Abstract
spo16 mutants in yeast were reported to have reduced map lengths, a high frequency of nondisjunction in the first meiotic division, and essentially unchanged coefficients of coincidence. Were all crossing over in yeast subject to interference, such data would suggest that the “designation” of recombination events to become crossovers is separable from the “implementation” of that crossing over. In the presence of coexisting interference and noninterference phases of crossing over, however, lack of change in the coefficient of coincidence may show only that spo16 reduces crossing over in the two phases by a similar factor.
Be careful. Coefficient of coincidence is a slippery concept.
A. H. Sturtevant to a young geneticist
Shinohara et al. (2008) report on spo16 strains of yeast with meiotic linkage map distances that are 0.32–0.73 of the wild-type values. These mutant strains had approximately wild-type values for indicators of interference, which, like the three-factor coefficient of coincidence (C3), assess the effect of crossing over in one interval on crossing over in an adjacent interval.
Because Shinohara et al. (2008) detected no appreciable change in the indicators of interference accompanying the spo16-induced reduction in linkage distances, they concluded that spo16 affects the “implementation” of crossing over and, in so doing, eliminates any hypothetical “assurance” of at least one crossover, without disturbing the “designation” of sites that, in wild type, would have received a crossover. They point out that their observation is compatible with a “stress” model for interference. This assertion caught our interest because of the implied possibility that the observation might be incompatible with a counting model for interference (e.g., Foss et al. 1993).
In this note we first indicate a version of a counting model (aka chi-square, gamma, or Erlang model) that, too, is compatible with the observation of Shinohara et al. (2008), and we then discuss the possibility that their observation has little bearing on models for interference or on issues of designation and implementation.
The expectations for spo16 under a counting model:
The counting model proposed by Foss et al. (1993) hypothesized that double-strand breaks (DSBs) occur independently of each other and that there will (ordinarily) be a fixed number of noncrossovers between adjacent crossovers. In one development of the model, counting is achieved by “sweeping” adjacent DSBs, or precursor structures, into clusters of fixed size, with one particular position in the cluster designated to yield a crossover (Stahl 1993; Stahl et al. 2004). Below, we show that the lack of a spo16-induced phenotype with respect to indicators of interference is as expected of such a system, as long as the spo16-induced reduction in linkage distances represents random loss of designated crossovers.
For simplicity, our indicator of interference in this section is the factor by which the presence of a crossover in interval 1 reduces the map length of adjacent interval 2 from its value in the total population, i.e., (crossover frequency in interval 2 among crossovers in interval 1)/(crossover frequency in interval 2 in the total population). (Since, in the spo16 mutant, DSBs are not diminished and appear to be repaired, we presume that a fraction of the DSBs that, in wild type, would have been repaired as crossovers will, in the mutant, be repaired as noncrossovers or by intrachromosomal repair.) For a spo16 mutant eliminating about half the crossovers, a DSB that had been destined by position in the cluster (or some other mechanism) to give an interhomolog crossover now has a probability of about one-half of actually doing so—crossover designation occurs but implementation fails about half the time. As long as the distribution of such failures among designated DSBs is random, i.e., independent of the presence or absence of a crossover in interval 1, the mutation will reduce the map length of interval 2 to about one-half that of wild type both among recombinants for interval 1 and in the total population, leaving the measured coefficient-of-coincidence-like indicator unchanged.
We conclude that the observation made by Shinohara et al. (2008) is compatible with such a version of the counting model, as it is likely to be with any model in which designation and implementation are separable events. However, as described below, the result may be irrelevant to all models for interference as well as to the conclusion that spo16 cripples implementation without affecting designation.
Evidence of two crossover phases complicates the interpretation of the coefficient of coincidence:
The concept of two wild-type phases (or pathways) for crossing over was based on the oft-reported lack of interference among the crossovers remaining in the zmm mutants msh4 and msh5 (reviewed in Stahl et al. 2004). Zalevsky et al. (1999) proposed that these crossovers represent a class of crossovers in wild-type yeast that serve to promote chromosome pairing. That proposal has received its most direct support from the observation of a class of crossovers in wild-type yeast, identifiable by their frequent evasion of mismatch repair, that lack positive interference and have a frequency that is independent of Msh4 (Getz et al. 2008).
We now show how the coexistence of interference (disjunction) and noninterference (pairing) phases of crossing over makes it plausible that the absence of detectable spo16-induced change in the coefficient of coincidence shows nothing about designation and implementation.
In this demonstration, our operational indicator of interference is C3, the familiar coefficient of coincidence for adjacent intervals 1 and 2; i.e., (observed frequency of double crossovers)/(frequency expected on the assumption of independence),
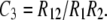 |
(1) |
Our algebra assumes linkage-map intervals that rarely involve more than one crossover, allowing us to approximate recombinant frequencies R1 and R2 with the probability of a single crossover. For each interval, R is the sum of the contributions from the two phases, RD and RP. To simplify the algebra, and with no serious loss of generality, we let the two intervals be of approximately equal length. Then, the frequency of double recombinants expected on the assumption of independence approaches (RD + RP)2. Expanding this binomial gives 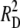 + 2RDRP +
+ 2RDRP + 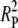 . These three terms describe three classes of double crossovers: one with both events derived from the disjunction phase, one with one event from each phase, and one with two events from the pairing phase, respectively.
. These three terms describe three classes of double crossovers: one with both events derived from the disjunction phase, one with one event from each phase, and one with two events from the pairing phase, respectively.
Because the two-pathway interpretation restricts interference to the disjunction phase [ignoring the possibility of negative interference between interfering and noninterfering crossovers (Getz et al. 2008)], the observed frequency of double crossovers becomes 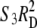 + 2RDRP +
+ 2RDRP + 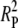 , where S3 is the coefficient of coincidence for double recombinants involving disjunction-phase crossovers in adjacent intervals, and the operational coefficient of coincidence is
, where S3 is the coefficient of coincidence for double recombinants involving disjunction-phase crossovers in adjacent intervals, and the operational coefficient of coincidence is
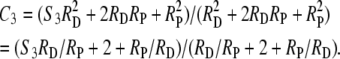 |
(2) |
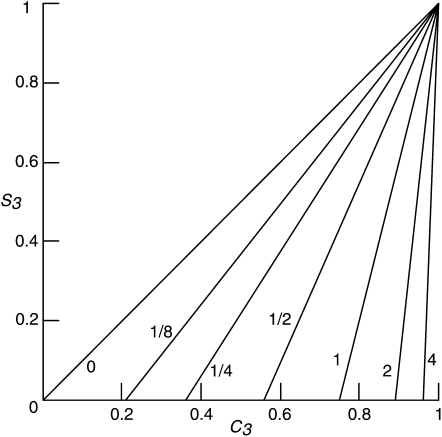
Since, for purely algebraic reasons, S3 can be expected to approach unity with increasing RD, experimental tests for interference appropriately involve intervals with small S3 values. Equation 2 implies that, when S3RD/RP is negligible compared to 2 + RP/RD, the value of S3 (the measure of interference between crossovers in the disjunction phase) is of little consequence for C3, so that an observed change, or lack of change, in C3 may be uninformative with respect to S3 and represent, instead, a change, or lack of change, in RP/RD. C3 vs. S3 is plotted in Figure 1 for select values of RP/RD.
Figure 1.—
S3 vs. C3 for select values of RP/RD, calculated from Equation 2, which assumes genetically short, adjacent intervals. RP/RD values are indicated by each curve.
Testing the data of Shinohara et al. (2008):
Whether it is legitimate to test the data presented in Tables S5 and S6 of Shinohara et al. (2008) for compatibility with C3 as presented in Equation 2 depends on the extent to which the experimental conditions match the hypothetical conditions stated above. A nearly inevitable difference between theory and experiment is that some of the interference data collected by Shinohara et al. (2008) involve intervals within which double exchanges, as assessed by nonparental ditype (NPD) tetrads, were detected, leading potentially to a data-based C3 value that is larger than the hypothetical value. A second potential issue is that the “C3's” calculated from the tetrad data by Shinohara et al. (2008) are not identical to the classical C3, which was defined for random meiotic products. However, they are easily related to C3, as shown below.
In Table S6, Shinohara et al. (2008) defined their indicator of interference as (observed frequency of tetrads that are not PD in either interval)/(frequency of tetrads not PD in interval 1)(frequency of tetrads not PD in interval 2), where PD means parental ditype tetrads. It is well known that this indicator, when applied to short distances under the presumption of no chromatid interference, is equivalent to C3 as written (Equation 1, and hence Equation 2). In Table S5, the authors used (map length in the presence of crossing over in an adjacent interval)/(map length in the absence of crossing over in the adjacent interval) as their indicator of interference, where map length is calculated according to Perkins (1949). This indicator is not equivalent to C3, but the tetrad data in that table may be used to calculate (map length in the presence of crossing over in an adjacent interval)/(map length in the total population). Since R and map length are equivalent for short intervals, this indicator, at short intervals, is equivalent to C3 in Equations 1 and 2. The C3 values in Table S6 are 0.51 and 0.8 for wild type and 0.55 and 0.65 for spo16, while those calculated from the data in Table S5 range from 0.54 to 0.85 for wild type and 0.31 to 0.65 for spo16. (The value of 0.55 for spo16 represents 3 observed double crossovers divided by 5.5 expected. Other values are more precise.)
The remaining obstacle to testing the possibility that the data of Shinohara et al. (2008) may reveal only the lack of spo16-induced change in RP/RD is the lack of means for determining values for S3, the “interesting” coefficient of coincidence. However, since Equation 2 was written for adjacent intervals within which double exchanges are rare, even in the face of some noninterfering crossovers, we may reasonably assume that S3 is essentially zero, as it is for short intervals in Drosophila (see Foss et al. 1993). When S3 = 0,
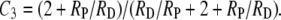 |
(3) |
Now we can ask whether the C3-like values, based on chromosome III data from Tables S5 and S6, are compatible with the S3-independent C3 values predicted by Equation 3. Values of the ratio of interfering to noninterfering crossovers, needed to evaluate Equation 3, may be obtained from data on zmm mutants such as msh4, and presumably zip1, where the density of residual crossovers reflects the density of noninterfering crossovers (Getz et al. 2008). According to the zip1 data of Shinohara et al. (2008) and the msh4 data of Stahl et al. (2004), the relative densities of the two types of crossovers on chromosome III appear to be close to 1/1, perhaps varying among intervals over a range from ∼1/3 to 3/1. The C3 values at these relative densities (Figure 1 at S3 = 0) indicate that, for both wild type and spo16, the C3 values reported by Shinohara et al. (2008), above, are compatible with values that are essentially independent of S3. Within the framework of our two-pathway analysis, this suggests that deletion of SPO16 reduced RD and RP on chromosome III by about the same factor.
Discussion:
In our first interpretation of the spo16 phenotypes, based on the assumption of a single recombination phase, we agreed with Shinohara et al. (2008) that the mutants apparently failed to implement some of the designated crossovers and proposed that such failures must have occurred in a random manner. As a result, the mutation certainly decreased the uniformity of intercrossover distances. Thus, even in a one-phase model, the lack of a significant spo16-induced change in the coefficient of coincidence reported by Shinohara et al. (2008) does not necessarily imply that the strength of interference in the mutant remains intact unless “interference” is redefined as designation, as indicated by Shinohara et al. (2008).
In the second example, we learned that the coexistence of an interference and a noninterference phase of recombination might render the coefficient of coincidence (C3) uninformative regarding strength of interference, especially when S3 is small. Under such conditions, C3 would reveal primarily the relative frequencies of interfering and noninterfering crossovers, as suggested by Kitani (1978), but nothing about designation or implementation. In fact, S3 might be reduced by the spo16 mutation, as would be expected for models (e.g., Foss et al. 1993) in which interference is determined by genetic linkage distance rather than physical distance, but the reduction need not register as a detectable change in C3.
The lack of a spo16-induced change in C3 for intervals on chromosome III suggests that the mutation lowered crossing over in the two phases of recombination by a comparable factor. (Because RP/RD is close to unity in these data, the underlying mechanism of the spo16 phenotype might be a reduction in number, rather than factor.) This phenotype differs from that of zmm mutants msh4 and msh5 (Getz et al. 2008) and, presumably, zip1, which are understood to lack disjunction-phase crossovers without suffering a loss of pairing-phase crossovers. A testable prediction of this view is that double mutants such as spo16 msh4 or spo16 zip1 would be reduced for crossing over to a somewhat greater degree than either single mutant. Shinohara et al. (2008) used isolated DNA to measure crossing over for the double mutant spo16 zip1 at the HIS-LEU construct and found a phenotype like that of zip1, in possible contradiction to our two-pathway analysis. However, quantification of the density of DNA bands in gels may lack the precision needed for that test. Furthermore, the sensitivity of yeast meiotic recombination to experimental conditions (Cotton et al. 2009) adds to the uncertainties of conclusions based on data derived from different strains sporulated under different regimes (Shinohara et al. 2008).
The Spo16 work has raised interesting questions that need more study (A. Shinohara, personal communication).
Acknowledgments
We are grateful to Akira Shinohara for helpful discussion. An “unidentified” referee contributed mightily to the usefulness of this note.
References
- Cotton, V. E., E. R. Hoffmann, M. F. F. Abdullah and R. H. Borts, 2009. The devil is in the details: the importance of genetic and environmental factors for yeast meiosis. Methods Mol. Biol. (in press). [DOI] [PubMed]
- Foss, E., R. Lande, F. W. Stahl and C. M. Steinberg, 1993. Chiasma interference as a function of genetic distance. Genetics 133 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz, T. J., S. A. Banse, L. S. Young, A. V. Banse, J. Swanson et al., 2008. Reduced mismatch repair of heteroduplexes reveals “non”-interfering crossing over in wild-type Saccharomyces cerevisiae. Genetics 178 1251–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani, Y., 1978. Absence of interference in association with gene conversion in Sordaria fimicola, and presence of interference in association with ordinary recombination. Genetics 89 467–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D. D., 1949. Biochemical mutants in the smut fungus Ustilago maydis. Genetics 34 607–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, M., S. D. Oh, N. Hunter and A. Shinohara, 2008. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40 299–309. [DOI] [PubMed] [Google Scholar]
- Stahl, F. W., 1993. Genetic recombination: thinking about it in phage and fungi, pp. 1–9 in The Chromosome, edited by J. S. Heslop-Harrison and R. B. Flavell. Bios, Oxford.
- Stahl, F. W., H. M. Foss, L. S. Young, R. H. Borts, M. F. F. Abdulla et al., 2004. Does crossover interference count in Saccharomyces cerevisiae? Genetics 168 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky, J., A. J. MacQueen, J. B. Duffy, K. J. Kemphues and A. M. Villeneuve, 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]