Abstract
Sister-chromatid separation at the metaphase–anaphase transition is regulated by a proteolytic cascade. Destruction of the securin Pds1p liberates the Esp1p separase, which ultimately targets the mitotic cohesin Mcd1p/Scc1p for destruction. Pds1p stabilization by the spindle or DNA damage checkpoints prevents sister-chromatid separation while mutants lacking PDS1 (pds1Δ) are temperature sensitive for growth due to elevated chromosome loss. This report examined the role of the budding yeast Pds1p in meiotic progression using genetic, cytological, and biochemical assays. Similar to its mitotic function, Pds1p destruction is required for metaphase I–anaphase I transition. However, even at the permissive temperature for growth, pds1Δ mutants arrest with prophase I spindle and nuclear characteristics. This arrest was partially suppressed by preventing recombination initiation or by inactivating a subset of recombination checkpoint components. Further studies revealed that Pds1p is required for recombination in both double-strand-break formation and synaptonemal complex assembly. Although deleting PDS1 did not affect the degradation of the meiotic cohesin Rec8p, Mcd1p was precociously destroyed as cells entered the meiotic program. This role is meiosis specific as Mcd1p destruction is not altered in vegetative pds1Δ cultures. These results define a previously undescribed role for Pds1p in cohesin maintenance, recombination, and meiotic progression.
MEIOSIS generates haploid gametes through a specialized cell division process that consists of one round of DNA replication followed by two nuclear divisions. The first meiotic division is unique to meiosis for two reasons. First, during the extended prophase I, homologous chromosomes synapse and undergo high levels of genetic recombination that is essential for the correct chromosome alignment at metaphase I (Kupiec et al. 1997). Second, following resolution of the recombination intermediates, the spindle makes monopolar attachments to the sister chromatids permitting the execution of meiosis I or the reductional division. Meiosis II resembles mitosis in that the replicated sister chromatids segregate to opposite poles.
Meiotic recombination establishes chromosome alignment essential for accurate segregation during the first meiotic division. It follows therefore that the first step in this process, i.e., the formation of double-strand breaks (DSBs), is also a critical event (Keeney et al. 1997). To date, in budding yeast, at least 10 proteins are required for this process (reviewed in Baudat and Keeney 2001; Arora et al. 2004; Borde 2007). Some of these proteins are meiosis specific whereas others also have roles in mitotically dividing cells. Significantly, apart from Spo11p, which initiates DSB formation (Keeney et al. 1997), little is known about the biochemical function of the individual components of this complex and how they are regulated.
The proper execution of recombination and other meiotic landmark events is governed by several checkpoint pathways (reviewed in Roeder 1997). The DNA damage checkpoint senses broken DNA ends and transduces the signal through the Rad9p kinase (Weber and Byers 1992; Lydall et al. 1996). The meiotic recombination checkpoint is more complex and can be divided into three different pathways depending on the signal that is generated (reviewed in Roeder and Bailis 2000; Hochwagen and Amon 2006). The rad50S checkpoint is triggered by unprocessed DSBs generated by the endonuclease Spo11p. The recombination (or dmc1) pathway is activated by resected, but not processed, DSB ends. Finally, the Zip1 checkpoint functions following strand invasion and is activated by an as-yet-undefined signal. Although the different checkpoint pathways monitor different steps in the recombination process, they share many components. For example, the various recombination DNA lesions are recognized by the Rad17-Ddc1-Mec3 clamp loader. However, different proteins are recruited depending on the checkpoint signal. For example, Tel1p is recruited by the rad50S complex but not the recombination pathway (Usui et al. 2001). Likewise, the chromosome structure proteins Red1p and Mek1p are not required for the DNA damage checkpoint but are involved in all three arms of the meiotic recombination checkpoint (reviewed in Hochwagen and Amon 2006). Mek1p is a meiotic kinase that, upon activation, phosphorylates Red1p, which in turn triggers a cascade of events that inhibits downstream effectors, including the Cdc28p cyclin-dependent kinase (Leu and Roeder 1999) and the transcription factor Ndt80p (Chu and Herskowitz 1998; Hepworth et al. 1998; Lindgren et al. 2000; Pak and Segall 2002). Ndt80p activates the “middle” set of meiotic genes that encode proteins necessary for establishment of the meiotic I spindle (Xu et al. 1995; Xie et al. 1999).
Cohesion between sister chromatids is essential for proper chromosome disjunction during meiosis (Revenkova et al. 2004). Dissolution of cohesion requires the ubiquitin ligase termed the anaphase-promoting complex/cyclosome (APC/C). The APC/C mediates the destruction of Pds1p (Cohen-Fix et al. 1996; Yamamoto et al. 1996a), thereby releasing Esp1p, which in turn triggers sister-chromatid separation by destroying the cohesin subunit Mcd1p/Scc1p (Visintin et al. 1997; Charles et al. 1998; Shirayama et al. 1998). Deleting PDS1 enables the cell to override the metaphase arrest imposed by apc mutations while a nondegradable form of Pds1p causes a metaphase arrest (Cohen-Fix et al. 1996; Yamamoto et al. 1996a). Upon entry into the meiotic program, Mcd1p provides connections between sister chromatids and helps establish domains for DSB initiation (Kateneva et al. 2005). These domains are accessible only for interhomolog recombination upon remodeling/removal of Mcd1p by Tid1p, a member of the SWI/SNF2 family of helicase-like chromatin-remodeling proteins (Zhang et al. 2005). Significantly, the removal of Mcd1p during meiotic prophase may be Esp1p independent (Kateneva et al. 2005) and, unlike its role during mitotic divisions (Guacci et al. 1997), is not essential for completion of the meiotic divisions (Klein et al. 1999). This nonessential role is probably in part due to the fact that sister-chromatid cohesion is maintained by Mcd1p's meiotic counterpart Rec8p (Klein et al. 1999; Watanabe and Nurse 1999; Buonomo et al. 2000). Similar to the mitotic strategy, Esp1p-mediated cleavage of Rec8p is required for sister separation in meiosis (Buonomo et al. 2000; Salah and Nasmyth 2000; Shonn et al. 2000), thereby making Rec8p essential for the proper completion of meiosis (Klein et al. 1999).
In addition to sequestering its protease activity, Pds1p also escorts Esp1p into the nucleus and to its final destination on the spindle (Jensen et al. 2001; Agarwal and Cohen-Fix 2002; Baskerville et al. 2008). Here Esp1 protease activity is required for spindle elongation independent of Mcd1p cleavage (Baskerville et al. 2008). Pds1p is also required for mitotic exit following DNA damage (Yamamoto et al. 1996b; Cohen-Fix and Koshland 1997; Sanchez et al. 1999; Hwang et al. 2001; Agarwal and Cohen-Fix 2002) and for double-strand break repair via single-strand annealing following radiation damage (Demase et al. 2005). Despite these multiple functions, Pds1p is not essential for mitotic cell division although pds1 cells are temperature sensitive for growth and exhibit high chromosome loss rates (Yamamoto et al. 1996a). In this study, we report that Pds1p is required for prophase I progression as mutants arrest prior to the nuclear divisions. This arrest is partially suppressed by eliminating DSB initiation, suggesting a role for Pds1p in recombination. Further studies revealed that Pds1p is indeed required for efficient DSB formation and recombination. Surprisingly, these phenotypes appear independent of Rec8p function. However, we observed premature cleavage of Mcd1p as pds1 cultures entering meiosis. This effect appears specific for meiotic cells as Mcd1p cleavage kinetics is not altered in vegetative cultures lacking PDS1 (Alexandru et al. 1999). These findings indicate a meiosis-specific role for Pds1p in preventing Mcd1p destruction prior to meiotic prophase.
MATERIALS AND METHODS
Strains, plasmids, and media:
The genotype, source, and background of the strains used in this study are listed in Table 1. Our wild-type parent strain (RSY335) is derived from SK1 and W303 parents. This strain sporulates to high levels similar to SK1 but does not enter meiosis prematurely. However, this strain background executes meiosis more slowly and asynchronously. The double-strand-break experiments and arg4 recombination measurements were performed in diploids KCY427 and KCY428, which were derived from haploid SK1 strains ORD7238-15C, ORD7238-26A, ORD7246-2B, and ORD7237-27A, respectively (a gift from A. Nicolas, Curie Institute, Paris). Diploids harboring the pds1Δ allele (KCY429 and KCY430) were generated by first deleting PDS1 in the haploid parents. Recombination at the his7 locus was monitored in strains RSY1343 and RSY1344 derived from REE223 and REE218 (a gift from R. E. Esposito, University of Chicago). The pds1Δ version of this strain was constructed by first deleting URA3 in REE223 using the marker swap plasmid M2660 (a gift from D. Stillman, University of Utah). Thereafter, PDS1 was deleted using pOC80 (a gift from O. Cohen-Fix, National Institutes of Health) in both haploids and crossed to form RSY1343. RSY1433 was made by using marker swap plasmids (pUL9 and pUT11; Cross 1997) to change pds1∷ura3 haploid strains to pds1∷ura3∷LEU2 and pds1∷ura3∷TRP1, respectively. RSY1433 was made by mating these strains. ndt80Δ (RSY1453 and RSY1456) and pch2Δ strains (RSY1536 and RSY1537) were made by integrating pTP89 or pSS53, respectively (gifts from S. Roeder, Yale University). The BUB2, MAD2, MEK1, SPO11, SPO13, and UBR1 disruptions were generated using oligonucleotide-directed recombination as described previously (Longtine et al. 1998). The RED1 disruption was made by transferring the red1∷KAN allele from the Research Genetics strain collection into our genetic background. Disruption of RAD17 or RAD9 was accomplished using pAAA19 or pAAA83, respectively (gifts from T. Weinert, University of Arizona). Successful integrations were verified by PCR analysis of genomic DNA. REC8-3HA strains were made as previously described (Shonn et al. 2002). The HA-tagged Pds1p expression construct pOC40 (CEN, URA3) was provided by O. Cohen-Fix. The PDS1-3FLAG expression plasmid was constructed by inserting the SalI/MfeI fragment from pCD11 into the SalI/EcoRI sites of YCplac111 (Gietz and Sugino 1988), forming pKC7000. Wild-type and a destruction box mutant of PDS1 were placed under the control of the AMA1 promotor by PCR amplification of the PDS1 ORF and subsequent substitution into the AMA1 genomic DNA (details available upon request) to form pMSC13. The PDS1-3HA destruction box mutation (RXXL → AXXA) contained in pVG279 under control of the AMA1 promotor and the CYC1 terminator in pRS426 (Christianson et al. 1992) formed pMSC14. The Rec8-HA high-copy plasmid (pKC7001) contains the REC8-3HA allele amplified from KCY447 inserted into YEplac181 (Gietz and Sugino 1988). The ESP1 high-copy plasmid (pKC7003) contains the KpnI/SacI fragment from pJN1 (ESP1-CEN plasmid, gift from Christine Pratt, Carnegie Institute, Baltimore) inserted into YEplac181. The ZIP1-GFP plasmid (Scherthan et al. 2007) was a gift from D. Kaback (UMDNJ). Yeast transformations were performed as described (Schiestl and Gietz 1989). Rich medium for yeast growth was either YPDA medium (2% dextrose, 2% peptone, 1% yeast extract supplemented with 10 mg/liter adenine or YPA [containing K acetate (1%) substituted for dextrose in YPDA]. Minimal medium for plasmid selection was either SD medium [0.17% yeast nitrogen base without amino acids, 0.5% ammonium sulfate, 2% dextrose, or synthetic acetate (K acetate 1% substituted for D-glucose, 1% phthalic acid, pH 5.5] supplemented with appropriate amino acid supplements. Liquid sporulation medium (SPM) for meiotic time courses contains 2% K acetate supplemented with uracil. Solid sporulation medium contains 2% K acetate, 0.1% dextrose, 0.25% yeast extract, and 1.5% agar. Both sporulation media were brought to pH 7.0 by addition of KOH.
TABLE 1.
Yeast strains used in this study
| Straina | Genotypeb | Background |
|---|---|---|
| RSY335a | MATa/MATα | SK1/W303 |
| RSY740a | MATa/MATα mad2∷TRP1 | SK1/W303 |
| RSY767a | MATa/MATα spo13∷TRP1 | SK1/W303 |
| RSY787a | MATa/MATα pds1∷URA3 | SK1/W303 |
| RSY795a | MATa/MATα pds1∷URA3 spo13∷TRP1 | SK1/W303 |
| RSY863a | MATa/MATα mad2∷TRP1 pds1∷URA3 rad17∷LEU2 | SK1/W303 |
| RSY864a | MATa/MATα mad2∷TRP1 pds1∷URA3 | SK1/W303 |
| RSY1343 | MATa/MATα ade1/ade2 cyh2/CYH2 his7-2/his7-1 leu1-12/leu1-c lys2-1/lys2-2 met13-d/met13-c trp5-d/trp5-c ura3/ura3∷LYS2 pds1∷URA3 | S288c |
| RSY1344 | MATa/MATα ade1/ade2 cyh2/CYH2 his7-2/his7-1 leu1-12/leu1-c lys2-1/lys2-2 met13-d/met13-c trp5-d/trp5-c ura3/ura3∷LYS2 | S288c |
| RSY1355a | MATa/MATα red1∷kanMX6 | SK1/W303 |
| RSY1356a | MATa/MATα mek1∷TRP1 | SK1/W303 |
| RSY1358a | MATa/MATα red1∷kanMX6 pds1∷URA3 | SK1/W303 |
| RSY1359a | MATa/MATα pds1∷URA3 mek1∷TRP1 | SK1/W303 |
| RSY1433a | MATa/MATα pds1∷ura3∷TRP1/pds1∷ura3∷LEU2 | SK1/W303 |
| RSY1453a | MATa/MATα ndt80∷LEU2/ndt80∷LEU2 pds1∷ura3∷TRP1/pds1∷URA3 | SK1/W303 |
| RSY1456a | MATa/MATα ndt80∷LEU2/ndt80∷LEU2 | SK1/W303 |
| RSY1536a | MATa/MATα pch2∷TRP1/pch2∷TRP1 | SK1/W303 |
| RSY1537a | MATa/MATα pds1∷ura3∷LEU2/pds1∷URA3 pch2∷TRP1/pch2∷TRP1 | SK1/W303 |
| KCY198a | MATa/MATα spo11∷kanMX6 | SK1/W303 |
| KCY207a | MATa/MATα spo11∷kanMX6 pds1∷URA3 | SK1/W303 |
| KCY257 | MATa/MATα his4/HIS4 leu2 trp1/TRP1 ura3 arg4(EV)/arg4(Bgl) | SK1 |
| KCY263a | MATa/MATα bub2∷TRP1 | SK1/W303 |
| KCY269a | MATa/MATα bub2∷TRP1 pds1∷URA3 | SK1/W303 |
| KCY274 | MATa/MATα his4/HIS4 leu2 trp1/TRP1 ura3 arg4(EV)/arg4(Bgl) pds1∷URA3 | SK1 |
| KCY345 | MATa/MATα leu2-3,112/LEU2 his3Δ200/HIS3 ura3-52 gal1/GAL1 | A364A |
| KCY348 | MATa/MATα leu2-3,112/LEU2 his3Δ200/HIS3 ura3-52 gal1/GAL1 pds1-1 | A364A |
| KCY385a | MATa/MATα rec8∷TRP1 | SK1/W303 |
| KCY392a | MATa/MATα REC8-3HA∷TRP1 | SK1/W303 |
| KCY398a | MATa/MATα rec8∷TRP1 spo11∷kanMX6 | SK1/W303 |
| KCY399a | MATa/MATα rec8∷TRP1 spo13∷TRP1 | SK1/W303 |
| KCY427 | MATa/MATα ura3 trp1 leu2 rad50S∷LEU2 | SK1 |
| KCY428 | MATa/MATα ura3 trp1 leu2 arg4(Nsp)/arg4(Bgl) | SK1 |
| KCY429 | MATa/MATα ura3 trp1 leu2 rad50S∷LEU2 pds1∷URA3 | SK1 |
| KCY430 | MATa/MATα ura3 trp1 leu2 arg4(Nsp)/arg4(Bgl) pds1∷URA3 | SK1 |
| KCY444a | MATa/MATα rad17∷LEU2 | SK1/W303 |
| KCY445a | MATa/MATα rad17∷LEU2 mad2∷TRP1 | SK1/W303 |
| KCY447a | MATa/MATα ubr1∷kanMX6 REC8-3HA∷TRP1 | SK1/W303 |
| KCY448a | MATa/MATα ubr1∷kanMX6 REC8-3HA∷TRP1 pds1∷URA3 | SK1/W303 |
| KCY392a | MATa/MATα REC8-3HA∷TRP1 | SK1/W303 |
| KCY472a | MATa/MATα REC8-3HA∷TRP1 pds1∷URA3 | SK1/W303 |
| KCY450a | MATa/MATα rad9∷LEU2 rad17∷LEU2 | SK1/W303 |
| KCY453a | MATa/MATα rad9∷LEU2 rad17∷LEU2 pds1∷URA3 | SK1/W303 |
Strain also harboring cyh2r-z ho∷LYS2 leu2∷hisG lys2 trp1∷hisG ura3 alleles.
All alleles listed are homozygous unless indicated otherwise. All strains were derived in this study except RSY335 (Cooper et al. 1997).
Meiotic time-course experiments:
Meiotic time-course experiments were conducted at 23° unless otherwise stated. Meiotic time-course experiments for Northern analysis, recombination assays (both physical and return-to-growth assays), chromatin pelleting, original pds1-1 and pds1Δ arrest characterization, and FACS analysis were conducted by growing cultures to 1 × 107 or 5 × 106 cells/ml for YPA and synthetic acetate cultures, respectively. Cells were harvested by centrifugation and washed in one-half culture volume sterile water. The resulting cell pellet was then resuspended in 1/5 original culture volume of SPM. Samples were collected by centrifugation and washed in 1/10 vol sterile water, and the resulting cell pellet was flash frozen in liquid nitrogen or in a dry ice/ethanol bath. Portions of each sample were removed prior to freezing and fixed in 70% ethanol [4′,6-diamidino-2-phenylindole (DAPI) staining] or 3.7% formaldehyde (tubulin indirect immunofluorescence) for nuclei and spindle visualization, respectively. Total RNA was prepared as previously described (Cooper et al. 1997) for Northern blot analysis. Western blot analysis was conducted with 100 μg total cell extract. Flag and HA-tagged proteins were visualized using α-Flag (M2 antibody, Sigma) and α-HA (Roche) mouse monoclonal antibodies, respectively, at a final concentration of 2 μg/ml concentration. Tub1p (tubulin) was detected using α-Tub1p rabbit polyclonal antibody at a final concentration of 0.1 μl/ml. Western blot signals were detected using goat anti-mouse secondary antibodies conjugated to alkaline phosphatase (Jackson ImmunoResearch Laboratories) and the CDP-Star chemiluminescence kit (Tropix, Bedford, MA). Blots were stripped and reprobed according to the manufacturer's instructions (Millipore).
Cellular analysis protocols:
Quantitation of meiosis I and II was achieved by analyzing DAPI-stained cells as described (Cooper and Strich 2002). At least 200 cells were counted per time point. For determining percentage sporulation of the various mutant strains, at least three independent isolates were sporulated as described above. After 24 hr at 23°, the cells were resuspended in 1 ml 70% ethanol for 1 hr, washed in 1 ml sterile water, and then stained with DAPI (0.05%) for 15 min. Cells were washed with water as before and examined by fluorescent microscopy. Cells were scored as follows: prophase I, spindle smaller than nucleus; metaphase I, spindle the same width as nucleus; pseudo-anaphase, spindle masses separate with elongated, but not separated, nuclei; fragmented nuclei, more than four DAPI-staining bodies, irregular-sized DAPI-staining bodies, and/or mis-oriented (more than two poles) spindle formation. The results presented are the mean of the three strains with standard deviation. FACS analysis was performed as previously described (Cooper et al. 2000). Chromatin pelleting was conducted as described (Guacci et al. 1997). Spindle morphology was determined using indirect immunofluorescence as previously described (Cooper et al. 2000). Likewise, indirect immunofluorescence was used to visualize the nucleolus using anti-Nop1 antibodies (Abcam). An Olympus PROVIS AX70 fluorescence microscope was used for all experiments except Zip1p localization where a Nikon eclipse 90i microscope was used. Z stacks were performed at 0.6-μm intervals using NIS elements software (Nikon). All microscopy is presented at ×1000 magnification except Figure 5B (×600).
Figure 5.—
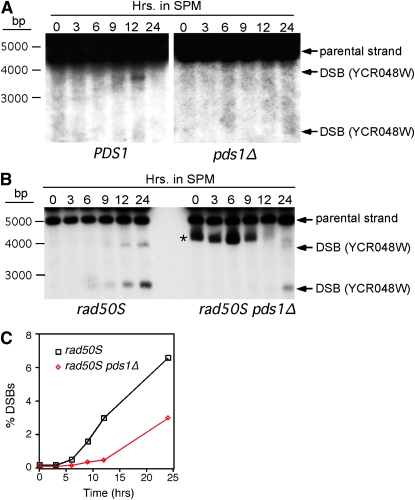
Pds1p is required for normal double-strand-break formation. (A) Physical detection of DSB at the YCR048W recombination hotspot in wild-type (KCY428) and pds1Δ (KCY430) SK1 derivatives. The strains were induced to enter meiosis at 23° and samples were taken at the time points indicated (in hours). DNA extracts were digested with AseI and probed with sequences specific to YCR048W as described (Borde et al. 2004). Arrows indicate predicted recombination–restriction enzyme double-strand-break fragments. The asterisk denotes a nonspecific cross-hybridizing band. (B) Same as in A except that rad50S (KCY427) and rad50S pds1Δ (KCY429) SKI strains were examined. (C) Graphic representation of quantified DSB percentages with respect to parental signal in B.
Determination of recombination frequencies:
Recombination frequencies in the return-to-growth assay were measured as described (Soustelle et al. 2002). Samples from the sporulation culture were taken at the times indicated in the text. The cells were lightly sonicated to disrupt clumps and directly counted by hemocytometer. Dilutions were made in sterile water and plated onto either complete minimal medium (to determine total viable cells plated) or minimal medium lacking arginine or histidine to identify recombinants. Recombination studies were done at least twice with two independent cultures each time. Plates were incubated for 4 days at 23° prior to cell count determinations. Only prototrophs observed above the 0-hr timepoint amount (indicative of mitotic recombination) were utilized in the calculations. Double-strand-break assays were performed essentially as described (Vedel and Nicolas 1999). Briefly, DNA was extracted in agarose plugs, digested with the appropriate restriction enzyme, separated on a 1% agarose gel, transferred to Hybond N membrane (Amersham), processed as described for Northern blot analysis (Cooper et al. 1997), and probed with sequences specific to YCR048W or CYS3. The YCR048W probe was made by amplifying wild-type DNA using PCR primers 5′-GGA TTT GTT GCA AGA CGA AG-3′ and 5′-TAA TGA TAC TGG GCC CTG AA-3′. The CYS3 probe was made by amplifying wild-type DNA using PCR primers 5′-GCT GAA TTC TTG GCA GAC AAG-3′ and 5′-TCT TAG GCA GGT AAT ACC TCG-3′.
RESULTS
Pds1p is required for progression through meiosis I:
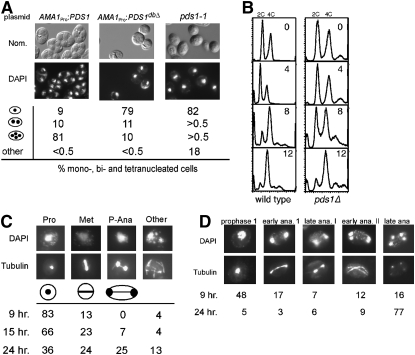
Previous studies have found that nondegradable Pds1p, when expressed from “early” meiotic gene promoters, induces a prophase I arrest (Shonn et al. 2000; Oelschlaegel et al. 2005). We confirmed these results by placing a nondegradable Pds1p mutant (PDS1dbΔ) under the control of the meiosis-specific AMA1 promoter (Cooper et al. 2000). Following 24 hr in liquid SPM, DAPI staining revealed that the wild-type diploid expressing AMA1pro-PDS1 exhibited an eightfold increase in tetra-nucleated cells compared with AMA1pro-PDS1dbΔ-expressing cells (Figure 1A). To further analyze the role of Pds1p during meiosis, we also followed meiotic progression in a pds1-1 strain. As pds1-1 cells are inviable at 30°, we performed these experiments at the permissive temperature for growth (23°). Surprisingly, the majority of the pds1-1 culture (82%) failed to execute either nuclear division compared to only 9% for the wild-type strain. Although normal bi- and tetra-nucleated cells were not observed (<0.5%), DAPI staining did reveal a significant number (18%) of cells demonstrating an abnormal fragmented nuclear morphology. These findings indicate that Pds1p is required for meiosis.
Figure 1.—
Pds1p regulates meiotic progression. (A) Pds1p destruction is required for meiotic progression. Wild-type culture (RSY335) harboring either wild-type or nondegradable Pds1p mutant (PDS1dbΔ) plasmids (pMSC13 and pMSC14, respectively) under the control of the meiosis-specific AMA1 promoter and pds1-1 cells (KCY348) were induced to enter meiosis at 23°. After 24 hr, nuclear divisions and spore formation were analyzed by DAPI staining (bottom) or Nomarski imaging (top), respectively. Percentages of the culture containing one, two or three-plus-four, or fragmented nuclei are indicated. Fragmented nuclei were scored as described in materials and methods. (B) Pds1p is not required for premeiotic S phase. Wild-type (RSY335) and pds1Δ (RSY787) diploid strains were induced to enter meiosis at 23° and samples were taken at the times indicated (in hours) and then analyzed by FACS. Peaks of 2C (unreplicated) and 4C (replicated) are indicated. (C and D) Terminal meiotic arrest phenotypes of pds1Δ mutants. Wild-type (RSY335, D) and pds1Δ (RSY787, C) diploid strains were induced to enter meiosis at 23° and samples were taken at the times indicated. Nuclear and spindle morphologies were determined by DAPI staining (top) and indirect immunofluorescence of tubulin (bottom). Cells were scored as described in materials and methods. For all morphology quantitations presented, the standard deviations were ≤6% for all values. Magnification is ×1000. (D) All strains used (except KCY348, which is derived from the A364A background) are isogenic to RSY335, our standard SK1/303 wild type.
We further characterized the requirement of Pds1p for meiosis by constructing a homozygous null mutant (pds1Δ) diploid in the RSY335 strain background (see Table 1). All meiotic time-course experiments were performed at the permissive temperature for vegetative growth (23°) unless otherwise indicated. Log-phase cultures were harvested, washed, and transferred to SPM. FACS analysis of samples taken at subsequent times following the shift revealed that pds1Δ cells completed meiotic S phase to the same extent as wild type, but did so with slower kinetics (Figure 1B). These findings indicate that the failure of pds1Δ mutants to execute the meiotic nuclear divisions was not due to a defect in meiotic entry.
To more precisely define the role of Pds1p in meiosis, spindle status of wild type and pds1Δ cultures following meiotic induction was determined by indirect immunofluorescence using Tub1p antibodies. The approach was the same as described for Figure 1B except the time course was extended to 24 hr. Following 9 hr in sporulation medium, 83% of the pds1Δ cells remained mononucleated with no recognizable spindle (Figure 1C). Most of the remaining pds1Δ cells (13%) contained short thick spindles diagnostic of metaphase I. Conversely, more than half of the wild-type culture had progressed past meiosis I or meiosis II by this time point (Figure 1D). The 15-hr time point revealed a reduction in cells arrested with a prophase spindle and an increase in the metaphase-like morphology. By 24 hr, 25% of the pds1Δ cells exhibited a morphology not normally observed during meiosis. These cells possessed a partially elongated spindle l containing a thin central region and elongated, but not segregated, nuclei. We classified this phenotype as pseudo-anaphase, since true anaphase I cells would have fully elongated spindles and completely separated DNA masses. The remaining 13% of pds1Δ cells exhibited multiple subnuclear-size DAPI-staining bodies with abnormal spindle formation, suggesting the presence of fragmented nuclei. None of the pds1Δ cells exhibited a recognizable spore wall, even following 36 hr in sporulation medium. Ninety-five percent of the wild-type population had undergone at least one nuclear division by 24 hr. A similar terminal phenotype was also observed with the pds1-1 homozygous diploid also at permissive temperature for growth (data not shown). The pds1-1 and pds1Δ strains were derived from A364A and SK1/W303 strains, respectively, demonstrating that this phenotype is not background specific.
The delayed appearance of the fragmented nuclei suggested two possibilities. They may represent nuclei that disintegrated in response to meiotic arrest. Alternatively, the fragmented nuclei may be the result of aberrant attempts to execute the meiotic divisions. To distinguish between these two models, we examined the terminal phenotype of ndt80Δ and ndt80Δ pds1Δ mutants. Ndt80p is a meiosis-specific transcription factor that is required for disassembly of the synaptonemal complex (SC) (reviewed in Zickler and Kleckner 1999). Importantly, ndt80 mutants arrest in late pachytene with synapsed chromosomes and are unable to enter into the meiotic divisions (Xu et al. 1995). As seen in supplemental Figure S1, ndt80Δ pds1Δ cells still display fragmented nuclei after 24 hr in sporulation medium, suggesting that the “fragmented nuclei phenotype” is due to deteriorating prophase I cells. To conclude, the pds1Δ fragmentation phenotype appears to be a secondary phenotype resulting from an extended prophase I arrest.
Double-strand-break formation is required for the pds1-dependent meiotic arrest:
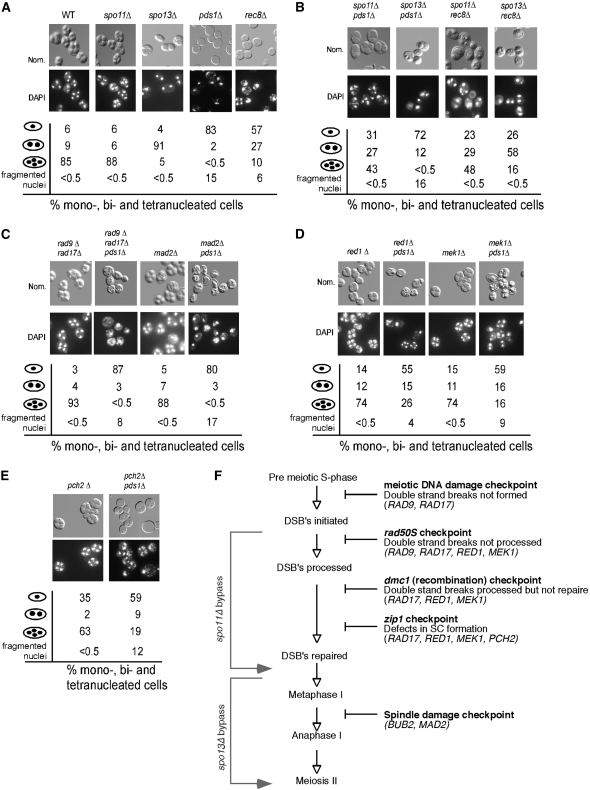
To better define the execution point of Pds1p during prophase I, we took advantage of previous suppressor bypass studies employing spo11 or spo13 mutations (see Figure 2F for summary). Spo13p prevents loss of centromere cohesion (Lee et al. 2004) and overexpression can prevent meiotic progression (McCarroll and Esposito 1994) Therefore, aberrant activation of Spo13p could lead to meiotic arrest. Mutants defective for Spo11p, the protein that initiates DSB formation (Keeney et al. 1997), bypass a meiotic arrest that occurs after recombination initiation (e.g., rad50S, mms4; Malone and Esposito 1981; Alani et al. 1990; De Los Santos et al. 2001). As mentioned above, pds1Δ cells arrest as both a mononucleated and a fragmentation phenotype, making the potential bypass scoring complicated. For clarification purposes, suppression was scored as the production of bi- or tetra-nucleated cells and spore wall assembly. Double-mutant pds1Δ spo11Δ and pds1Δ spo13Δ strains were constructed and their ability to complete meiosis and spore formation was tested (see materials and methods for details). After 24 hr in liquid sporulation medium, partial suppression was observed with 39% of the spo11Δ pds1Δ mutants completing both meiotic divisions and producing spores, indicating that DSB formation was required for at least a part of the pds1Δ-dependent arrest (Figure 2B). As expected, microdissection of these asci revealed that the spores were inviable, indicating that the requirement for recombination to allow normal meiosis I division is retained. A reduced, but reproducible suppression (12% vs. <0.5% dyads) was observed when SPO13 was deleted in the pds1Δ mutant background (Figure 2B). Taken together, the partial suppression by spo13 and spo11 alleles of the pds1Δ meiotic defect suggests that Pds1p might be involved in some aspect of recombination.
Figure 2.—
Recombination initiation is required for the pds1Δ-induced meiotic arrest. (A) The meiotic progression and spore-wall assembly of wild-type (RSY335), spo11Δ (KCY198), spo13Δ (RSY767), pds1Δ (RSY787), and rec8Δ (KCY385) strains were assayed after 24 hr at 23° by DAPI analysis (bottom) and Nomarski imaging (top), respectively. Population percentages of mono-, bi-, tri-, and tetra-nucleated cells as well as irregular nuclei (scored as fragmented nuclei) in each culture are indicated. (B) Same as in A except that spo11Δ pds1Δ (KCY207), spo13Δ pds1Δ (RSY795), spo11Δ rec8Δ (KCY398), and spo13Δ rec8Δ (KCY399) strains were assayed for nuclear division and spore formation. (C) Same as in A except that rad9Δ rad17Δ (KCY450), rad9Δ rad17Δ pds1Δ (KCY453), mad2Δ (RSY740), and mad2Δ pds1Δ (RSY864) strains were assayed for nuclear division and spore formation. (D) Same as in A except that red1Δ (RSY1355), red1Δ pds1Δ (RSY1358), mek1Δ (RSY1356), and mek1Δ pds1Δ (RSY1359) strains were examined. (E) Same as in A except that pch2Δ (RSY1536) and pch2Δ pds1Δ (RSY1537) strains were examined. For all morphology quantitations presented, the standard deviations were ≤9% for all values. Magnification is ×1000. All the strains used are isogenic to RSY335, the SK1/W303 parent. (F) Diagram depicting the meiotic checkpoints tested with the bypass function of the genes assayed indicated.
The pds1Δ prophase I arrest is partially dependent on the recombination checkpoint-signaling pathway:
Recombination-defect-induced meiotic arrest is mediated by three checkpoint pathways (rad50S, dmc1, and zip1) collectively referred to as the meiotic recombination checkpoint (see Figure 2F). These pathways are abrogated by inactivating genes that sense the damage signal (e.g., RAD17, RAD24, MEC1, DDC1, and MEC3) or by those mediating the arrest signal (e.g., RED1 and MEK1; reviewed in Hochwagen and Amon 2006). To determine if the pds1Δ-induced arrest requires the checkpoint pathways, bypass experiments were performed with RAD17, a gene common to all three pathways (see Figure 2F). Surprisingly, deleting RAD17 failed to suppress the pds1Δ arrest phenotype (data not shown). As this result was unexpected, we also determined whether the DNA damage checkpoint pathway was being evoked in pds1Δ cells (see Figure 2F). However, a rad9Δ null allele did not suppress the arrest, indicating that the DNA damage pathway was not involved (data not shown). As only a partial suppression was observed when SPO11 was deleted, this may suggest that multiple pathways are involved in the pds1Δ arrest. To test this possibility, a triple-mutant diploid (rad9Δ rad17Δ pds1Δ) was constructed. This strain also displayed no suppression (Figure 2C). Taken together, these results suggest that neither the DNA damage nor the recombination checkpoint pathway is required for the pds1Δ-dependent meiotic arrest.
We next asked if mutations in the genes that regulate or generate the arrest signal (RED1 or MEK1) would bypass the terminal meiotic pds1Δ phenotype. As mentioned above, these proteins perform a function different from that of RAD17, providing a framework for the activation of the recombination checkpoint. Deleting either RED1 or MEK1 in a pds1Δ strain still resulted in about one-half of the population arresting prior to meiosis I (Figure 2D). However, a significant fraction of the pds1Δ red1Δ or pds1Δ mek1Δ strains completed one or both divisions (41 and 32% bi- and tetra-nucleated cells, respectively) and formed spore walls. In addition, the production of fragmented nuclei in these double mutants was also reduced. Although the appearance of bi- and tetra-nucleated cells first occurred at a similar time in the red1 and red1 pds1 mutants (supplemental Figure S2), the accumulation of these cells was slower in the double mutant. This observation suggests that Pds1p has an additional role in meiotic progression independent of Red1p. A similar result was obtained when PCH2, a gene required for the meiotic arrest in cells with defective SC or lacking DMC1 (San-Segundo and Roeder 1999) was deleted in combination with the pds1Δ null allele (Figure 2E). To conclude, these data suggest that pds1Δ cells activate the recombination checkpoint. However, these results suggest that the pds1Δ-induced signal does not go through Rad17p as normally observed. Rather, this damage signal may contact Mek1p and Red1p more directly.
The pds1Δ prophase I arrest is not dependent on the spindle-checkpoint-signaling pathway:
Our initial characterizations of the pds1Δ arrest revealed abnormal spindle formations including a pseudo-anaphase configuration and fragmented nuclei. These findings suggested either a spindle defect or a problem with chromosome cohesion. To test whether the kinetochore or spindle checkpoint pathways were responsible for the pds1Δ-associated arrest, each checkpoint was inactivated by mutating BUB2 (Gardner and Burke 2000) or MAD2 (Shonn et al. 2000). However, neither double mutant executed the first division, indicating that these pathways were not solely responsible for the meiotic arrest (Figure 2C and data not shown). In addition, a pds1Δ rad17Δ mad2Δ triple mutant also arrested prior to meiosis I, suggesting that the recombination and spindle checkpoints were not performing redundant roles in mediating the pds1Δ arrest.
Pds1p is required for the transcription of mid–late meiotic genes:
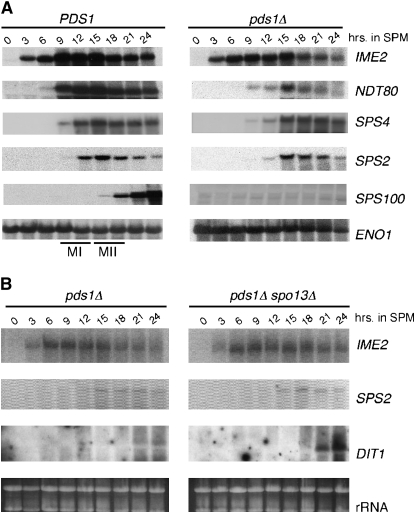
Our data indicate that the pds1Δ arrest is partially dependent on the signaling, but not sensing, components of the recombination checkpoint pathways. Activation of the recombination checkpoint results in meiotic arrest through inhibition of middle gene transcription (Roeder and Bailis 2000). One possible explanation of our results is that activation of Red1p and Mek1p in a pds1Δ mutant induces a similar loss in middle gene expression. To test this possibility, a meiotic time course with wild type and pds1Δ cultures was performed at 23° and total RNA preparations from the samples were subjected to Northern blot analysis. The blot was probed for transcripts from genes belonging to the “early” (IME2), “mid-early” (NDT80), “middle” (SPS4, SPS2), and “late” (SPS100) expression classes. This study revealed that the induction kinetics of the IME2 mRNA appeared similar in both cultures (Figure 3A) although the overall levels were reduced in the mutant strain. NDT80 transcript levels were more significantly reduced and peak levels were delayed in the pds1Δ mutant compared to wild type. Paradoxically, the transcript levels of SPS2 and SPS4, two middle genes whose transcription requires Ndt80p (Hepworth et al. 1998; Lindgren et al. 2000), are similar to wild type although a delay in their induction of ∼3 hr was observed. These results suggest that sufficient Ndt80p is being synthesized to promote nearly normal levels of middle gene expression. Consistent with our suppression studies, the reduction, but not elimination, of NDT80 transcription suggests that the pds1Δ arrest partially activates the recombination checkpoint.
Figure 3.—
The pds1Δ prophase arrest does not inhibit middle meiotic gene expression. (A) Wild-type (RSY335) and pds1Δ (RSY787) SK1/W303 cells were induced to enter meiosis at 23° and samples were taken at the times indicated (in hours). Total RNA was prepared and the transcript expression profiles of IME2, NDT80, SPS2, SPS4, and SPS100 were analyzed by Northern analysis. The approximate times of meiosis I (MI) and meiosis II (MII) as determined by DAPI analysis are indicated in the wild-type strain. ENO1 serves as a loading control. (B) Northern analysis of IME2, SPS2, and DIT1 mRNA expression in pds1Δ (RSY787) and pds1Δ spo13Δ (RSY795) SK1/W303 strains. (Bottom) Ethidium-bromide-stained rRNA is shown as a loading control.
Given the robust expression of the two middle genes in the pds1Δ mutant, it was surprising to find that the mRNA levels of the late gene SPS100 were below the limits of detection. It is unlikely that this absence is due to the delay observed in pds1Δ strains as SPS100 transcripts are present by 18 hr in the control. These results suggested two possibilities. First, Pds1p may be directly required for late gene expression. Alternatively, the expression loss could be an indirect effect of the meiotic arrest associated with the pds1Δ allele. To test these possibilities, a meiotic time course was performed with a pds1Δ spo13Δ strain that allows partial suppression of the pds1Δ arrest (see Figure 2C). In this experiment, the expression of early (IME2) and middle (SPS2) genes was identical in both strains. However, similarly to SPS100, DIT1, another late gene that is also required for spore-wall assembly (Briza et al. 1990), transcripts were not detectable in the pds1Δ mutant (Figure 3B). However, DIT1 was transcribed in the double-mutant strain. These findings suggest that the defect in late gene expression is an indirect effect associated with the pds1Δ-induced meiotic arrest.
Pds1p is required for normal meiotic recombination:
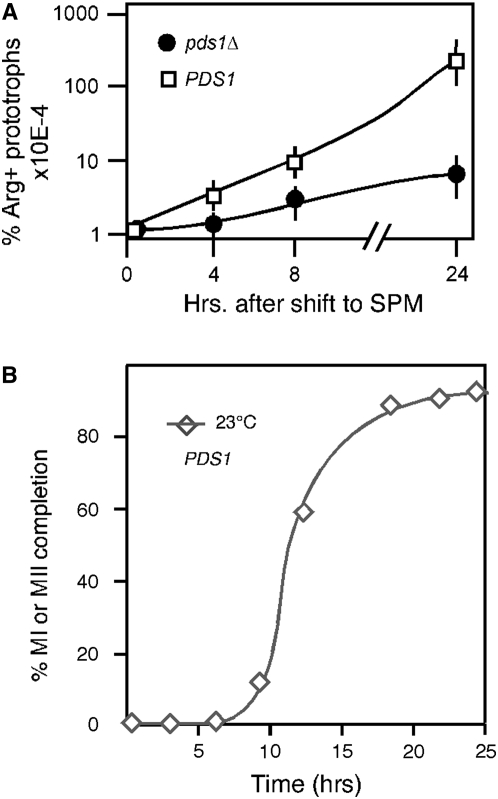
The ability of a spo11Δ mutation to suppress the pds1Δ meiotic arrest suggested a role for Pds1p in recombination. To test this possibility, heteroallelic recombination was measured in pds1Δ strains using return-to-growth assays (see materials and methods). Wild-type and pds1Δ SK1 strains containing arg4 heteroalleles (arg4-Erv and arg4-Ebg; Nicolas et al. 1989) were induced to enter meiosis at 23°. Samples, taken at several time points following the shift to SPM, were assayed for the presence of Arg+ prototrophs. The wild-type strain exhibited a 100-fold increase in recombination frequency 8 hr following shift to SPM, reaching a final level of 6.2% (Figure 4A). These values are similar but lower than those observed in other studies (10%) examining the same strain (Soustelle et al. 2002). This may be due to the different temperatures (23° vs. 30°) under which these experiments were conducted. Significantly, ∼100-fold fewer Arg+ prototrophs were observed for the pds1Δ diploid (Figure 4A). This decrease in recombination efficiency was not the result of decreased pds1Δ diploid viability over the course of the experiment as determined by return-to-growth studies (data not shown). The timing of recombination commitment occurs just prior to the appearance of bi- and tetra-nucleated cells observed in this strain at 23° (see KCY428; Figure 4B). To determine if the defect in recombination was also observed at another locus, recombination was also monitored using his7 heteroalleles in the S288C strain background. This locus gives a significantly lower recombination frequency in the wild-type strain (supplemental Table S1). Similar to our findings with recombination at the arg4 locus, the number of His+ prototrophs was reduced at least two orders of magnitude in the pds1Δ strain. These results indicate that Pds1p is required for normal levels of meiotic recombination.
Figure 4.—
Pds1p is required for meiotic recombination. (A) Three individual cultures of wild-type (KCY257) and pds1Δ (KCY274) SK1 strains containing heteroalleles at the arg4 locus were induced to enter meiosis at 23°. Samples were taken at the time points indicated following meiotic induction, serial diluted, and plated onto SD medium lacking arginine or complete SD medium. The resulting colonies were counted and the percentage of Arg+ prototrophs recovered was calculated (see supplemental Table S1). Plating dilutions for all experiments were adjusted on the basis of known recombination frequencies in wild-type strains at these loci. (B) Appearance of bi- and tetra-nucleated cells in wild-type cells in the SKI background at 23°.
Pds1p is required for normal double-strand-break formation:
To investigate the requirement of Pds1p for recombination, we first asked whether the mutant strain was able to form DSBs, the initiating step in this process. In wild-type diploid SK1, the PDS1 gene was deleted and the resulting wild-type and pds1Δ strains were induced to enter meiosis at 23°. Samples were collected at various times and total DNA was prepared. The appearance of DSBs was examined at the YCR048W locus (Liu et al. 1995; Baudat and Nicolas 1997) by Southern blot analysis (see materials and methods). In the wild type, the expected transient accumulation of DSBs was detected peaking in the 9- to 12-hr time point (Figure 5A). This is consistent with the commitment to recombination studies at this temperature. Significantly, little or no DSBs were detected in the pds1Δ mutant strain. This result is consistent with the ∼100-fold reduction in recombination frequency.
This very low level of DSBs in pds1Δ could be due to a decrease in DSB initiation, accelerated repair of the breaks, or both. To distinguish among these possibilities, we examined DSB accumulation in an SK1 strain harboring the rad50S allele that permits DSB accumulation but not its repair (Alani et al. 1990). In the rad50S strain, the appearance of the DSB-induced fragment was observed 6 hr after exposure to sporulation medium (Figure 5B) and reached an endpoint of 6.8% at 24 hr (Figure 5C), which is similar to previously reported values (Baudat and Nicolas 1997). However, DSB formation in the pds1Δ rad50S strain was at the limits of detection prior to the terminal time point with a final level of 3%. Identical results were also obtained if a different restriction enzyme was used to monitor the same DSB locus (supplemental Figure S3A) or at a different locus entirely (CYS3; supplemental Figure S3B). These results indicate that Pds1p is required for normal DSB formation.
Pds1p is required for synaptonemal complex formation:
The SC is a proteinacious structure that forms along homologous chromosomes following normal DSB initiation and processing (Roeder 1997). Therefore, if Pds1p plays a role in DSB formation, it would be predicted that pds1Δ strains would exhibit aberrant or no SC formation. To address this possibility, wild-type (RSY335) and pds1Δ (RSY1433) cells were transformed with a single-copy functional GFP-ZIP1 expression plasmid (Scherthan et al. 2007). Zip1, a component of the SC (Sym et al. 1993), is commonly used to monitor SC formation. Fluorescent microscopic analysis of GFP-Zip1p in live cells revealed the expected SC filament formation in wild-type cells (Figure 6A). However, a bright crescent-shaped signal was observed in the pds1Δ mutants. This crescent-shaped signal was further characterized using indirect immunofluorescence of fixed meiotic cells (Figure 6B). These studies revealed that GFP-Zip1p formed a crescent-shaped aggregate at the nuclear periphery but independently of the nucleolus as determined by Nop1p staining (Figure 6C). This GFP-Zip1p aggregate may be similar to the polycomplex described in spo11 mutants using chromosome spreads (Henderson and Keeney 2004). These data are consistent with a role for Pds1p in normal DSB formation and subsequent recombination.
Figure 6.—
Pds1p is required for normal SC formation. (A) Live-cell overlay images (GFP and Nomaski) of wild type (RSY335) and pds1Δ (RSY1433) strains expressing GFP-Zip1p 6 hr after transfer to SPM. The finger-like structure present in the wild-type cells represents individual chromosomes with formed SCs. (B) Fixed pds1Δ cells (RSY1433) harboring the GFP-ZIP1 expression plasmid 7.5 hr after transfer to SPM stained with DAPI (blue) examined under the fluorescent microscope. An expanded region indicated by the box is provided on the right (×600 magnification). (C) The cells described in B were subjected to indirect immunofluorescence staining for the nucleolar protein Nop1p. The images shown represent deconvoluted Z-stacks (0.6 μm slices) of Nomarski, GFP-Zip1p (green), Nop1p (red), and nuclei (blue). Magnification is ×1000.
Pds1p does not associate with the chromatin during recombination:
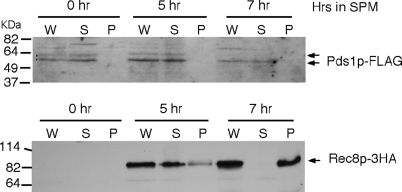
Previous studies in vegetative cells revealed that Pds1p regulates sister-chromatid cohesion indirectly via Esp1p activity (Koshland and Guacci 2000). The studies described above indicate a new role for Pds1p in DSB formation. To address whether Pds1p functioned at the chromatin or in an indirect manner, the presence of Pds1p was examined in chromatin pellets isolated through a sucrose cushion (see materials and methods for details). A wild-type culture (RSY335) harboring epitope-tagged alleles of PDS1 (PDS1-FLAG) and REC8 (REC8-3HA) was induced to enter meiosis with time points taken 5 and 7 hr following the switch to SPM. As this experiment was performed at 30°, these time points were chosen as 50% of this strain background (SK1/W303) completes the meiotic divisions by 9 hr (data not shown). Consistent with its meiosis-specific expression pattern, Rec8p-3HA was not detected in whole-cell extracts (“W” in Figure 7) at T = 0 but readily observed by 5 hr (Figure 7, bottom). Fractionation of this extract revealed that Rec8p-3HA was found primarily in the supernatant (“S” in Figure 7), indicating that it was not bound to the chromatin in the pellet (“P” in Figure 7). Similarly to proteins required for DSB formation (Kee et al. 2004) or components of the SC (Smith and Roeder 1997; Dong and Roeder 2000), Rec8p-3HA localized almost exclusively with the chromatin fraction by 7 hr. Conversely, Pds1p-FLAG was not associated with the chromatin during mitotic cell division (0 hr) or in either of the meiotic samples (Figure 7, top). These results indicate that Pds1p most likely plays an indirect role in promoting recombination.
Figure 7.—
Pds1p does not associate with chromatin during recombination. REC8-HA-tagged SK1/W303 strain (KCY392) harboring the Pds1p-FLAG expression plasmid (pKC7000) was grown to mid-log phase at 30° in synthetic acetate medium and then transferred to SPM and incubated at the same temperature. Samples were taken at the time points indicated (in hours) and whole-cell lysates (W) fractionated through a sucrose cushion to generate a chromatin bound pellet (P) and unbound supernatant (S) were analyzed by Western blotting and probed for the presence of Pds1p-FLAG (arrows, top) or Rec8p-HA (arrow, bottom). Molecular weight markers (in kilodaltons) are given on the left.
Rec8p cleavage is not altered in the pds1Δ mutant:
In meiotic cells, Pds1p protects the cohesin Rec8p from separase-dependent destruction, thus preventing the onset of anaphase I (Klein et al. 1999). Similarly to pds1 strains, rec8 mutants are also defective for recombination (Klein et al. 1999). Therefore, one possible explanation of our results is that deleting PDS1 causes premature cleavage of Rec8p. In this scenario, the pds1 and rec8 null alleles should share similar phenotypes. The analysis of a SK1/W303 rec8Δ diploid during meiosis revealed a similar, but not identical, phenotype compared to the pds1Δ strain. Although a similar level of mononucleated cells was found in the rec8Δ strain compared to the pds1Δ mutant, a significant percentage of the population was able to execute one or both meiotic divisions (Figure 2A). This finding is similar to that reported for rec8 mutants in a SK1 background (Klein et al. 1999). Similar to pds1Δ strains, mutating SPO11 suppressed the rec8 meiotic defect. However, a spo13 mutation significantly suppressed the rec8 phenotype compared to pds1Δ (58–12% binucleates). Taken together, these results indicate that pds1 and rec8 exhibit similar, but not identical, phenotypes.
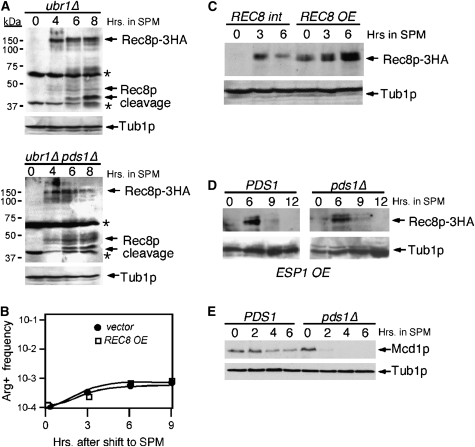
We next tested whether Rec8p cleavage was altered in meiotic cultures lacking Pds1p. The chromosomal allele of REC8 was tagged with three repeats of the HA epitope (see materials and methods). This allele was functional as determined by sporulation assays (data not shown). Rec8p-3HA cleavage was monitored in a strain background mutated for the ubiquitin ligase Ubr1p to allow accumulation of the cleaved intermediates (Buonomo et al. 2000). This strain, and the isogenic pds1Δ ubr1Δ double mutant, were induced to enter meiosis at 23° and time points taken for Western blot analysis. Rec8p-3HA accumulation was observed in both strains by 4 hr following the shift to SPM although the steady-state levels appear lower in the pds1Δ strain (Figure 8A). The appearance of the Rec8p cleavage products also occurred with similar timing (6 hr). Taken together, these studies indicate that Pds1p is not essential for proper Rec8p cleavage but may play a subtle role in its total accumulation (see below). To investigate this issue further, REC8 was placed on a high-copy plasmid and introduced into the pds1Δ SK1 strain used to measure recombination at the arg4 locus. The rationale for this experiment was that if the small reduction in Rec8p full-length levels in the pds1Δ mutant was sufficient to cause a recombination phenotype, this defect should be rescued by increasing gene dosage. However, no difference in recombination was observed with the pds1Δ strain harboring the REC8 high-copy plasmid (Figure 8B) even though Rec8p levels were increased above vector control levels (Figure 8C). These findings suggest that the requirement for Pds1p during recombination is not linked to the timing of Rec8p cleavage or its overall levels.
Figure 8.—
Mcd1p is cleaved prematurely in pds1Δ strains. (A) ubr1Δ REC8-3HA (KCY447) and ubr1Δ pds1Δ REC8-3HA (KCY448) SK1/W303 strains were induced to enter meiosis at 23° and samples were taken for Western analysis at the time points indicated (in hours). Full-length Rec8p-3HA and cleavage products are indicated by arrows. Asterisks indicate nonspecific cross-reactive bands observed with the HA antibody. The blot was stripped and reprobed for the presence of Tub1p for a loading control. (B) Return-to-growth recombination experiments as described in Figure 3 were performed on SK1/W303 pds1Δ culture (KCY274) at 23° harboring either Rec8p-HA expressed from a high-copy plasmid (pKC7001) or vector control (YEplac181). (C) Increasing REC8 gene dosage increases Rec8p production. Western blot analysis of Rec8p in a pds1Δ strain harboring either a single-copy (REC8 int) or a multi-copy (REC8 OE) HA-epitope-tagged REC8 allele following transfer to SPM medium as indicated (in hours). Tub1p serves as a loading control. (D) Increased gene dosage of ESP1 does not alter Rec8p degradation. Wild-type (RSY392) and pds1Δ (RSY448) SK1/W303 diploid strains containing the high-copy ESP1 plasmid (pKC7003) were induced to enter meiosis at 23° and samples were taken at the times indicated (in hours). Rec8p-3HA levels were monitored by Western blot analysis of total protein extracts. Tub1p serves as a loading control. (E) Mcd1p is prematurely destroyed in pds1Δ cells. Wild-type (RSY392) and pds1Δ (RSY448) SK1/W303 diploid strains were induced to enter meiosis at 23° and samples were taken for Western analysis at the time points indicated (in hours). Full-length Mcd1p was visualized by Western blot probing with antibodies directed against Mcd1p. The blot was stripped and reprobed for Tub1p for a loading control.
Pds1p also ensures proper localization of Esp1 to the nucleus and spindle in mitotic cells (Jensen et al. 2001). This latter role may also be critical as overexpression of ESP1 is able to partially bypass the temperature-sensitive growth phenotype associated with pds1-1 alleles (Ciosk et al. 1998). To address whether ESP1 overexpression can also relieve the meiotic requirement for Pds1p, a pds1Δ diploid (KCY472) containing either a high-copy ESP1 plasmid (pKC7003) or a control vector was induced to enter the meiotic program. Unlike the vegetative cultures, no differences in the terminal phenotype of pds1Δ cells were observed between the ESP1 overexpression construct and a vector control (data not shown) even at the permissive temperature (23°). Introduction of pKC7003 was able to suppress the pds1-1 vegetative lethality at restrictive temperature (supplemental Figure S3), indicating that our overexpression system was functioning as anticipated. Finally, we monitored Rec8p destruction in a wild-type and pds1Δ strain overexpressing Esp1p. No change in Rec8p degradation kinetics was observed in these two strains (Figure 8D) although in this experiment the kinetics of Rec8p cleavage was faster than experiments performed in the ubr1Δ background (Figure 8A). These data indicate that Esp1p-dependent destruction of Rec8p is not altered in a pds1Δ mutant and are consistent with a model that Pds1p performs a role during meiosis independently of Rec8p function.
Pds1p prevents the premature destruction of Mcd1p during meiosis:
During meiosis, it has been proposed that Mcd1p colocalizes with sites of DSB formation. These domains remain refractory for DSB initiation until Tid1p is recruited to these domains. This model suggests that the loading of Tid1p leads to the separation, rather than the alignment, of sister chromatids, thereby promoting interhomolog exchange (Kateneva et al. 2005). As Pds1p forms a complex with Esp1 and in turn Esp1 cleaves Mcd1p during mitotic cell division, the kinetics of Mcd1p cleavage was examined in wild-type and pds1Δ strains upon entry into the meiotic program. As previously described (Klein et al. 1999), Mcd1p levels are maintained in the wild-type culture until meiotic S phase at which point Rec8p is synthesized and loaded onto the chromosome (Figure 8E). However, in pds1Δ cells, Mcd1p is prematurely destroyed within 2 hr following shift to sporulation medium, approximately the start of meiS. These data indicate that Pds1p is required to maintain Mcd1p on the chromosomes as cells enter the meiotic program. Interestingly, this role occurs only in meiotic cells as Mcd1p levels are not affected in vegetative cells (T = 0) in these experiments.
DISCUSSION
This study reports new roles for Pds1p in meiotic progression and recombination. Similar to mitotic cells, Pds1p destruction is required for the metaphase–anaphase transition at meiosis I. However, mutants lacking Pds1p arrest in prophase I although extended exposure to sporulation medium produces cells with abnormal DNA masses and spindle formation. This meiotic defect is partially suppressed by preventing DSB formation or by inactivating the signaling portion of the recombination checkpoint system (Red1p and Mek1p). Mutants lacking Pds1p exhibit reduced DSB initiation, resulting in lower recombination efficiency and defective SC formation. The role of Pds1p in recombination is independent of its function prohibiting Rec8p cleavage as the expression profile of this meiotic cohesin appears normal. However, Pds1p prevents Mcd1p destruction as the cells enter meiosis. Significantly, this role is meiosis specific, as the kinetics of Mcd1p cleavage is not affected in pds1 during mitotic cell division (Alexandru et al. 1999). Taken together, these data indicate that Pds1p has additional functions during meiosis independent of its role as an anaphase inhibitor. In addition, these studies suggest an early role for chromosome cohesion for normal recombination and meiotic progression.
Our data indicate that Mcd1p is prematurely destroyed in a pds1 mutant background. However, this finding is unlikely to be the sole cause of all the pds1Δ-associated phenotypes as strains lacking Mcd1p/Scc1p execute both meiotic divisions (Klein et al. 1999). However, an important role has been reported in several species for Mcd1p and cohesion in DSB repair (Sonoda et al. 2001; Strom et al. 2004; Unal et al. 2004). Therefore, it is possible that the recombination defect observed in pds1Δ strains may be a consequence of early Mcd1p destruction. Upon entry into the meiotic program, Mcd1p not only maintains cohesion between newly replicated sister chromatids, but also has been hypothesized to serve as a guide for recruiting Tid1p onto chromosomes (Kateneva et al. 2005). In the absence of Pds1p, premature cleavage of Mcd1p may reduce or delay Tid1p loading onto chromosomes. As a consequence, Tid1p, and potentially Dmc1p, which has been shown to interact with Tid1p (Shinohara et al. 2000), may not be able to effectively promote homolog-directed repair. If this model is correct, then one prediction is that pds1 phenotypes would mimic those of tid1Δ strains. This is indeed the case with respect to some tid1Δ phenotypes. Both pds1 and tid1 strains arrest primarily as mononucleates that are partially suppressed by also deleting spo11 or spo13. In addition, recombination in tid1 mutants appears reduced and delayed, similar to the pds1 strain. However, many important differences exist. For example, tid1 mutants display normal SC assembly, which is lacking in the pds1 mutant. Therefore, Pds1p appears to have functions independent of Tid1p in directing meiotic recombination that may not involve Mcd1p regulation.
This model is consistent with an additional recombination phenotype associated with loss of Pds1p activity. We demonstrate that, in pds1Δ strains, DSBs are decreased only 2.5-fold whereas interhomolog exchange is decreased 100-fold. These findings suggest the possibility that DSBs are being repaired by sister chromatids rather than by homologs. In support of this idea is the observation that deletions of Red1p and Mek1p, two checkpoint proteins that prevent sister-chromatid exchange (reviewed in Hochwagen and Amon 2006), weakly bypass the requirement for Pds1p during meiosis. Previous studies have found that Dmc1p is required for the interhomolog bias observed for recombination (Schwacha and Kleckner 1997). However, we found that Dmc1p is able to load onto chromosomes with similar efficiency (supplemental Figure S5), suggesting that loss of homolog bias is not due to a defect in Dmc1p function. It is possible that Dmc1p does not load properly or in a productive manner in the pds1 mutant that would not be discerned by chromatin spreads. Further study into Dmc1p activity in the pds1 mutant strain would be required to distinguish between these models.
Pds1p is required for the normal transcript accumulation of the mid-early gene NDT80 and two loci (SPS100 and DIT1) that are expressed late in meiosis. Both of these results may be instructive for understanding the meiotic defect associated with loss of Pds1p activity. NDT80 transcription is under control of the recombination/DNA damage checkpoint pathways. When activated, these pathways inhibit NDT80 transcription, thus preventing meiotic progression. In the pds1Δ mutant, we observed a reduced, but not eliminated, NDT80 transcription. These findings may suggest that partial activation of the checkpoint pathway(s) are occurring. Although eliminating the checkpoint systems did not restore normal meiosis and sporulation to pds1 mutant strains, a partial rescue was observed (e.g., mek1 or red1 mutants). These results may suggest that Pds1p has multiple roles in meiosis that are manifested in a complex phenotype in the mutant strain. The requirement of Pds1p in late meiotic gene transcription was more surprising. Two models could explain this result. Either Pds1p is directly required for the transcription of this late expression class or the defect was due to the earlier meiotic arrest. The finding that late gene transcription was restored in the pds1 spo13 double mutant suggests that the latter explanation is more likely. If late gene transcription is dependent on the execution of earlier events, then a system that permits the execution of late events only after the successful completion of earlier ones must exist. As Pds1p is involved in several early processes, the nature of this signal is not clear. However, these results do suggest that the checkpoint pathways linking meiotic progression to spore morphogenesis may be more complex than previously appreciated.
Acknowledgments
We thank Orna Cohen-Fix, Phillip Hieter, David Kaback, Scott Keeney, Christine Pratt, Shirleen Roeder, David Stillman, Ted Weinert, Edward Winter, and Wolfgang Zachariae for strains, plasmids, and antibodies. We particularly thank Alain Nicolas for the haploid parents of strain KCY257 and KCY314 and Valerie Borde for help with the double-strand-break assays. This research was supported by American Cancer Society grant RSG03-249-01CCG to K.F.C., National Institutes of Health (NIH) grant CA099003 and March of Dimes grant 1-FY01-240 to R.S., and NIH grant GM62178 to V.G.
References
- Agarwal, R., and O. Cohen-Fix, 2002. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 16 1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani, E., R. Padmore and N. Kleckner, 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61 419–436. [DOI] [PubMed] [Google Scholar]
- Alexandru, G., W. Zachariae, A. Schleiffer and K. Nasmyth, 1999. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 18 2707–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, C., K. Kee, S. Maleki and S. Keeney, 2004. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13 549–559. [DOI] [PubMed] [Google Scholar]
- Baskerville, C., M. Segal and S. I. Reed, 2008. The protease activity of yeast separase (esp1) is required for anaphase spindle elongation independently of its role in cleavage of cohesin. Genetics 178 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat, F., and S. Keeney, 2001. Meiotic recombination: making and breaking go hand in hand. Curr. Biol. 11 R45–R48. [DOI] [PubMed] [Google Scholar]
- Baudat, F., and A. Nicolas, 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA 94(10): 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde, V., 2007. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 15 551–563. [DOI] [PubMed] [Google Scholar]
- Borde, V., W. Lin, E. Novikov, J. H. Petrini, M. Lichten et al., 2004. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell 13 389–401. [DOI] [PubMed] [Google Scholar]
- Briza, P., M. Breitenbach, A. Ellinger and J. Segall, 1990. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 4 1775–1789. [DOI] [PubMed] [Google Scholar]
- Buonomo, S. B., R. K. Clyne, J. Fuchs, J. Loidl, F. Uhlmann et al., 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103 387–398. [DOI] [PubMed] [Google Scholar]
- Charles, J. F., S. L. Jaspersen, R. L. Tinker-Kulberg, L. Hwang, A. Szidon et al., 1998. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 8 497–507. [DOI] [PubMed] [Google Scholar]
- Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero and P. Hieter, 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110 119–122. [DOI] [PubMed] [Google Scholar]
- Chu, S., and I. Herskowitz, 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1 685–696. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchenko, M. Mann et al., 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93 1067–1076. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix, O., and D. Koshland, 1997. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc. Natl. Acad. Sci. USA 94 14361–14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix, O., J. M. Peters, M. W. Kirschner and D. Koshland, 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10 3081–3093. [DOI] [PubMed] [Google Scholar]
- Cooper, K. F., and R. Strich, 2002. Saccharomyces cerevisiae C-type cyclin Ume3p/Srb11p is required for efficient induction and execution of meiotic development. Eukaryot. Cell 1 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, K. F., M. J. Mallory, J. S. Smith and R. Strich, 1997. Stress and developmental regulation of the yeast C-type cyclin UME3 (SRB11/SSN8). EMBO J. 16 4665–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, K. F., D. E. Egeland, M. J. Mallory, M. Jarnik and R. Strich, 2000. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl. Acad. Sci. USA 97 14548–14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R., 1997. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast 13 647–653. [DOI] [PubMed] [Google Scholar]
- de los Santos, T., J. Loidl, B. Larkin and N. M. Hollingsworth, 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMase, D., L. Zeng, C. Cera and M. Fasullo, 2005. The Saccharomyces cerevisiae PDS1 and RAD9 checkpoint genes control different DNA double-strand break repair pathways. DNA Repair 4 59–69. [DOI] [PubMed] [Google Scholar]
- Dong, H., and G. S. Roeder, 2000. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J. Cell Biol. 148 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, R. D., and D. J. Burke, 2000. The spindle checkpoint: two transitions, two pathways. Trends Cell Biol. 10(4): 154–158. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and A. Sugino, 1988. Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base-pair restriction sites. Gene 74 527–534. [DOI] [PubMed] [Google Scholar]
- Guacci, V., D. Koshland and A. Strunnikov, 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, K. A., and S. Keeney, 2004. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 101 4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth, S. R., H. Friesen and J. Segall, 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol Cell Biol 18 5750–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen, A., and A. Amon, 2006. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr. Biol. 16 R217–R228. [DOI] [PubMed] [Google Scholar]
- Hwang, M. S., Y. N. Yum, J. H. Joo, S. Kim, K. K. Lee et al., 2001. Inhibition of c-erbB-2 expression an activity in human ovarian carcinoma cells by hypericin. Anticancer Res. 21 2649–2655. [PubMed] [Google Scholar]
- Jensen, S., M. Segal, D. J. Clarke and S. I. Reed, 2001. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J. Cell Biol. 152 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kateneva, A. V., A. A. Konovchenko, V. Guacci and M. E. Dresser, 2005. Recombination protein Tid1p controls resolution of cohesin-dependent linkages in meiosis in Saccharomyces cerevisiae. J. Cell Biol. 171 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee, K., R. U. Protacio, C. Arora and S. Keeney, 2004. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J. 23 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375–384. [DOI] [PubMed] [Google Scholar]
- Klein, F., P. Mahr, M. Galova, S. B. Buonomo, C. Michaelis et al., 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98 91–103. [DOI] [PubMed] [Google Scholar]
- Koshland, D. E., and V. Guacci, 2000. Sister chromatid cohesion: the beginning of a long and beautiful relationship. Curr. Opin. Cell Biol. 12 297–301. [DOI] [PubMed] [Google Scholar]
- Kupiec, M., B. Byers, R. E. Esposito and A. P. Mitchell, 1997. Meiosis and sporulation in Saccharomyces cerevisiae, pp. 889–1036 in The Molecular and Cellular Biology of the Yeast Saccharomyces, edited by J. R. Pringle, J. R. Broach and E. W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Lee, B. H., B. M. Kiburz and A. Amon, 2004. Spo13 maintains centromeric cohesion and kinetochore coorientation during meiosis I. Curr. Biol. 14 2168–2182. [DOI] [PubMed] [Google Scholar]
- Leu, J. Y., and G. S. Roeder, 1999. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell 4 805–814. [DOI] [PubMed] [Google Scholar]
- Lindgren, A., D. Bungard, M. Pierce, J. Xie, A. Vershon et al., 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19 6489–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., T. C. Wu, M. Lichten and N. C. I. B. M. D. U. S. A. Laboratory of Biochemistry, 1995. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 14(18): 4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Lydall, D., Y. Nikolsky, D. K. Bishop and T. Weinert, 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383 840–843. [DOI] [PubMed] [Google Scholar]
- Malone, R. E., and R. E. Esposito, 1981. Recombinationless meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll, R. M., and R. E. Esposito, 1994. SPO13 negatively regulates the progression of meitotic and meiotic nuclear division in Sacchromyces cerevisiae. Genetics 138 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, A., D. Treco, N. P. Shultes and J. W. Szostak, 1989. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature 338 35–39. [DOI] [PubMed] [Google Scholar]
- Oelschlaegel, T., M. Schwickart, J. Matos, A. Bogdanova, A. Camasses et al., 2005. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell 120 773–788. [DOI] [PubMed] [Google Scholar]
- Pak, J., and J. Segall, 2002. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22 6417–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova, E., M. Eijpe, C. Heyting, C. A. Hodges, P. A. Hunt et al., 2004. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 6(6): 555–562. [DOI] [PubMed] [Google Scholar]
- Roeder, G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11 2600–2621. [DOI] [PubMed] [Google Scholar]
- Roeder, G. S., and J. M. Bailis, 2000. The pachytene checkpoint. Trends Genet. 16 395–403. [DOI] [PubMed] [Google Scholar]
- Salah, S. M., and K. Nasmyth, 2000. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma 109 27–34. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., J. Bachant, H. Wang, F. Hu, D. Liu et al., 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286 1166–1171. [DOI] [PubMed] [Google Scholar]
- San-Segundo, P. A., and G. S. Roeder, 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97 313–324. [DOI] [PubMed] [Google Scholar]
- Scherthan, H., H. Wang, C. Adelfalk, E. J. White, C. Cowan et al., 2007. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 104 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl, R. H., and R. D. Gietz, 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16 339–346. [DOI] [PubMed] [Google Scholar]
- Schwacha, A., and N. Kleckner, 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90 1123–1135. [DOI] [PubMed] [Google Scholar]
- Shinohara, M., S. L. Gasior, D. K. Bishop and A. Shinohara, 2000. Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. USA 97 10814–10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama, M., W. Zachariae, R. Ciosk and K. Nasmyth, 1998. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonn, M. A., R. McCarroll and A. W. Murray, 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289 300–303. [DOI] [PubMed] [Google Scholar]
- Shonn, M. A., R. McCarroll and A. W. Murray, 2002. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev. 16 1659–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A. V., and G. S. Roeder, 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 136(5): 957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda, E., C. Morrison, Y. M. Yamashita, M. Takata and S. Takeda, 2001. Reverse genetic studies of homologous DNA recombination using the chicken B-lymphocyte line, DT40. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soustelle, C., M. Vedel, R. Kolodner and A. Nicolas, 2002. Replication protein A is required for meiotic recombination in Saccharomyces cerevisiae. Genetics 161 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom, L., H. B. Lindroos, K. Shirahige and C. Sjogren, 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16 1003–1015. [DOI] [PubMed] [Google Scholar]
- Sym, M., J. A. Engebrecht and G. S. Roeder, 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72 365–378. [DOI] [PubMed] [Google Scholar]
- Unal, E., A. Arbel-Eden, U. Sattler, R. Shroff, M. Lichten et al., 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16 991–1002. [DOI] [PubMed] [Google Scholar]
- Usui, T., H. Ogawa and J. H. Petrini, 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7 1255–1266. [DOI] [PubMed] [Google Scholar]
- Vedel, M., and A. Nicolas, 1999. CYS3, a hotspot of meiotic recombination in Saccharomyceces cerevisiae: effects of heterozygosity and mismatch repair functions on gene conversion and recombination intermediates. Genetics 151 1245–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., S. Prinz and A. Amon, 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278 460–463. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., and P. Nurse, 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400 461–464. [DOI] [PubMed] [Google Scholar]
- Weber, L., and B. Byers, 1992. A RAD9-dependent checkpoint blocks meiosis of cdc13 yeast cells. Genetics 131 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter et al., 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18 6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., M. Ajimura, R. Padmore, C. Klein and N. Kleckner, 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., V. Guacci and D. Koshland, 1996. a Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J. Cell Biol. 133 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., V. Guacci and D. Koshland, 1996. b Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J. Cell Biol. 133 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Q. Ren, H. Yang, M. N. Conrad, V. Guacci et al., 2005. Budding yeast PDS5 plays an important role in meiosis and is required for sister chromatid cohesion. Mol. Microbiol. 56 670–680. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner, 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33 603–754. [DOI] [PubMed] [Google Scholar]