Abstract
Over one-third of human genome sequence is a product of non-LTR retrotransposition. The retrotransposon that currently drives this process in humans is the highly abundant LINE-1 (L1) element. Despite the ubiquitous nature of L1's in mammals, we still lack a complete mechanistic understanding of the L1 replication cycle and how it is regulated. To generate a genetically amenable model for non-LTR retrotransposition, we have reengineered the Zorro3 retrotransposon, an L1 homolog from Candida albicans, for use in the budding yeast Saccharomyces cerevisiae. We found that S. cerevisiae, which has no endogenous L1 homologs or remnants, can still support Zorro3 retrotransposition. Analysis of Zorro3 mutants and insertion structures suggest that this is authentic L1-like retrotransposition with remarkable resemblance to mammalian L1-mediated events. This suggests that S. cerevisiae has unexpectedly retained the basal host machinery required for L1 retrotransposition. This model will also serve as a powerful system to study the cell biology of L1 elements and for the genetic identification and characterization of cellular factors involved in L1 retrotransposition.
NON-LTR retrotransposons are ancient genetic elements that have persisted in eukaryotic genomes for hundreds of millions of years (Eickbush and Malik 2002). A phylogenetic analysis groups the non-LTR retrotransposons into several distinct clades, one of which (the L1 clade) consists of the mammalian LINE elements (Malik et al. 1999). These elements comprise 17% of human DNA (Lander et al. 2001), are still transpositionally active (Kazazian et al. 1988), can generate disease alleles by insertional mutagenesis (Kazazian et al. 1988; Babushok and Kazazian 2007), and are responsible for a significant proportion (up to ∼30%) of genome structural variation between human individuals (Korbel et al. 2007; Kidd et al. 2008). Unregulated retrotransposition of L1 elements may have catastrophic consequences to the host organism, leading to germ line cell death and sterility (Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008; Soper et al. 2008). Thus, it is likely that the host has multiple pathways to tightly regulate retrotransposition.
A typical full-length, functional member of the L1 family consists of two open reading frames, ORF1 and ORF2 (Figure 1A). ORF1 encodes a protein with nucleic acid binding properties and nucleic acid chaperone activity (Hohjoh and Singer 1996, 1997; Martin and Bushman 2001; Kolosha and Martin 2003), both of which are important for L1 activity (Kulpa and Moran 2005; Martin et al. 2005). ORF2 encodes endonuclease (Feng et al. 1996) and reverse transcriptase activity (Mathias et al. 1991), also important for L1 function (Moran et al. 1996). ORF1 and ORF2 proteins assemble with L1 RNA into a ribonucleoprotein (RNP) complex (Martin 1991), which is presumably transported into the nucleus (Kinsey 1990; Kubo et al. 2006). The endonuclease of ORF2 nicks a chromosomal target site, and the resulting free 3′ DNA end serves to prime reverse transcription of L1 RNA. This process is termed target-primed reverse transcription (TPRT) (Luan et al. 1993). The subsequent steps of replication/integration are not well understood. It is believed that host factors are intimately involved in the regulation of L1 elements and perhaps directly in the integration process (Moran and Gilbert 2002). A simple genetic system to identify these factors and study their interactions with non-LTR retrotransposons would be ideal.
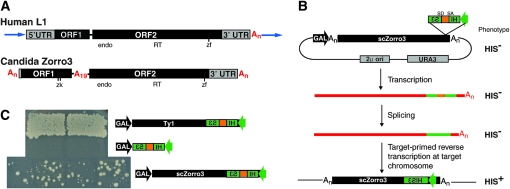
Figure 1.—
scZorro3 retrotransposition in S. cerevisiae. (A) Schematic diagram of full-length human L1 and C. albicans Zorro3. zk, zinc knuckle motif; endo, endonuclease domain; RT, reverse transcriptase domain; zf, zinc finger motif. Blue arrows are target-site duplications. (B) Retrotransposition assay. scZorro3 is controlled by the GAL1 inducible promoter, and an antisense reporter (mHIS3AI) interrupted with an intron on the scZorro3 sense strand is placed in the 3′-UTR. Only after scZorro3 transcription, splicing, and reverse trascription/integration does the marker produce functional HIS3 protein. This assay is based on previously described assays in yeast, mouse, and human (Heidmann et al. 1988; Curcio and Garfinkel 1991; Moran et al. 1996). (C) mHIS3AI-tagged Ty1, empty vector, or scZorro3 on 2μ vectors were transformed into strain GRF167 and individual clones were induced on SC −URA +galactose plates for 3 days at 23°, then replica plated to SC −HIS plates. Ty1 is a highly active endogenous yeast retrotransposon that serves as a positive control. scZorro3 produced no HIS+ colonies when grown on SC −URA +glucose plates (not shown).
One of the most powerful model organisms for genetics is the budding yeast Saccharomyces cerevisiae. However, non-LTR retrotransposons have never been found in S. cerevisiae. Although S. cerevisiae has been used to assay the enzymatic activity of ORF2 (Mathias et al. 1991; Dombroski et al. 1994; Teng et al. 1996; Clements and Singer 1998; Martin et al. 1998; Naas et al. 1998), to our knowledge no one has demonstrated non-LTR retrotransposition in this model system. Because non-LTR retrotransposons are vertically inherited (Malik et al. 1999) and are present in a variety of fungal species (Goodwin et al. 2001; Goodwin and Poulter 2001; Casaregola et al. 2002), we reasoned that an ancestor of S. cerevisiae harbored an element similar to L1, which was subsequently lost during the course of evolution. If required host factors for non-LTR retrotransposition have been fortuitously preserved, introduction of a heterologous element could lead to active retrotransposition. On the basis of this assumption, we took advantage of the discovery of Zorro3, a member of the L1 clade from the distantly related Candida albicans that is known to be active for retrotransposition in its host (Goodwin et al. 2007). Zorro3 has the same general features as a human L1 element, including putative endonuclease, reverse transcriptase (RT), and zinc finger domains in ORF2 (Figure 1A). Distinguishing features of Zorro3 (as compared to the human element) are a polydeoxyadenosine [poly(A)] tract in the 5′-untranslated region (UTR), a 19-bp poly(A) tract in the interORF region, and two zinc knuckle motifs in ORF1. In this study, we have generated a synthetic C. albicans Zorro3 for retrotransposition and demonstrate, for the first time, authentic non-LTR retrotransposition in S. cerevisiae.
MATERIALS AND METHODS
Yeast strains:
Strains were derived from GRF167 (Boeke et al. 1985). JHY85 is an isogenic MATa derivative of GRF167 generated by mating-type switching with the plasmid pGAL-HO as described previously (Herskowitz and Jensen 1991). JHY146 and JHY148 were made by PCR-based deletion (Baudin et al. 1993) of the LYS2 open reading frame followed by selection on α-aminoadipate plates. The templates for these PCRs were gel-purified FseI/EagI fragments of pSCmHIS3AI and pSCZorro3mHIS3AI, respectively. Approximately 500 bp of LYS2 flanking sequence were added on both ends by PCR to increase the efficiency of integration. The resulting mHIS3AI and Zorro3mHIS3AI loci of JHY146 and JHY148 were completely sequenced. To generate all other strains, JHY146 or JHY148 was converted to diploids by mating to JHY85. These diploids were used to knock out SPT3 and RAD52 by PCR-mediated disruption (Baudin et al. 1993; Wach et al. 1994) and to convert Zorro3mHIS3AI integrants to various mutants (ORF1mut, etc.) by two-step gene replacement using pRS406-derived (Sikorski and Hieter 1989) plasmids (supplemental Table S2). These resulting strains were sporulated and the corresponding MATα haploids (supplemental Table S1) were used for retrotransposition assays.
Plasmids:
All plasmids were generated with standard molecular biology techniques. All PCR-derived cloning products were generated with Phusion polymerase (New England Biolabs), and products were completely sequenced. The plasmid pSC was derived from pJEF724 (Boeke et al. 1985) with the following changes: (1) All Ty1 sequence is deleted, (2) a polylinker containing XhoI/HpaI/SalI/BamHI sites was inserted downstream of the GAL1 promoter, (3) the CYC1 terminator was placed downstream of the polylinker, and (4) the entire GAL-Cyc1 expression cassette was flanked by FseI/EagI restriction sites. pSCmHIS3AI was made by subcloning an mHIS3AI (Curcio and Garfinkel 1991) PCR product into the XhoI/SalI sites of pSC. pSCZorro3mHIS3AI was made by subcloning an XhoI–scZorro3–BamHI fragment (synthesized by DNA2.0) into pSC, followed by subcloning an mHIS3AI PCR product into the SalI site of the scZorro3 3′-UTR. Zorro3 mutants were generated by site-directed mutagenesis. pRS406FE is a derivative of pRS406 (Sikorski and Hieter 1989) with the addition of a unique FseI site in the polylinker. All Zorro3 pRS integrating plasmids were generated by subcloning an FseI/EagI fragment from the corresponding pSC plasmid into pRS406FE. pBSmHIS3 probe was made by subcloning a JH162/JH163 PCR product of HIS3 into the XhoI/SacII sites of pBluescript II KS(−) (Stratagene). pBS5′Z3 probe was made by subcloning an XhoI/SacII fragment of scZorro3 into the XhoI/SacI sites of pBluescript II KS(−). Complete nucleotide sequences of all of these plasmids are available upon request.
Retrotransposition assays:
To quantitate retrotransposition, strains were inoculated in 4 ml SC +glucose or SC +galactose and incubated with mixing for 72 hr at 23°. After induction, the concentrations of cells were normalized to OD600 2.5, and 3 ml were concentrated then plated on SC −HIS plates. Dilutions of the same cultures were plated on rich YPD plates to normalize for cells plated/viability. Plasmid-based retrotransposition assays were done identically except induction was done in SC −URA. All inductions were done at 23° since inductions at 16°, 30°, or 37° led to lower retrotransposition activity, similar to previously reported results in C. albicans (Goodwin et al. 2007).
For qualitative retrotransposition (e.g. Figure 1C), cells were patched on SC glucose or SC galactose plates and incubated at 23° for 72 hr. Patched cells were then replica plated to SC −HIS plates. To isolate stable independent HIS+ insertions, strains were patched on SC glucose or SC galactose plates, grown at 23° for 3–5 days, replica plated to YPD, grown for 2 days, then replica plated to SC −HIS plates.
Cloning of scZorro3 retrotransposition events:
Genomic DNA was prepared as described previously (Amberg et al. 2005). Ligation-mediated PCR, based on a previously described protocol (Dupuy et al. 2001), was used to identify 3′ flanking sequence. Briefly, 0.25 μg genomic DNA was digested with EcoRI, a JH1/JH122 linker was ligated to the ends, and PCR was performed with JH4 (hybridizes to linker) and JH102 (hybridizes to 3′ end of scZorro3mHIS3AI). The resulting PCR products were sequenced with an equimolar mixture of primers JH176-JH178. Once flanks were identified, primers were designed to amplify individual insertions (see supplemental Table S3). All amplifications were done with ExTaq DNA Polymerase (Takara). Insertions were TOPO-TA cloned (Invitrogen) and sequenced. For endo− insertions at non-poly(A) tracts, primers were designed to amplify, TOPO clone, and sequence the 5′ junctions.
Northern blot:
Total yeast RNA was isolated with acid phenol as described previously (Amberg et al. 2005). Five micrograms of each sample were run on a 0.8% agarose/formaldehyde gel and blotted to positively charged nylon membrane (Millipore). Hybridization was performed in Ultrahyb (Ambion) with ∼20 ng/ml of probe at 65°. Riboprobes were biotin-16-UTP labeled T7 in vitro transcription products of XhoI-digested pBSmHIS3 probe (His probe) or CD15/CD16 PCR product of genomic DNA (tubulin probe). Detection was performed with a Phototope-Star detection kit (New England Biolabs). Membranes were stripped in 1% SDS, 0.1 × SSC at 100°.
Southern blot:
One microgram of genomic DNA was digested with HaeII and run on a 0.8% agarose gel and blotted in 0.4 m NaOH to positively charged nylon membrane (Millipore). Hybridization conditions were as above except incubations were performed at 42°. Riboprobes were biotin-16-UTP T7 in vitro transcription products of XhoI digested pBS5′Z3 probe (Zorro probe) or the same tubulin probe as described above. Membranes were stripped in 0.4 m NaOH, 0.1% SDS.
RESULTS AND DISCUSSION
A budding yeast model for non-LTR retrotransposition:
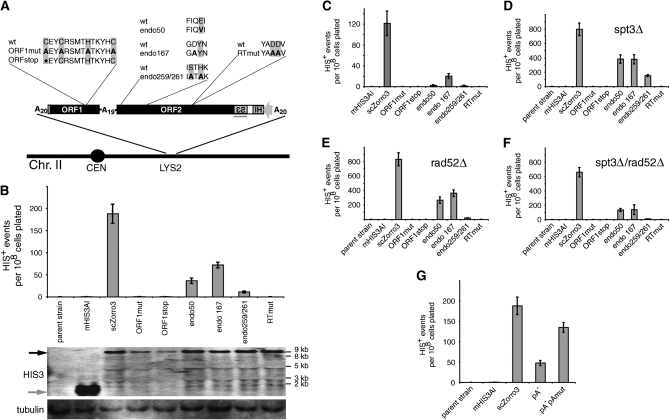
Because the genetic code of C. albicans differs from the universal genetic code, we redesigned and synthesized Zorro3 sequence, converting all CUG codons to UCU to generate a S. cerevisiae-compatible element, scZorro3 (GenBank accession no. EU597266). Placing the mHIS3AI cassette (Curcio and Garfinkel 1991) in the 3′-UTR allowed us to track retrotransposition from a plasmid (for a description of the assay, see Figure 1B). When placed on a high-copy plasmid and introduced into S. cerevisiae, this led to scZorro3-dependent HIS+ colony formation (Figure 1C), indicating that retrotransposition occurred. To examine scZorro3 retrotransposition from a more “natural” habitat, we integrated scZorro3mHIS3AI into chromosome II, along with mutants predicted to abolish the function of activities required for L1 retrotransposition (Figure 2A). Under the conditions of our assay, wild-type scZorro3 retrotransposed with a frequency of ∼2 × 10−6 events/cells plated (Figure 2B). Nonsense or missense mutations within the first zinc knuckle motif of ORF1 eliminated retrotransposition activity. Although these zinc knuckle motifs are not present in mammalian ORF1, they can be found in other members of the L1 clade (Schwarz-Sommer et al. 1987; Garrett et al. 1989; Leeton and Smyth 1993; Wright et al. 1996) and may represent the functional surrogate of the known RNA binding activity of human/mouse ORF1. Similarly, a reverse transcriptase missense mutation abolished retrotransposition. Three endonuclease mutations produced a significant reduction of scZorro3 activity, although not to the extent of the ORF1 and RT mutations. This could be due to “leaky” mutations that retain residual endonuclease activity, endonuclease-independent events that occur at preexisting chromosome nicks/breaks (Morrish et al. 2002), or a combination of both. scZorro3 RNA levels in ORF1/ORF2 mutants did not fluctuate in concordance with scZorro3 activity, suggesting that the defects are due to lack of ORF1 or ORF2 protein function (Figure 2B). These functional requirements for ORF1 and ORF2 imply that scZorro3 is using the same fundamental mechanism for retrotransposition as mammalian LINE elements. This is the case whether the donor element is on a plasmid or integrated in a chromosome (Figure 2C). Assays done with integrated scZorro3 have slightly higher overall retrotransposition frequencies as compared to the plasmid-based assay. This may reflect the difficulty in maintaining plasmids expressing high levels of retrotransposon proteins, even under selection (Boeke et al. 1985; Han and Boeke 2004).
Figure 2.—
Requirements for scZorro3 retrotransposition in S. cerevisiae. (A) Amino acid comparison of wild-type and mutant scZorro3's used in this study. These sequences were integrated into the LYS2 locus in B, D, E, F, and G. Thin bars represent the high and low range of experiments. (B) Retrotransposition of integrated Zorro3/marker variants in wt (GRF167) cells. Total RNA blot for the corresponding strains shown in the bottom sections. Solid and shaded arrows indicate expected positions of scZorro3 transcripts and mHIS3AI-only transcripts, respectively. The location of the HIS3 probe is shown as a shaded line in A. Tubulin, loading control. (C) 2μ plasmid-based retrotransposition in wt (GRF167) cells. (D–F) Retrotransposition of integrated Zorro3/marker variants in spt3Δ cells, rad52Δ cells, and spt3Δrad52Δ cells, respectively. (G) Effect of scZorro3 poly(A) tract deletion/mutation in wt cells. See supplemental Figure S3 for frequencies of individual experiments.
We considered other possible mechanisms of scZorro3 mobilization. Previous work had demonstrated that pseudogene formation in S. cerevisiae can occur via trans-mobilization of mRNAs mediated by Ty1, an active endogenous yeast retrotransposon (Derr et al. 1991; Dombroski et al. 1994). A possible role for Ty1 in these retrotransposition events was especially of concern because analysis of preexisting and de novo Zorro3 events in C. albicans indicate that the target sequence for Zorro3 integration is a poly(A) tract (Goodwin et al. 2007). Reverse transcription of full-length scZorro3 mRNA by Ty1 RT would generate a cDNA flanked by poly(A) tracts (see Figure 1A). This would presumably be a potential substrate for homologous recombination with poly(A) target sites in the genome. To examine this possibility we performed retrotransposition assays in spt3Δ (Figure 2D) and rad52Δ (Figure 2E) strains, which are defective in Ty1 activity (Winston et al. 1984) and homologous recombination (Symington 2002), respectively. scZorro3 activity was robust in these strains, as well as in an spt3Δrad52Δ double mutant (Figure 2F). All strains showed the same relative pattern of ORF1, endonuclease, and RT dependence when compared to wild-type background. Retrotransposition activities were globally increased in spt3Δ, rad52Δ, and spt3Δ/rad52Δ strains, which may be an indirect consequence of altered gene expression profiles and/or general phenotypic alterations in spt3 or rad52 strains (Eisenmann et al. 1992; Grant et al. 1997; Dudley et al. 1999; Larschan and Winston 2001; Bhaumik and Green 2002; Steinmetz et al. 2002; Wilson et al. 2002; Deutschbauer et al. 2005). In addition, it has recently been shown that rad52Δ strains have an increase in Ty1 mobility (Nyswaner et al. 2008)—a similar effect may be causing the increase in scZorro3 mobility.
To further rule out homologous recombination as a possible mechanism for scZorro3 mobilization, we created mutants of scZorro3 with the entire 5′ poly(A) tract deleted (pA−) or both the 5′ poly(A) tract deleted and the interORF poly(A) tract extensively mutagenized (pA−pAmut, see supplemental material). Although retrotransposition frequencies were reduced in comparison to wild-type scZorro3 (Figure 2G), both the pA− and pA−pAmut elements were nevertheless able to retrotranspose, suggesting that recombination via Zorro3-encoded poly(A) tracts is not essential for Zorro3 function, although the poly(A) tracts likely play a role in facilitating the TPRT reaction (see discussion below).
Cloning and analysis of scZorro3 retrotransposition events:
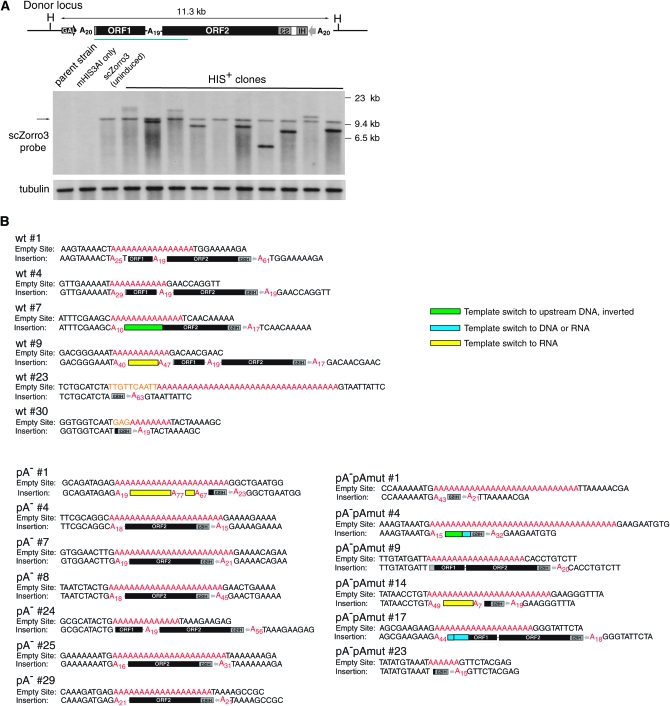
Southern analysis of 10 scZorro3-induced, stable histidine prototrophs showed in most instances a new Zorro3 insertion in addition to the original donor (Figure 3A). To verify that scZorro3 is undergoing L1-like retrotransposition to a new chromosome, we identified the insertion locations from 6 scZorro3, 7 scZorro3pA−, and 6 scZorro3pA−pAmut retrotransposition events by ligation-mediated PCR, then cloned and sequenced each (supplemental Fig S1). The structure of insertions revealed features consistent with non-LTR retrotransposition (Figure 3B and Table 1; for greater detail, see supplemental Figure S2). A variety of chromosomes were targeted. All insertions targeted a preexisting poly(A) tract (ranging from 8 to 36 bp in length), and reverse transcription started within the scZorro3 poly(A) tail, consistent with de novo events in C. albicans (Goodwin et al. 2007). Most insertions were 5′ truncated. There were also examples of untemplated nucleotides and genomic deletions, both of which have been well documented with mammalian L1 elements (Gilbert et al. 2002, 2005; Symer et al. 2002). Within these 20 insertions, as with all previously cloned insertions in C. albicans (Goodwin et al. 2007), there were no clear examples of target site duplications (TSDs). This could simply reflect the absence of TSD formation during Zorro3 retrotransposition. However, another plausible explanation is that during Zorro3 retrotransposition, both bottom and top strand cleavages occur within the target site poly(A) tract. This would generate a pure poly(A) TSD, extending the number of A's at the 5′ insertion junction. Because we do not know where the first and second strand cleavages occur, we cannot distinguish between these two possibilities on the basis of our sequence data alone. Further investigation of this question will likely require biochemical analysis to determine where the top and bottom strand cleavages occur.
Figure 3.—
Characterization of scZorro3 retrotransposition events in S. cerevisiae. (A) New scZorro3 insertions after galactose induction. Top: Donor scZorro3 integrated at the LYS2 locus in an spt3Δ/rad52Δ strain (JHY339). H, HaeII. Blue bar indicates the position of the Zorro3 probe. Bottom: Southern analysis of galactose-induced scZorro3 HIS+ clones. Total genomic DNA was digested with HaeII and probed with indicated probes. (B) General structures of empty sites and insertions of cloned scZorro3 retrotransposition events (summarized in Table 1). Poly(A) tracts are highlighted in red and the subscript represents the average poly(A) length when multiple clones were sequenced. Deleted sequences are highlighted in orange.
TABLE 1.
Characteristics of cloned de novo scZorro3, scZorro3pA−, and scZorro3pA−pAmut retrotransposition events
| Insertion | Chromosome | Poly(A) tail | 5′ Poly(A) tract | 5′ truncation | Microhomology at 5′ Zorro3 jxn | Inversion | Target-site deletion | Untemplated nucleotides | Additional template switch | TSD |
|---|---|---|---|---|---|---|---|---|---|---|
| wt no. 1 | V | Y | Y | Y | N | N | N | Y | N | CND |
| wr no. 4 | VII | Y | Y | N | Y | N | N | N | N | CND |
| wt no. 7 | III | Y | Y | Y | Y | Y (upstream gDNA) | N | N | Y | CND |
| wt no. 9 | X | Y | Y | N | N | N | N | Y | Y | CND |
| wt no. 23 | IV | Y | N | Y | Y | N | Y, 10 bp | N | N | N |
| wt no. 30 | II | Y | N | Y | Y | N | Y, 3 bp | N | N | N |
| pA- no. 1 | VIII | Y | Y | Y | Y | N | N | Y | Y | CND |
| pA- no. 4 | VIII | Y | Y | Y | Y | N | N | N | N | CND |
| pA- no. 7 | XV | Y | Y | Y | Y | N | N | N | N | CND |
| pA- no. 8 | IV | Y | Y | Y | Y | N | N | N | N | CND |
| pA- no. 24 | XIV | Y | N | Y | Y | N | N | N | N | N |
| pA- no. 25 | XIII | Y | Y | Y | Y | N | N | N | N | CND |
| pA- no. 29 | IV | Y | Y | Y | Y | N | N | N | N | CND |
| pA-pAmut no. 1 | XV | Y | Y | Y | N | N | N | N | N | CND |
| pA-pAmut no. 4 | I | Y | Y | Y | Y | Y (upstream gDNA) | N | N | Y | CND |
| pA-pAmut no. 9 | VII | Y | N | N | Y | N | N | N | N | N |
| pA-pAmut no. 14 | XIII | Y | Y | Y | N | N | N | Y | Y | CND |
| pA-pAmut no. 17 | III | Y | Y | Y | N | N | N | Y | Y | CND |
| pA-pAmut no. 23 | XIV | Y | N | Y | Y | N | N | N | N | N |
Junctions where a poly(A) tract of scZorro3 meets a poly(A) tract from the target site were scored “Y” for microhomology. Y, yes; N, no; CND, could not determine; TSD, target-site duplication.
Endonuclease independent retrotransposition events:
Human LINE elements have the ability to retrotranspose via an endonuclease(endo)-independent mechanism (Morrish et al. 2002). Features of human L1 endo-independent insertions include integration at atypical target sites, target-site deletions, incorporation of cellular cDNA sequences, and initiation of reverse transcription of L1 RNA internally. How these unusual features are produced is poorly understood, but they are believed to be the result of reverse transcription priming from naturally occurring DNA nicks/breaks in the chromosome, followed by resolution using host DNA repair pathways (Morrish et al. 2002). To determine if scZorro3 can undergo endo-independent retrotransposition, we determined the insertion locations from seven scZorro3endo50 retrotransposition events. PCR amplification and sequencing of 3′ junctions indicated that four of seven insertions occurred at long poly(A) tracts, ranging from 18- to 24-nucleotides long (Figure 4A). Since poly(A) tracts are the canonical Zorro3 target sites, we did not perform addition analysis on these four insertions on the assumption that they are conventional Zorro3 insertions. We further characterized the remaining three insertions by amplifying the 5′ junctions (Figure 4B). As might be predicted for endo-independent events, none of these three events integrated at long poly(A) tracts. Endo50 no. 3 and endo50 no. 10 had large (750 nucleotides and 181 nucleotides, respectively) TSDs. For L1 retrotransposition events generated in mammalian tissue culture, this would not be unusual. However, of the >50 naturally occurring or cloned de novo wild-type Zorro/scZorro3 integrations (this study and (Goodwin et al. 2007), there are no cases of obvious, long, TSDs composed of uniquely identifiable sequence (e.g. not poly(A) tract). Endo50 no. 5 contained a 101-nucleotide target-site deletion. These are commonly found in mammalian endo-independent retrotransposition events (Morrish et al. 2002). In endo50 no. 10, we also found an example of a 3′ truncated element, where reverse transcription appears to prime internally on the scZorro3 sequence, near the 5′-UTR. After reverse transcribing the 5′-UTR and 5′ poly(A) tract, the resultant poly(T) first strand cDNA appears to template jump to the poly(A) tract of scZorro3 (either the same RNA/DNA or a new RNA/DNA; see the next section for discussion on template switching). Internal priming of L1 elements is another hallmark of mammalian endo-independent L1 insertions (Morrish et al. 2002). Taken together, our data suggest that a large fraction of scZorro3endo50-derived retrotransposition events have features atypical of wild-type scZorro3-derived insertions, with some (but not all) features shared with mammalian endo-independent L1 insertions. The lack of perfect concordance between the features of human vs. yeast endo-independent insertions may be due to differences in the nature/distribution of endogenous nicks or double-strand breaks (the presumptive primers for endo-independent insertions) in yeast vs. mammalian cells, and/or differences in the host DNA repair pathways that resolve the retrotransposition intermediate. However, since scZorro3endo mutants give rise to integrations at noncanonical Zorro3 endonuclease target sites and have some features atypical for wild-type Zorro3 integrants, we propose that a fraction of scZorro3endo integration events represent endo-independent retrotransposition.
Figure 4.—
Characterization of scZorro3endo50 retrotransposition events. (A) Endo50 retrotransposition events at canonical Zorro3 target sites. 3′ junctions from each event were cloned and sequenced. Poly(A) tracts are highlighted in red and the subscript represents the average poly(A) length when multiple clones were sequenced. (B) Endo50 retrotransposition events at noncanonical target sites. 3′ and 5′ junctions for each event were cloned and sequenced. Purple nucleotides represent target-site duplications. Orange nucleotides represent target-site deletions. Underlined nucleotides indicate regions of temple switch microhomology. Bottom arrows and top arrows indicate possible sites of bottom and top strand cleavage, respectively.
Frequent template switching by scZorro3:
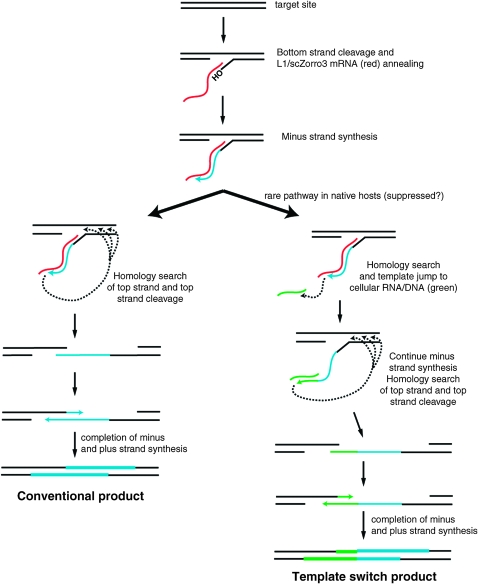
To resolve TPRT intermediates, L1 element minus strand synthesis must switch templates from L1 mRNA to the target chromosome insertion site (Figure 5). This often occurs at regions of microhomology (Symer et al. 2002; Zingler et al. 2005), and may be assisted by ORF1 (Martin and Bushman 2001) or host DNA repair pathways (Zingler et al. 2005). In some instances, minus strand synthesis can jump to a different template (DNA or RNA) other than the target site, generating deletions (Gilbert et al. 2002, 2005; Symer et al. 2002) or chimeras of L1 mRNA and another cellular RNA (Buzdin et al. 2002; Gilbert et al. 2005; Garcia-Perez et al. 2007). The former appears to occur with native Zorro3 elements in C. albicans, as 6/30 isolated C. albicans insertions are chimeras with preexisting Zorro3 elements in the genome (with the region between the target site and preexisting Zorro3 presumed to be deleted) (Goodwin et al. 2007). In vitro biochemistry with the Bombyx mori R2 non-LTR element reverse transcriptase has also directly shown template switching between RNAs, which is mediated by nontemplated nucleotide addition to the minus strand 3′ end to generate microhomology, followed by annealing and the continuation of minus strand synthesis (Bibillo and Eickbush 2004).
Figure 5.—
Model for L1/scZorro3 retrotransposition. During minus strand synthesis, the reverse transcription complex searches for sequences with homology to the minus strand to enable template jumping. In native hosts (e.g., humans, C. albicans), this search is largely restricted to regions around the target site (left pathway). In S. cerevisiae, this search space is relaxed and template jumps can occur to other RNAs/DNAs (green) at a higher frequency (right pathway). For simplicity, target-site deletions and 5′ inversions are omitted from this figure.
One striking characteristic of our isolated scZorro3 insertions is the high occurrence of additional template switches (Figure 3B, Table 1, and supplemental Figure S2) in addition to the standard template jumps to and from the target site DNA. These additional template jumps appear to occur on adjacent DNA, or cellular RNA that is fortuitously in proximity to the reverse transcription reaction. Since we examined events pre- and post-integration (as is the case for all mammalian L1 studies), we can only hypothesize whether the template switch was to DNA or RNA. Thus, although we classified the template switches on the basis of what we believe to be the most plausible mechanism, further investigation may alter these classifications in the future.
We classified template switches to sequences upstream of the target site on the same chromosome as jumps to DNA [wild-type (wt) no. 7 and pA−pAmut no. 4; see supplemental Figure S2]. In some instances (wt no. 9, pA− no. 1, and pA−pAmut no. 14) there was a poly(A) tract added at the template switch site between Zorro3 sequence (the template switch donor) and the template switch recipient sequence. This poly(A) tract was not encoded by Zorro3 sequence or the corresponding chromosomal sequence of the template switch recipient. The presence of a non-DNA-encoded poly(A) tract can be explained if the template switch was not to genomic DNA, but rather to a polyadenylated RNA. The existence of RNA corresponding to any particular segment of DNA in the yeast genome would not be surprising since the majority of the yeast genome is transcribed (David et al. 2006). In addition, RNA polyadenylation is not simply restricted to mRNAs, as polyadenylation is an important step in general RNA degradation mechanisms in yeast (Wyers et al. 2005). Alternatively, the non-DNA encoded poly(A) tracts could result from nontemplated addition of long poly(A) runs by the scZorro3 reverse transcriptase. We favor the former hypothesis since we are not aware of viral/retrotransposon reverse transcriptases that add such long (up to 89 nucleotide) nontemplated homopolymers. For this reason, we have classified the additional template switches of wt no. 9, pA− no. 1, and pA−pAmut no. 14 as template switches to RNA. Finally, we classified additional template switches to sequences on chromosomes different than the target site, and without a nontemplated poly(A) tract at the switch junction, as jumps to RNA or DNA (wt no. 7, pA−pAmut no. 4, and pA−pAmut no. 17). Although some of these recipient templates do not correspond to annotated RNAs, as mentioned above it is becoming increasingly apparent that a large portion of the yeast genome is transcribed as nonconventional or cryptic transcripts (David et al. 2006).
In some cases there are microhomologies at scZorro3 template switch sites (underlined in blue in supplemental Figure S2 and Figure 4). Where we do not see microhomology, we cannot differentiate between direct nonhomologous template jumping or nontemplated nucleotide addition to generate microhomology. This is also true for human L1 integrants in the human genome, where only 50–65% of 5′ truncated elements have homology between the 5′ truncated end and the insertion site (Zingler et al. 2005). The most striking instances of homology at template switch sites is when Zorro3 RT jumps from the target site to Zorro3 RNA during first strand synthesis initiation, and when Zorro3 RT jumps from the Zorro3 5′-UTR back to the target site near the end of first strand synthesis. Since the target sites are poly(A) tracts, these template-jump events are predicted to consist of poly(T) minus strand annealing to poly(A) recipient template, as described by the proposed model for Zorro3 mobilization (Goodwin et al. 2007). It is noteworthy that in our study, deleting the 5′ poly(A) tract of scZorro3 results in a preference for insertions to truncate at the interORF poly(A) tract (supplemental Figure S2). We propose that in these cases the long interORF poly(T) tract synthesized as the minus strand jumps to the poly(A) target site to end first strand synthesis. This is simply a variation of the proposed Zorro3 integration model (Goodwin et al. 2007). It is not clear why this tends to occur only when the 5′ poly(A) tract is absent. Perhaps the 5′ poly(A) tract of wt Zorro3 RNA is anchored to the poly(T) DNA at the target site, giving the 5′ end better access to make the jump to the target-site top strand. When the 5′ poly(A) tract is absent, the interORF poly(A) may be free to occupy this position. Further studies will be required to address this question.
Conclusions:
Overall, our data argue that we have used scZorro3 to faithfully recapitulate the non-LTR retrotransposition process in S. cerevisiae. It is interesting to note that there are two major differences between Zorro3/C. albicans retrotransposition and scZorro3/S. cerevisiae retrotransposition: Zorro3/C. albicans gives predominantly full-length insertions, and Zorro3/C. albicans does not frequently template switch to other cellular RNAs. We propose that during minus strand synthesis, the 3′ end of the minus strand can probe nearby sequences (RNA or DNA that is fortuitously in proximity to the reverse transcription complex) for homology to resolve the 5′ insertion junction (Figure 5). In C. albicans, Zorro3 DNA or the target site poly(A) are preferred substrates for this homology search, since all cloned insertions from C. albicans template jump back to the target site or to an already existing Zorro3 in the Candida genome (Goodwin et al. 2007). In S. cerevisiae, this preference appears to be relaxed considerably. This may reflect the presence of additional factor(s) in C. albicans that enhance the ability of Zorro3 to generate full-length copies and inhibit chimera formation with other RNAs (which would likely produce nonactive elements). Even if this is the case, such factors are not essential, since retrotransposition still occurs in our system. It is also possible that our alteration of Zorro3 sequence (scZorro3 is 1.6% divergent from the consensus Zorro3 sequence) affected RNA secondary structure and/or Zorro3 RNA-protein interactions, and this could account for some of the differences that we see.
The S. cerevisiae system we present here has striking similarities to models for mammalian L1 retrotransposition (Figure 5) and is probably using the same general L1 replication mechanism. This suggests that a common ancestor of S. cerevisiae had L1-related elements, and the cellular requirements for LINE-1-like retrotransposition have been conserved to the present day. Since S. cerevisiae at present does not normally contain LINE elements and therefore is not subjected to the consequent evolutionary pressure, we do not expect that all, or even most, of the regulatory mechanisms that control LINE elements in mammals will also be present in yeast. Thus it will be important to inquire whether Zorro3 interacting components and the roles they play in retrotransposition are conserved and play similar roles in mammals, since undoubtedly there will be species-specific differences uncovered upon further investigation. Alternatively, the ability of scZorro3 to mobilize in a non-LTR retrotransposon “naive” host such as S. cerevisiae also raises the intriguing (but unlikely) possibility that the ORF1/ORF2 TPRT reaction is largely autonomous and requires no contributions from the host.
In conclusion, our model system presents a unique opportunity to use the tremendous resources and speed of yeast genetics to dissect core LINE host components and possibly other regulatory pathways. The frequency of retrotransposition under the conditions of our assay (∼1 event in 106 cells) allows us to do quick qualitative assessments of relative retrotransposition activity on plates with patches of different yeast strains (that contain ∼108 cells/patch). This can be easily adapted for collections such as the yeast knockout collection (Giaever et al. 2002). Finally, this S. cerevisiae system gives us the opportunity to study the cell biology of a single copy of a non-LTR retrotransposition in a genomic landscape completely free of all other non-LTR retrotransposons, eliminating any potential interactions or dominant effects of other endogenous, wild-type elements.
Acknowledgments
We thank Jef D. Boeke for discussions and generous sharing of resources supported by National Institutes of Health grant CA16519; Wenfeng An, Jef Boeke, Don Brown, Joe Gall, Doug Koshland, Maxine Singer, and Allan Spradling for their review of the manuscript; the Koshland lab for discussions; Allison Pinder for sequencing; and Willie Garvin for miscellaneous assistance. This work was supported by the Carnegie Institution of Washington (J.S.H.).
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. EU597266.
References
- Amberg, D. C., D. J. Burke and J. N. Strathern, 2005. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Babushok, D. V., and H. H. Kazazian, Jr., 2007. Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 28 527–539. [DOI] [PubMed] [Google Scholar]
- Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute and C. Cullin, 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik, S. R., and M. R. Green, 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22 7365–7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibillo, A., and T. H. Eickbush, 2004. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J. Biol. Chem. 279 14945–14953. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., D. J. Garfinkel, C. A. Styles and G. R. Fink, 1985. Ty elements transpose through an RNA intermediate. Cell 40 491–500. [DOI] [PubMed] [Google Scholar]
- Buzdin, A., S. Ustyugova, E. Gogvadze, T. Vinogradova, Y. Lebedev et al., 2002. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics 80 402–406. [DOI] [PubMed] [Google Scholar]
- Carmell, M. A., A. Girard, H. J. van de Kant, D. Bourc'his, T. H. Bestor et al., 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12 503–514. [DOI] [PubMed] [Google Scholar]
- Casaregola, S., C. Neuveglise, E. Bon and C. Gaillardin, 2002. Ylli, a non-LTR retrotransposon L1 family in the dimorphic yeast Yarrowia lipolytica. Mol. Biol. Evol. 19 664–677. [DOI] [PubMed] [Google Scholar]
- Clements, A. P., and M. F. Singer, 1998. The human LINE-1 reverse transcriptase: effect of deletions outside the common reverse transcriptase domain. Nucleic Acids Res. 26 3528–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio, M. J., and D. J. Garfinkel, 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, L., W. Huber, M. Granovskaia, J. Toedling, C. J. Palm et al., 2006. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA 103 5320–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr, L. K., J. N. Strathern and D. J. Garfinkel, 1991. RNA-mediated recombination in S. cerevisiae. Cell 67 355–364. [DOI] [PubMed] [Google Scholar]
- Deutschbauer, A. M., D. F. Jaramillo, M. Proctor, J. Kumm, M. E. Hillenmeyer et al., 2005. Mechanisms of haploinsufficiency revealed by genomewide profiling in yeast. Genetics 169 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombroski, B. A., Q. Feng, S. L. Mathias, D. M. Sassaman, A. F. Scott et al., 1994. An in vivo assay for the reverse transcriptase of human retrotransposon L1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 14 4485–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, A. M., C. Rougeulle and F. Winston, 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy, A. J., S. Fritz and D. A. Largaespada, 2001. Transposition and gene disruption in the male germline of the mouse. Genesis 30 82–88. [DOI] [PubMed] [Google Scholar]
- Eickbush, T. H., and H. S. Malik, 2002. Origins and evolution of retrotransposons, pp. 1111–1144 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. ASM Press, Washington, DC.
- Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney and F. Winston, 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6 1319–1331. [DOI] [PubMed] [Google Scholar]
- Feng, Q., J. V. Moran, H. H. Kazazian, Jr. and J. D. Boeke, 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87 905–916. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez, J. L., A. J. Doucet, A. Bucheton, J. V. Moran and N. Gilbert, 2007. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 17 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, J. E., D. S. Knutzon and D. Carroll, 1989. Composite transposable elements in the Xenopus laevis genome. Mol. Cell. Biol. 9 3018–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387–391. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., S. Lutz-Prigge and J. V. Moran, 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110 315–325. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., S. Lutz, T. A. Morrish and J. V. Moran, 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25 7780–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, T. J., and R. T. Poulter, 2001. The diversity of retrotransposons in the yeast Cryptococcus neoformans. Yeast 18 865–880. [DOI] [PubMed] [Google Scholar]
- Goodwin, T. J., J. N. Busby and R. T. Poulter, 2007. A yeast model for target-primed (non-LTR) retrotransposition. BMC Genomics 8 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, T. J., J. E. Ormandy and R. T. Poulter, 2001. L1-like non-LTR retrotransposons in the yeast Candida albicans. Curr. Genet. 39 83–91. [DOI] [PubMed] [Google Scholar]
- Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell et al., 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11 1640–1650. [DOI] [PubMed] [Google Scholar]
- Han, J. S., and J. D. Boeke, 2004. A highly active synthetic mammalian retrotransposon. Nature 429 314–318. [DOI] [PubMed] [Google Scholar]
- Heidmann, T., O. Heidmann and J. F. Nicolas, 1988. An indicator gene to demonstrate intracellular transposition of defective retroviruses. Proc. Natl. Acad. Sci. USA 85 2219–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz, I., and R. E. Jensen, 1991. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 194 132–146. [DOI] [PubMed] [Google Scholar]
- Hohjoh, H., and M. F. Singer, 1996. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15 630–639. [PMC free article] [PubMed] [Google Scholar]
- Hohjoh, H., and M. F. Singer, 1997. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 16 6034–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian, Jr., H. H., C. Wong, H. Youssoufian, A. F. Scott, D. G. Phillips et al., 1988. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332 164–166. [DOI] [PubMed] [Google Scholar]
- Kidd, J. M., G. M. Cooper, W. F. Donahue, H. S. Hayden, N. Sampas et al., 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey, J. A., 1990. Tad, a LINE-like transposable element of Neurospora, can transpose between nuclei in heterokaryons. Genetics 126 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosha, V. O., and S. L. Martin, 2003. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1). J. Biol. Chem. 278 8112–8117. [DOI] [PubMed] [Google Scholar]
- Korbel, J. O., A. E. Urban, J. P. Affourtit, B. Godwin, F. Grubert et al., 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, S., M. C. Seleme, H. S. Soifer, J. L. Perez, J. V. Moran et al., 2006. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl. Acad. Sci. USA 103 8036–8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa, D. A., and J. V. Moran, 2005. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 14 3237–3248. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa, S., T. Watanabe, K. Gotoh, Y. Totoki, A. Toyoda et al., 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody et al., 2001. Initial sequencing and analysis of the human genome. Nature 409 860–921. [DOI] [PubMed] [Google Scholar]
- Larschan, E., and F. Winston, 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeton, P. R., and D. R. Smyth, 1993. An abundant LINE-like element amplified in the genome of Lilium speciosum. Mol. Gen. Genet. 237 97–104. [DOI] [PubMed] [Google Scholar]
- Luan, D. D., M. H. Korman, J. L. Jakubczak and T. H. Eickbush, 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72 595–605. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., W. D. Burke and T. H. Eickbush, 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16 793–805. [DOI] [PubMed] [Google Scholar]
- Martin, S. L., 1991. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 11 4804–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. L., and F. D. Bushman, 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE- 1 retrotransposon. Mol. Cell. Biol. 21 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. L., M. Cruceanu, D. Branciforte, P. Wai-Lun Li, S. C. Kwok et al., 2005. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J. Mol. Biol. 348 549–561. [DOI] [PubMed] [Google Scholar]
- Martin, S. L., J. Li, L. E. Epperson and B. Lieberman, 1998. Functional reverse transcriptases encoded by A-type mouse LINE-1: defining the minimal domain by deletion analysis. Gene 215 69–75. [DOI] [PubMed] [Google Scholar]
- Mathias, S. L., A. F. Scott, H. H. Kazazian, Jr., J. D. Boeke and A. Gabriel, 1991. Reverse transcriptase encoded by a human transposable element. Science 254 1808–1810. [DOI] [PubMed] [Google Scholar]
- Moran, J. V., and N. Gilbert, 2002. Mammalian LINE-1 retrotransposons and related elements, pp. 836–869 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. ASM Press, Washington, DC.
- Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke et al., 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87 917–927. [DOI] [PubMed] [Google Scholar]
- Morrish, T. A., N. Gilbert, J. S. Myers, B. J. Vincent, T. D. Stamato et al., 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31 159–165. [DOI] [PubMed] [Google Scholar]
- Naas, T. P., R. J. DeBerardinis, J. V. Moran, E. M. Ostertag, S. F. Kingsmore et al., 1998. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 17 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyswaner, K. M., M. A. Checkley, M. Yi, R. M. Stephens and D. J. Garfinkel, 2008. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178 197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., L. Leclercq, E. Gobel and H. Saedler, 1987. Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J. 6 3873–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper, S. F., G. W. van der Heijden, T. C. Hardiman, M. Goodheart, S. L. Martin et al., 2008. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev. Cell 15 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz, L. M., C. Scharfe, A. M. Deutschbauer, D. Mokranjac, Z. S. Herman et al., 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 31 400–404. [DOI] [PubMed] [Google Scholar]
- Symer, D. E., C. Connelly, S. T. Szak, E. M. Caputo, G. J. Cost et al., 2002. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110 327–338. [DOI] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S. C., B. Kim and A. Gabriel, 1996. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature 383 641–644. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wilson, W. A., Z. Wang and P. J. Roach, 2002. Systematic identification of the genes affecting glycogen storage in the yeast Saccharomyces cerevisiae: implication of the vacuole as a determinant of glycogen level. Mol. Cell Proteomics 1 232–242. [DOI] [PubMed] [Google Scholar]
- Winston, F., K. J. Durbin and G. R. Fink, 1984. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39 675–682. [DOI] [PubMed] [Google Scholar]
- Wright, D. A., N. Ke, J. Smalle, B. M. Hauge, H. M. Goodman et al., 1996. Multiple non-LTR retrotransposons in the genome of Arabidopsis thaliana. Genetics 142 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour et al., 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121 725–737. [DOI] [PubMed] [Google Scholar]
- Zingler, N., U. Willhoeft, H. P. Brose, V. Schoder, T. Jahns et al., 2005. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res. 15 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]