions or reduces fitness out of desperation for survival due to the predominantly deleterious nature of mutation.
Presently, the E. coli umuDC system remains the most widely studied example of MDR. However, enteric bacteria are not routinely exposed to UVR-induced DNA damage, the best recognized elicitor of MDR, making them unsuitable for investigating the ecological or evolutionary implications of MDR. Alternatively, the umuDC homolog rulAB has been identified on an indigenous plasmid in the plant pathogen Pseudomonas syringae (Sundin et al. 1996). Epiphytic plant pathogens like P. syringae require large populations on host leaf surfaces for successful infection. Since leaves are optimized for solar UVR exposure, MDR-mediated UVR tolerance provides a critical ecological advantage to these organisms (Sundin and Murillo 1999). Plant-associated Pseudomonas species harboring rulAB display varied phenotypic levels of UVR tolerance and mutability, suggesting a beneficial but likely not essential function (Sundin et al. 2000). In a survey of plant-associated Pseudomonas isolates, P. cichorii 302959, an epiphytic pathogen of celery, has emerged as exceptionally UVR mutable owing to a chromosomal copy of rulAB (Zhang and Sundin 2004a). P. cichorii 302959 rulAB provides an ecologically relevant context and a pronounced transient mutator phenotype ideal for investigating the potential role of inducible mutability in adaptive evolution.
We have designed a bacterial lineage experiment modeled after the E. coli Long-Term Experimental Evolution Project (Lenski et al. 1991), using the highly mutable P. cichorii 302959. The Pseudomonas MDR determinant rulAB within the experimental evolution model provides a controllable system that is both ecologically relevant and amenable to genetic manipulation in which to observe the adaptive influence of MDR. Here we report a long-term study to test our hypothesis that regular activation of MDR has a positive influence on adaptation to stressful environments without reducing fitness in a control environment in P. cichorii 302959. We maintained parallel cultures in a serial transfer regime with and without a daily dose of UVR and quantified changes in fitness relative to the ancestor. Among those lineages exposed to UVR, we observed distinct colony morphotypes, each of which exhibited increased fitness under the experimental conditions despite a transiently elevated mutation rate due to MDR induction.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and general molecular biology techniques:
The ancestral “round” (R) strain of P. cichorii 302959 was originally isolated in Japan (Zhang and Sundin 2004a). All evolved isolates were derived from the ancestral genotype following 500 generations of selection in the lineage experiment described below. Evolved isolates are denoted by lineage replicate and morphotype (e.g., 25R refers to an isolate from lineage 25 that exhibits the round colony morphotype).
Strains of P. cichorii were cultured at 28° in Luria–Burtani (LB) broth (Difco, Detroit), in Davis Minimal broth supplemented with 25 mg liter−1 glucose (DM25) (Difco), or on King's medium B (KB) agar (King et al. 1954). E. coli DH5α was cultured at 37° in LB broth or on LB agar. Antibiotics were used where appropriate at the following concentrations: ampicillin 50 μg ml−1, carbenicillin 50 μg ml−1, gentamicin 10 μg ml−1, rifampicin 250 μg ml−1.
Competent cell preparation was performed according to Sambrook et al. (1989) for E. coli and according to Choi et al. (2006) for P. cichorii. Plasmid DNA was isolated from E. coli and P. cichorii, using either QIAprep (QIAGEN, Valencia, CA) or Wizard Plus SV Miniprep (Promega, Madison, WI) kits and genomic DNA was isolated with the DNeasy Tissue kit (QIAGEN). Transformation by electroporation, standard agarose gel electrophoresis, and other recombinant DNA techniques were performed according to Sambrook et al. (1989).
PCR amplification and DNA sequencing:
Primers were designed on the basis of a consensus composed of P. aeruginosa, P. cichorii, P. fluorescens, P. putida, and P. syringae sequences for 16S rDNA (small ribosomal RNA subunit), atpD (catalytic subunit of F0F1-ATPase), and recA (SOS recombinase) genes (Table 1). PCR amplifications were performed in a 20-μl reaction volume containing 1× PCR buffer, 2 mm MgCl2, 125 μm of each deoxynucleoside triphosphate (Invitrogen, Carlsbad, CA), 0.4 μm of each primer, 1.0 unit Taq polymerase (Invitrogen), and 40 ng genomic DNA. PCR amplifications were carried out as follows: one cycle at 94° for 3 min; 35 cycles at 94° for 1 min, 55° for 16S rRNA or 59° for recA and atpD for 1 min, and 72° for 1.5 min; and a final extension at 72° for 7 min. PCR products were purified using the PCR Clean-Up kit (QIAGEN), and the purified fragments were sequenced at the Research Technology Support Facility at Michigan State University using the PCR primers listed in Table 1.
TABLE 1.
Pseudomonas PCR primers
| Gene | Sequencea | Fragment length (bp) |
|---|---|---|
| 16S | 5′-GAGCGGCGGACGGGTGAGTAATG | 1400 |
| 5′-AGGTGATCCAGCCGCAGGTTCC | ||
| atpD | 5′-AAGGGCGGTAAAGTCGGTCTGTTC | 650 |
| 5′-GAGCGGTGTCGTAGTGTTCCTG | ||
| recA | 5′-GATGCCGACCACGCACTGGAC | 520 |
| 5′-TGCCCTTGCCGTAAAGAATCTGGA |
Consensus based on aligned P. aeruginosa, P. cichorii, P. fluorescens, P. putida, and P. syringae gene sequences.
Long-term bacterial lineage experiment:
Sixteen parallel lineages of P. cichorii 302959 were founded from a single colony and propagated in a manner similar to the E. coli Long-Term Experimental Evolution Project (Lenski et al. 1991). Cultures were maintained in 10 ml DM25 in 50-ml Erlenmeyer flasks and incubated at 28° and 150 rpm in the dark. These culture conditions resulted in a stationary phase density of ∼5 × 108 colony-forming units/ml. Every 24 hr, 8 UVR lineages (numbered 25–32) were individually mixed 1:1 with saline (0.85% NaCl) in a glass petri dish and exposed to ∼40 J m−2 of UVC (254 nm) radiation from an XX-15 UV lamp (UVP Products, San Gabriel, CA) placed horizontally at a fixed height above the cell suspension. The lamp was turned on 15 min prior to use to allow for stabilization of the UV output. The energy output of the lamp was monitored with a UV-X radiometer fitted with a UV-25 sensor (UVP Products) and determined to be 1.5 J m−2 s−1. Following UVR irradiation, cultures were diluted 1:100 into fresh media and incubated under dark conditions to minimize photoreactivation. The other 8 non-UVR lineages (numbered 33–40) were diluted 1:100 in saline and then transferred 1:10 into fresh media without UVR. Both transfer strategies result in 1000-fold daily growth of each population, representing ∼10 generations of binary fission. Cultures were grown in this manner with daily transfers for ∼500 generations. Population samples from each lineage were preserved in a nonevolving state in 10% glycerol at −80° every 10 generations for the first 200 generations and then every 50 generations thereafter for later analysis.
UVR mutability assays:
Three lineages, two UVR and one non-UVR, were chosen at random, and preserved population samples at 100-generation intervals were thawed and ∼108 cells were inoculated into 5 ml LB medium. Following overnight growth, 2 ml of culture was pelleted, washed with 1 ml saline, and resuspended in 1 ml saline on ice. Cell suspensions were mixed with 9 ml saline in a glass petri dish and irradiated with a single dose of ∼40 J m−2 as described above. Before irradiation, cell suspensions were plated on KB and KB + rifampicin to determine the frequency of rifampicin-resistant (RifR) cells in each population. Following irradiation, 1 ml of cell suspension was mixed with 1 ml 2× LB broth, incubated overnight in the dark, and plated on KB and KB + rifampicin to determine UVR-inducible mutability. The number of spontaneous RifR mutants in the absence of UVR was subtracted and the frequency of mutation to RifR due to UVR-inducible mutability was calculated as the number of RifR mutants per 108 surviving cells.
Competition experiments and relative fitness calculations:
The relative fitness of isolates from each evolved lineage was determined by direct competition with the ancestor under the same non-UVR and UVR conditions as the long-term lineage experiment. Strain differentiation was accomplished by using a marker plasmid encoding catechol 2,3-dioxygenase (xylE) and gentamicin acetyltransferase-3-1 (aacC1) genes. Catechol dioxygenase-expressing colonies turn yellow when sprayed with 0.5 m catechol for easy discrimination on agar media. The marker plasmid was constructed by inserting the xylE-aacC1 cassette from pX1918GT (Schweizer and Hoang 1995) into the multiple cloning site of pUCP19 (Schweizer 1991), resulting in pMRW2. A control plasmid, pMRW3, was constructed by removing an EcoRV restriction fragment required for both xylE and aacC1 gene function. In preparation for competition assays, clones of each isolate were transformed with either the marker or the control plasmid. Six replicates of each competition assay were conducted with three replicates of each marker pair (e.g., three replicates of ancestor/pMRW2 paired with 25R/pMRW3 and three replicates of ancestor/pMRW3 paired with 25R/pMRW2).
To ensure that competing isolates were comparably acclimated to the competition environment, isolates were simultaneously removed from the freezer, individually grown in LB broth overnight, and then individually grown for 24 hr in the competition environment. Under non-UVR conditions, competitors were mixed at a 1:1 volumetric ratio, diluted into fresh DM25, and incubated in the dark at 28° and 150 rpm for 24 hr. Appropriate dilutions were plated on KB agar in triplicate at 0 and 24 hr to estimate the initial and final density of each competitor. Relative fitness (W) under non-UVR conditions was calculated as the ratio of the Malthusian parameters (m) of the two competing strains as determined by the equations
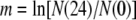 |
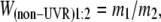 |
where N(0) and N(24) are the population sizes of the strains at 0 and 24 hr, respectively (Lenski et al. 1991). For competitions under UVR conditions, individual cultures were acclimated as described above, mixed at a 1:1 volumetric ratio, and then diluted in saline, irradiated, and transferred to fresh medium as described for the long-term lineage experiment. Dilutions were plated before UVR irradiation (0 hr) and after 24 hr of growth. Because the 0-hr density is measured before dilution into fresh medium, W must be calculated as the ratio of each competitor's realized (net) growth rate (r),
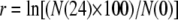 |
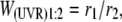 |
where N(0) and N(24) still represent the population sizes of the strains at 0 and 24 hr, respectively. This was necessary to account for the 100-fold dilution into fresh medium following UVR (Sleight and Lenski 2007). When the two strains are equally fit in the competition environment, W = 1.
The six replicate relative fitness measurements were analyzed by two-tail, independent t-tests against the hypothesis of mean equal fitness (W = 1). Fitness measurements of each isolate under non-UVR and UVR conditions were compared by a two-tailed paired t-test to determine any adaptation to daily UVR. Two-tailed t-tests were used because it was not known beforehand if UVR-induced mutability would produce an increase or a reduction in relative fitness.
Similarly, the relative fitness of population samples from evolved UVR lineages was determined under both non-UVR and UVR conditions with a few modifications. Preserved lineage samples were thawed and ∼108 cells were inoculated into 5 ml LB medium and grown overnight before acclimation to the competition environment. To maintain diversity, population samples were not transformed with either pMRW2 or pMRW3 and all six replicate competitions were conducted with the ancestor bearing pMRW2.
UVR tolerance assays:
Bacterial isolates were grown overnight in LB broth and 2 ml of culture were pelleted, washed with 1 ml saline, and resuspended in 1 ml saline on ice. The cell suspensions were mixed with 9 ml saline in a glass petri dish and irradiated as described above. Increases in the radiation dose delivered were accomplished by increasing the time of exposure. Cell suspensions were mixed continuously while receiving UVR doses to eliminate survival as a result of shading. Following irradiation, surviving cells were enumerated by dilution plating conducted under dark conditions.
RESULTS
UVR mutability:
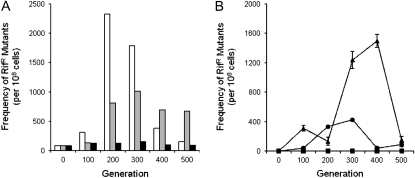
To confirm MDR activity during lineage growth, preserved population samples from three lineages taken at 100-generation intervals were thawed and the frequency of UVR-induced mutation to rifampicin resistance was measured in each population. The results illustrated in Figure 1A show that the rate of UVR-inducible mutability fluctuated within UVR lineages 27 and 31 over the course of the lineage experiment. Conversely, non-UVR lineage 35 exhibited a constant rate of UVR-induced mutation to rifampicin resistance that was comparable to that of the ancestral population (Figure 1A). The total frequency of RifR cells within these three lineages is plotted in Figure 1B. RifR cells appeared and varied in frequency within UVR lineages 27 and 31 but were not present in non-UVR lineage 35.
Figure 1.—
(A) UVR-inducible mutability of P. cichorii 302959 lineages 27 (open bars), 31 (shaded bars), and 35 (solid bars) assessed from culture samples taken at 100-generation intervals during the long-term lineage experiment. The UVR dose applied was 40 J m−2. The number of spontaneous mutations conferring rifampicin resistance in the absence of UVC irradiation has been subtracted. (B) Frequency of rifampicin-resistant mutants in P. cichorii 302959 lineages 27 (•), 31 (▴), and 35 (▪). Values are means and error bars represent standard error of the mean. If error bars are not visible it is because they are smaller than the size of the marker.
Emergence of a “fuzzy” morphotype in long-term bacterial lineages:
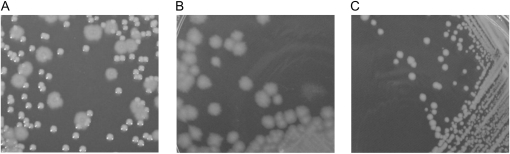
To track population structure and identify possible contamination, each lineage population was plated daily on KB agar. After 60 generations, all eight of the UVR lineages contained a rare fuzzy (F) colony morphotype (morph) in addition to the ancestral R colonies (Figure 2A). Colonies with the F morphotype exhibited enlarged but variable size, a grainy surface, and rough edges when grown on KB agar (Figure 2B) but not when grown on LB agar (Figure 2C).
Figure 2.—
Phenotypic diversification among colonies of P. cichorii 302959 evolved in UVR lineages. (A) A “fuzzy” (F) colony morphotype emerged reproducibly within the first 60 generations and coexisted with the ancestral “round” morphotype in all lineages exposed to daily UVR. Colonies with the F morphology exhibited enlarged but variable size, grainy surface, and rough edges when plated on (B) KB agar but not on (C) LB agar.
PCR amplification and fragment sequencing were completed with the three housekeeping sequences 16S rDNA, atpD, and recA to determine if the F colonies were the result of a systematic contamination or represented a new morphotype of P. cichorii 302959. Aligned sequence fragments from R and F morphs from each lineage and the ancestor shared 100% sequence identity (data not shown). These F morphs never appeared in any of the non-UVR lineages or in a sterile medium control that was irradiated and serially transferred in parallel. We never detected any contamination in this sterile medium control throughout the duration of the experiment.
Relative abundance of the fuzzy morphotype:
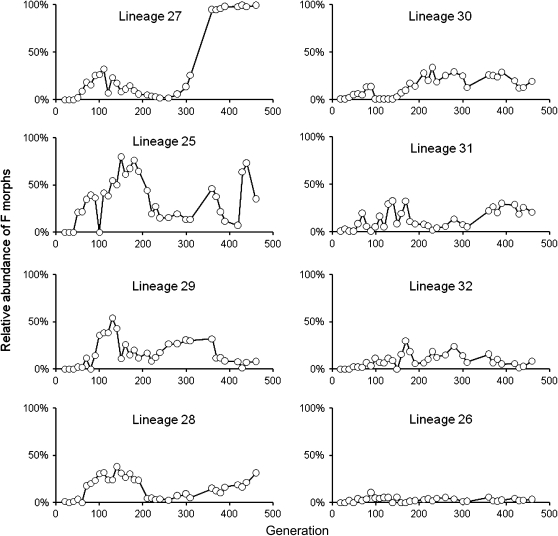
Daily dilution plating of lineage populations on KB agar allowed for tracking of F morph abundance relative to ancestral R morphs. Figure 3 shows the percentage of total colonies that exhibited the F morphotype in the eight UVR lineages over the course of the long-term experiment. The relative abundance of the F morphotype fluctuated stochastically over time, exhibiting a mixture of local peaks and short periods of apparent stable population distribution. However, F morph abundance never exceeded 10% in lineage 26 and appeared to reach a plateau at ∼94% in lineage 27.
Figure 3.—
Relative abundance of “fuzzy” (F) morphs in the eight replicate UVR lineages over the course of the experiment. Lineages are sorted in descending order by highest recorded abundance of F morphs. Sampling was conducted every 10 generations with the exception of a gap between generations 310 and 360. Although F morphs emerged reproducibly within the first 60 generations of all eight UVR lineages, their abundance fluctuated relative to the ancestral “round” (R) type.
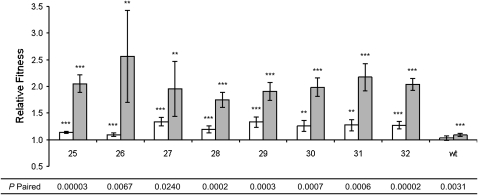
Relative fitness of isolates from evolved lineages:
Single-colony representatives of R and F morphs were isolated from UVR lineages and R morphs were isolated from non-UVR lineages. Relative fitness of these 24 isolates was determined by competition with the P. cichorii 302959 ancestor and the results are illustrated in Figure 4. The average relative fitness of the ancestor competed against itself under both the non-UVR and the UVR conditions was not significant (both P > 0.32, two-tailed independent t-tests, d.f. = 5, μ = 1), confirming that pMRW2 and pMRW3 did not influence competitive growth (data not shown).
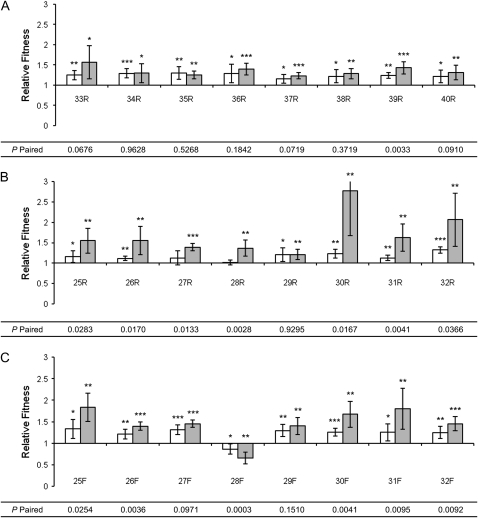
Figure 4.—
Relative fitness of isolates from evolved lineages of P. cichorii 302959 after 500 generations. Fitness values were measured in competition with the ancestor under non-UVR (open bars) and UVR (shaded bars) conditions. (A) Lineages 33–40 were maintained without UVR and contained only the “round” (R) morphotype. Lineages 25–32 received daily doses of UVR and gave rise to both (B) R and (C) “fuzzy” (F) morphs. Relative fitness values are means and error bars represent 95% confidence intervals. Values are significant (two-tailed independent t-test, d.f. = 5, α = 0.05) where indicated (*P < 0.05, **P < 0.01, ***P < 0.001). P-paired values are from two-tailed paired t-tests (d.f. = 5, α = 0.05) between non-UVR and UVR relative fitness values for each isolate.
The R morphs isolated from non-UVR lineages exhibited increased fitness under non-UVR conditions (all P < 0.038, two-tailed independent t-test, d.f. = 5, μ = 1) and UVR conditions (all P < 0.022, two-tailed independent t-test, d.f. = 5, μ = 1) relative to the ancestor (Figure 4A). When comparing fitness gains under the two conditions by two-tailed paired t-test, only the relative fitness of isolate 39R varied significantly, showing an increase in fitness under UVR conditions that exceeded the gain under non-UVR conditions (Figure 4A). Isolates from the other seven non-UVR lineages did not exhibit significantly different increases in fitness between the non-UVR and UVR conditions (Figure 4A).
Six of the eight R morphs isolated from UVR lineages gained fitness under non-UVR conditions (all P < 0.038, two-tailed independent t-test, d.f. = 5, μ = 1) and all eight gained fitness under UVR conditions (all P < 0.009, two-tailed independent t-test, d.f. = 5, μ = 1) as seen in Figure 4B. The relative fitness of R morphs from UVR lineages, with the exception of isolate 29R, was significantly different under the two conditions when compared by two-tailed paired t-tests (Figure 4B).
F morphs emerged in all eight UVR lineages and these isolates also demonstrated significant changes in fitness relative to the ancestor under both non-UVR (all P < 0.037, two-tailed independent t-test, d.f. = 5, μ = 1) and UVR (all P < 0.009, two-tailed independent t-test, d.f. = 5, μ = 1) conditions (Figure 4C). One F morph isolate (28F) lost fitness relative to the ancestor when competed under both conditions. With the exception of isolates 27F and 29F, F morphs exhibited significantly different changes in relative fitness between the two growth conditions (Figure 4C). R and F morphs from UVR lineages exhibited significantly greater gains in fitness under the UVR conditions in which they evolved but neither morphotype as a group exhibited higher relative fitness compared to the other. The reduced fitness of 28F relative to the ancestor under both non-UVR and UVR conditions was unique among all isolates.
UVR tolerance of isolates from evolved lineages:
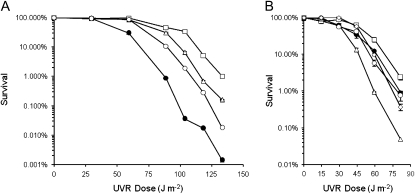
The UVR tolerance of isolates from a subset of evolved lineages was compared to that of the P. cichorii 302959 ancestor and the results are shown in Figure 5. Competitions under UVR conditions indicated that isolates 30R, 30F, and 32R all gained fitness relative to the ancestor (Figure 4) and these isolates displayed corresponding increases in UVR tolerance (Figure 5A). Isolate 40R was taken from a non-UVR lineage and exhibited a UVR tolerance phenotype comparable to the ancestor (Figure 5B). Isolates 26R and 32F were both taken from UVR lineages and displayed significantly higher fitness under UVR conditions (Figure 4), but did not exhibit increased UVR tolerance (Figure 5B). Isolate 28F displayed reduced fitness relative to the ancestor (Figure 4C) and also demonstrated reduced UVR tolerance (Figure 5B).
Figure 5.—
UVR tolerance of “round” (R) and “fuzzy” (F) isolates from evolved lineages of P. cichorii 302959 after 500 generations. (A) Survival of P. cichorii 302959 ancestor (•), 30R (□), 30F (▵), and 32R (○). (B) Survival of P. cichorii 302959 ancestor (•), 26R (□), 28F (▵), 32F (○), and 40R (◊). Values are means from three replicates and bars represent standard error of the mean. If error bars are not visible it is because they are smaller than the size of the marker.
Relative fitness of populations from evolved lineages:
Fitness changes in lineages were also assessed at the population level. Preserved population samples from the eight UVR lineages were thawed and subcultured. The relative fitness of these populations was determined by competition with the P. cichorii 302959 ancestor and the results are displayed in Figure 6. All eight UVR lineage populations exhibited increased fitness under non-UVR (all P < 0.0012, two-tailed independent t-test, d.f. = 5, μ = 1) and UVR (all P < 0.0055, two-tailed independent t-test, d.f. = 5, μ = 1) conditions relative to the ancestor. When compared by two-tailed paired t-tests, the relative fitness of these populations under UVR conditions was significantly greater than under non-UVR conditions.
Figure 6.—
Relative fitness of populations from evolved UVR lineages of P. cichorii 302959 after 500 generations. Fitness values were measured in competition with the ancestor marked with pMRW2 under non-UVR (open bars) and UVR (shaded bars) conditions. Relative fitness values are means and error bars represent 95% confidence intervals. Values are significant (two-tailed independent t-test, μ = 1, d.f. = 5, α = 0.05) where indicated (*P < 0.05, **P < 0.01, ***P < 0.001). P-paired values are from two-tailed paired t-tests (d.f. = 5, α = 0.05) between non-UVR and UVR relative fitness values for each population.
DISCUSSION
The results of this long-term study suggest that inducible mutability can contribute to gains in fitness and diversification in a stressful environment. Using the highly UVR-mutable P. cichorii 302959, we maintained parallel cultures in a serial transfer regime with and without UVR exposure. The relative fitness of isolates and diverse populations from evolved lineages and the emergence of a new colony morphotype were investigated to evaluate the influence of MDR on adaptive evolution.
Before looking closely at the outcome of the long-term lineages, it was necessary to confirm the activity of UVR-inducible mutability in UVR lineages over the course of our experiment. To this end, we have observed the frequency of RifR cells following UVR irradiation in population samples taken from three randomly chosen lineages. UVR lineages 27 and 31 exhibited increased and varied rates of UVR-inducible mutability observed as a higher frequency of rifampicin resistance following UVR exposure (Figure 1A). Not only did UVR lineages maintain inducible MDR activity, but also mutability to rifampicin resistance increased following repeated activation. The total number of RifR cells within UVR lineages 27 and 31 fluctuated over time (Figure 1B), similar to studies conducted previously (Zhang and Sundin 2004a). Because the total frequency of RifR cells varied rather than simply accumulating over time, this suggests the presence of other linked mutations that dictated the relative frequency of rifampicin resistance in the population. Conversely, non-UVR lineage 35 maintained a constant level of UVR mutability similar to that of the ancestor (Figure 1A) and a very low overall frequency (<3 per 108 cells) of RifR cells (Figure 1B). This indicates that within non-UVR lineages inducible mutability does not decline in the absence of activation while maintaining a lower mutation rate compared to UVR lineages.
During the course of the long-term experiment, all 16 lineages were plated daily to monitor for potential contamination. The emergence of the F morphotype in our UVR lineages was unexpected and originally believed to be a contaminant resulting from our daily UVR transfer scheme (Figure 2). The competitive-exclusion principle dictates that the number of coexisting genotypes cannot exceed the number of limiting resources and the evolution of asexual populations in an environment containing a single, limiting resource is expected to proceed by a series of clonal replacements (Rainey et al. 2000). However, the emergence and maintenance of diversity by adaptive radiation is well documented in evolution experiments with bacteria in simple environments (Turner et al. 1996; Rozen and Lenski 2000). The diversification of P. fluorescens SBW25 after 3 days of growth in a spatially heterogeneous environment represents the most notable example (Rainey and Travisano 1998). Rainey and Travisano (1998) found three morphotypes occupying different physical niches and maintaining their balanced coexistence through negative frequency-dependent selection.
Our serial transfer regime, although spatially homogenous, exhibited temporal resource heterogeneity as nutrient availability fluctuated during each 24-hr growth cycle. A stable coexistence can emerge in such an environment provided one genotype has an advantage when resources are abundant and the other has an opposing advantage when resources are limited (Rainey et al. 2000; Kassen and Rainey 2004). Additionally, organisms themselves are capable of creating metabolic heterogeneity within a well mixed, and otherwise homogeneous, environment to relieve resource competition (Rozen and Lenski 2000; Rozen et al. 2005). The F morphotype emerged rapidly and reproducibly in all eight UVR lineages, suggesting both strong selection pressure in the environment and high adaptive potential of the ancestral P. cichorii 302959 genotype. Such divergent natural selection produces diversity as a general feature of adaptive evolution within a heterogeneous environment (MacLean 2005).
Although the F morph emerged reproducibly, abundance fluctuated in all UVR lineages (Figure 3), suggesting the F determinant itself may not carry a strong selective advantage. Fluctuations in F morph abundance likely correspond to relative changes in fitness between the R and F subpopulations as beneficial mutations occur at alternate loci. Linkage with a highly beneficial mutation likely led to the high relative abundance of F and R morphs in UVR lineages 27 and 26, respectively (Figure 3). Interestingly, in these two extreme cases the lesser abundant morphotype is never eliminated, suggesting a frequency-dependent relationship between the two subpopulations.
We have not pursued the underlying genetic loci responsible for the F morphotype. Although studies in P. fluorescens SBW25 have identified the genetic basis for the “wrinkly spreader” (Spiers et al. 2002), the genetic basis of the “fuzzy spreader” morph, which bears likeness to our P. cichorii 302959 F morph, remains unknown. Further characterization of the phenotype may provide candidate genes for investigation. The disappearance of the phenotype during growth on LB agar suggests a nutritional requirement, and previous studies of MDR in P. syringae identified a link between diversification in colony morphology and motility (Zhang and Sundin 2004b).
Our primary method for evaluating adaptation was through measuring changes in relative fitness. Non-UVR lineages were expected to gain relative fitness through adaptation to the culture conditions on the basis of the results from similar studies conducted previously (Lenski et al. 1991). The increased relative fitness of isolates from non-UVR lineages (Figure 4A) indicates that 500 generations of growth by P. cichorii 302959 in this simple environment are adequate to produce measurable and statistically significant changes and, therefore, validate our experimental system. These isolates displayed comparable fitness gains under non-UVR and UVR conditions such that any gains in relative fitness can be attributed to adaptation to the culture medium only.
The accumulation of deleterious mutations reduces organismal fitness in asexual lineages (Muller's ratchet) (Muller 1964) and presents a critical opposition to an argument for the evolutionary advantage of MDR. The pleiotropic hypothesis suggests that inducible mutagenesis is simply the unfortunate by-product of a last-ditch repair system designed to enhance survival following DNA damage (Tenaillon et al. 2004). If this is true, the increased mutability associated with long-term induction of MDR, as seen in Figure 1, could result in population extinction due to mutation accumulation. The results in Figure 4, B and C, indicate that isolates from evolved lineages of P. cichorii 302959, with the exception of 28F, exhibited increased fitness relative to the ancestor after ∼500 generations of growth with daily induction of MDR. Similarly, relative fitness gains by theoretical and experimental mutator populations have been reported previously (Sniegowski et al. 1997; Taddei et al. 1997). To our knowledge, this is the first report of changes in relative fitness following prolonged activation of inducible mutagenesis in the form of MDR. Isolates from UVR lineages (Figure 4, B and C) not only gained relative fitness through adaption to the culture medium, as seen under non-UVR conditions, but also most exhibited greater increases in relative fitness under UVR conditions. These results suggest a specificity of adaptation in the eight UVR lineages that includes the UVR treatment.
Measures of relative fitness indicate the overall advantage of one strain over another but tell nothing of the potential underlying physiological differences. In a similar experimental evolution study with E. coli in a stressful freeze–thaw environment, Sleight and Lenski (2007) investigated changes in growth dynamics to gain insight into changes in relative fitness. Improved freeze–thaw survival, faster physiological recovery from freeze–thaw stress, and faster exponential-phase growth were identified as distinct components of overall gains in fitness. By competing isolates from UVR lineages with the ancestor under both non-UVR and UVR conditions, we have observed an adaptive specificity toward UVR irradiation (Figure 4, B and C). To investigate the underlying physiology of fitness gains under UVR conditions, we compared the UVR tolerance of isolates from evolved lineages. Isolates 30R, 30F, and 32R displayed both greater relative fitness under UVR conditions (Figure 4, B and C) and higher UVR tolerance (Figure 5A). However, isolates 26R and 27F, as well as others (data not shown), displayed UVR tolerance levels similar to those of the ancestor (Figure 5B) despite increased relative fitness under UVR conditions (Figure 4, B and C). Therefore, increased UVR tolerance may contribute to fitness gains under UVR conditions for some, but not all, isolates from UVR lineages. Isolate 28F displayed reduced fitness relative to the ancestor under UVR conditions (Figure 4C) and exhibited a corresponding decrease in UVR tolerance (Figure 5A). However, isolate 28F also displayed reduced fitness under non-UVR conditions (Figure 4C), suggesting other factors likely contributed to lower relative fitness in this isolate. This suggests that changes in UVR tolerance can contribute to altered relative fitness under UVR conditions but further characterization of growth dynamics is required as other components to fitness likely exist.
Colony morphology provides an underestimate of diversity and we can be certain that our UVR lineages harbor a much greater level of genetic diversity not reflected in colony morphology variation. Therefore, population level measures of fitness in UVR lineages were also performed. Measurements of relative fitness under non-UVR conditions at the population level (Figure 6) were comparable to R and F isolate fitness measurements with the exception of lineage 28 (Figure 4, B and C). However, under UVR conditions, the relative fitness of population samples from UVR lineages (Figure 6) was greater than or equal to the relative fitness of individual R and F isolates (Figure 4, B and C). The design of our long-term experiment permitted the maintenance of population diversity within a lineage by nature of the large number of cells transferred during each cycle. The structure and diversity of these populations may influence relative fitness gains in UVR lineages such that individual isolates sampled from the population are not as fit. Additionally, these UVR populations exhibited the same specificity of adaptation as UVR lineage isolates observable as significantly greater gains in fitness under UVR conditions compared to non-UVR conditions.
In summary, the transient rise in mutation rate associated with MDR activation may provide an opportunity for adaptive evolution only if the advantage of an increased supply of beneficial mutations can outweigh the cost of accumulating deleterious mutations. We have tested the hypothesis that regular activation of MDR has a positive influence on adaption to stressful environments without reducing fitness in a control environment. Our results suggest that inducible mutability can contribute to gains in relative fitness and diversification in a stressful environment. Isolates from UVR lineages exhibited significantly greater gains in relative fitness under UVR conditions and this adaptive specificity cannot be attributed to changes in UVR tolerance alone. In tracking the frequency of two mutations we have shown that complex interactions between these mutations, their genomic backgrounds, and sympatric genotypes dictate the fluctuations in their abundance.
The emergence and maintenance of genotypic diversity in UVR lineage populations is a general feature of adaptive evolution within a heterogeneous environment and a relevant strategy for pathogenic bacteria. In microbial communities assembled by adaptive radiation, fitness relies on the ecological interactions of sympatric genotypes (MacLean 2005). The ecological interactions between genotypes in this study significantly influenced relative fitness under UVR conditions such that diverse populations were more fit than individual isolates. Additional components of relative fitness gains in evolved isolates and populations of P. cichorii 302959 and fitness relationships between isolates within lineages are currently being addressed. In the future, we will continue our investigation of the potential role of MDR in adaptive evolution with the use of this and other genetic systems.
Acknowledgments
The authors thank Rich Lenksi, Tom Schmidt, and two anonymous reviewers for helpful comments and Sean Sleight for statistical insight. This work was supported by the U.S. Department of Agriculture National Research Initiative and the Michigan Agricultural Experiment Station.
References
- Choi, K. H., A. Kumar and H. P. Scheizer, 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64 391–397. [DOI] [PubMed] [Google Scholar]
- De Visser, J. A. G. M., 2002. The fate of microbial mutators. Microbiology 148 1247–1252. [DOI] [PubMed] [Google Scholar]
- De Visser, J. A. G. M., and D. E. Rozen, 2005. Limits to adaptation in asexual populations. J. Evol. Biol. 18 779–788. [DOI] [PubMed] [Google Scholar]
- De Visser, J. A. G. M., C. W. Zeyl, P. J. Gerrish, J. L. Blanchard and R. E. Lenski, 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283 404–406. [DOI] [PubMed] [Google Scholar]
- Erill, I., S. Campoy and J. Barbe, 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31 637–656. [DOI] [PubMed] [Google Scholar]
- Giraud, A., M. Radman, I. Matic and R. Taddei, 2001. The rise and fall of mutator bacteria. Curr. Opin. Microbiol. 4 582–585. [DOI] [PubMed] [Google Scholar]
- Goodman, M. F., 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71 17–50. [DOI] [PubMed] [Google Scholar]
- Hall, L. M. C., and S. K. Henderson-Begg, 2006. Hypermutable bacteria isolated from humans – a critical analysis. Microbiology 152 2505–2514. [DOI] [PubMed] [Google Scholar]
- Jarosz, D. F., P. J. Beuning, S. E. Cohen and G. C. Walker, 2007. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 15 70–77. [DOI] [PubMed] [Google Scholar]
- Kassen, R., and P. B. Rainey, 2004. The ecology and genetics of microbial diversity. Annu. Rev. Microbiol. 58 207–231. [DOI] [PubMed] [Google Scholar]
- King, E. O., M. K. Ward and D. C. Raney, 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44 301–307. [PubMed] [Google Scholar]
- Lenski, R. E., M. R. Rose, S. C. Simpson and S. C. Tadler, 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138 1315–1341. [Google Scholar]
- MacLean, R. C., 2005. Adaptive radiation in microbial microcosms. J. Evol. Biol. 18 1376–1386. [DOI] [PubMed] [Google Scholar]
- Michel, B., 2005. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 3: e255. [DOI] [PMC free article] [PubMed]
- Muller, H. J., 1964. The relation of recombination to mutational advantage. Mutat. Res. 1 2–9. [DOI] [PubMed] [Google Scholar]
- Ohmori, H., E. C. Friedberg, R. P. P. Fuchs, M. F. Goodman, F. Hanaoka et al., 2001. The Y-family of DNA polymerases. Mol. Cell 8 7–8. [DOI] [PubMed] [Google Scholar]
- Radman, M., 2001. Fidelity and infidelity. Nature 413 115. [DOI] [PubMed] [Google Scholar]
- Rainey, P. B., 1999. Evolutionary genetics: the economics of mutation. Curr. Biol. 9 R371–R373. [DOI] [PubMed] [Google Scholar]
- Rainey, P. B., and M. Travisano, 1998. Adaptive radiation in a heterogeneous environment. Nature 394 69–72. [DOI] [PubMed] [Google Scholar]
- Rainey, P. B., A. Buckling, R. Kassen and M. Travisano, 2000. The emergence and maintenance of diversity: insights from experimental bacterial populations. Trends Ecol. Evol. 15 243–247. [DOI] [PubMed] [Google Scholar]
- Rattray, A. J., and J. N. Strathern, 2003. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu. Rev. Genet. 37 31–66. [DOI] [PubMed] [Google Scholar]
- Rozen, D. E., and R. E. Lenski, 2000. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am. Nat. 155 24–35. [DOI] [PubMed] [Google Scholar]
- Rozen, D. E., D. Schneider and R. E. Lenski, 2005. Long-term experimental evolution in Escherichia coli. XIII. Phylogenetic history of a balanced polymorphism. J. Mol. Evol. 61 171–180. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schlacher, K., and M. F. Goodman, 2007. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat. Rev. Mol. Cell Biol. 8 587–594. [DOI] [PubMed] [Google Scholar]
- Schweizer, H. P., 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97 109–121. [DOI] [PubMed] [Google Scholar]
- Schweizer, H. P., and T. T. Hoang, 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158 15–22. [DOI] [PubMed] [Google Scholar]
- Sleight, S. C., and R. E. Lenski, 2007. Evolutionary adaptation to freeze-thaw-growth cycles in Escherichia coli. Physiol. Biochem. Zool. 80 370–385. [DOI] [PubMed] [Google Scholar]
- Sniegowski, P. D., P. J. Gerrish and R. E. Lenski, 1997. Evolution of high mutation rates in experimental populations of E. coli. Nature 387 703–705. [DOI] [PubMed] [Google Scholar]
- Sniegowski, P. D., P. J. Gerrish, T. Johnson and A. Shaver, 2000. The evolution of mutation rates: separating causes from consequences. BioEssays 22 1057–1066. [DOI] [PubMed] [Google Scholar]
- Spiers, A. J., S. G. Kahn, J. Bohannon, M. Travisano and P. B. Rainey, 2002. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin, G. W., and J. Murillo, 1999. Functional analysis of the Pseudomonas syringae rulAB determinant in tolerance to ultraviolet B (290–320 nm) radiation and distribution of rulAB among P. syringae pathovars. Environ. Microbiol. 1 75–87. [DOI] [PubMed] [Google Scholar]
- Sundin, G. W., and M. R. Weigand, 2007. The microbiology of mutability. FEMS Microbiol. Lett. 277 11–20. [DOI] [PubMed] [Google Scholar]
- Sundin, G. W., S. P. Kidambi, M. Ullrich and C. L. Bender, 1996. Resistance to ultraviolet light in Pseudomonas syringae: sequence and functional analysis of the plasmid-encoded rulAB genes. Gene 177 77–81. [DOI] [PubMed] [Google Scholar]
- Sundin, G. W., J. L. Jacobs and J. Murillo, 2000. Sequence diversity of rulA among natural isolates of Pseudomonas syringae and effect on function of rulAB-mediated UV radiation tolerance. Appl. Environ. Microbiol. 66 5167–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, M. D., B. T. Smith, V. G. Godoy and G. C. Walker, 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34 479–497. [DOI] [PubMed] [Google Scholar]
- Taddei, R., M Radman, J. Maynard-Smith, B. Toupance, P. H. Gouyon et al., 1997. Role of mutator alleles in adaptive evolution. Nature 387 700–702. [DOI] [PubMed] [Google Scholar]
- Tenaillon, O., B. Toupance, H. Le Nagard, F. Taddei and B. Godelle, 1999. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics 152 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, O., F. Taddei, M. Radman and I. Matic, 2001. Second order selection in bacterial evolution: selection acting on mutation and recombination rates in the course of adaptation. Res. Microbiol. 152 11–16. [DOI] [PubMed] [Google Scholar]
- Tenaillon, O., E. Ednamur and I. Matic, 2004. Evolutionary significance of stress-induced mutagenesis in bacteria. Trends Microbiol. 12 264–270. [DOI] [PubMed] [Google Scholar]
- Turner, P. E., V. Souza and R. E. Lenski, 1996. Tests of ecological mechanisms promoting the stable coexistence of two bacterial genotypes. Ecology 77 2119–2129. [Google Scholar]
- Zhang, S., and G. W. Sundin, 2004. a Mutagenic DNA repair potential in Pseudomonas spp., and characterization of the rulABPc operon from the highly mutable strain Pseudomonas cichorii 302959. Can. J. Microbiol. 50 29–39. [DOI] [PubMed] [Google Scholar]
- Zhang, S., and G. W. Sundin, 2004. b Long-term effect of mutagenic DNA repair on accumulation of mutation in Pseudomonas syringae B86–17. J. Bacteriol. 186 7807–7810. [DOI] [PMC free article] [PubMed] [Google Scholar]