Abstract
The control of mRNA degradation and translation are important for the regulation of gene expression. mRNA degradation is often initiated by deadenylation, which leads to decapping and 5′–3′ decay. In the budding yeast Saccharomyces cerevisae, decapping is promoted by the Dhh1 and Pat1 proteins, which appear to both inhibit translation initiation and promote decapping. To understand the function of these factors, we identified the ribosome binding protein Stm1 as a multicopy suppressor of the temperature sensitivity of the pat1Δ strain. Stm1 loss-of-function alleles and overexpression strains show several genetic interactions with Pat1 and Dhh1 alleles in a manner consistent with Stm1 working upstream of Dhh1 to promote Dhh1 function. Consistent with Stm1 affecting Dhh1 function, stm1Δ strains are defective in the degradation of the EDC1 and COX17 mRNAs, whose decay is strongly affected by the loss of Dhh1. These results identify Stm1 as an additional component of the mRNA degradation machinery and suggest a possible connection of mRNA decapping to ribosome function.
CONTROL of mRNA translation and degradation are important points of regulation of eukaryotic gene expression. In eukaryotic cells there are two general mechanisms for the degradation of mRNAs, both of which initiate with deadenylation, leading either to 3′–5′ exonucleolytic degradation or to decapping followed by 5′–3′ exonucleolytic destruction of the mRNA (reviewed in Parker and Song 2004; Garneau et al. 2007; Shyu et al. 2008). In Saccharomyces cerevisiae, the major pathway of mRNA decay involves decapping followed by 5′–3′ decay (Coller and Parker 2004), with removal of the poly(A) tail predominantly promoted by the Ccr4/Pop2/Not deadenylase complex (Decker and Parker 1993; Muhlrad et al. 1994; Tucker et al. 2001). The 5′ m7G cap is then removed by the Dcp1/2 decapping enzyme and 5′–3′ decay is performed by the exonuclease Xrn1 (Hsu and Stevens 1993; Beelman et al. 1996; Dunckley and Parker 1999).
Decapping is a critical step in this decay pathway as it permits destruction of the mRNA and is a site of numerous control inputs. Moreover, many observations indicate that decapping and translation in yeast cells are intertwined processes that are often in competition. For example, when mRNAs are maintained in association with ribosomes by the inhibition of translation elongation using cycloheximide, the rate of decapping is reduced (Beelman and Parker 1994). Conversely, mRNAs poorly translated because of cis elements, such as secondary structures in the 5′ untranslated region or a poor AUG context, are decapped faster than their well-translated counterparts (Muhlrad et al. 1995; Lagrandeur and Parker 1999). Moreover, mutation of initiation factors such as eIF-4E, the cap binding protein, or Prt1 (part of the eIF3 complex) lead to faster degradation of mRNAs (Schwartz and Parker 2000). Consistent with this competition, eIF-4E has been shown to inhibit the decapping enzyme in vitro (Schwartz and Parker 1999). Thus, a key step in mRNA decapping is exchanging translation initiation factors for the mRNA decapping machinery.
The balance between translation and decay also correlates with the type of mRNP formed and its subcellular localization. When mRNAs exit translation, they form nontranslating mRNPs, which can undergo decapping and degradation and/or accumulate in cytoplasmic foci referred to as P-bodies (Sheth and Parker 2003). P-bodies are cytoplasmic foci that accumulate translationally repressed mRNA along with the decay machinery and translational repressors (reviewed in Eulalio et al. 2007; Parker and Sheth 2007). Analyses of P-bodies provide additional evidence for an inverse relationship between translation and formation of mRNPs capable of mRNA decapping. For example, blocking translation initiation using mutations in initiation factors leads to an increase in the P-body size and number along with accelerated decay rates. Conversely, inhibition of translation elongation and trapping of the mRNAs in polysomes lead to the loss of P-bodies (Sheth and Parker 2003; Teixeira et al. 2005).
An important question is the mechanism by which mRNAs cease translation initiation and form nontranslating mRNPs capable of decapping and accumulation in P-bodies. In yeast, the Dhh1 and Pat1 proteins appear to be involved in this transition from translation to the nontranslating mRNP (Coller and Parker 2005). Dhh1 and Pat1 appear to act, at least partially, independently of each other. Strains lacking either Dhh1 or Pat1 show reductions in decapping rates, while strains lacking both proteins are severely blocked for decapping (Coller and Parker 2005). Moreover, overexpression of either Dhh1 or Pat1 causes global translational repression, as seen by a decrease in polysomes and an increase in size and number of P-bodies in a manner independent of each other (Coller and Parker 2005). Finally, Dhh1 has been shown to directly repress translation in vitro (Coller and Parker 2005).
Although these general translation repressors have been identified, much remains to be understood about their mode of action. One major unresolved issue is understanding how Dhh1 and Pat1 interact with the translation machinery to promote translation repression and/or target mRNAs for decapping. We have approached this issue by using genetic methods to try to find proteins that could link Dhh1 and/or Pat1 to the translation machinery. In this work we identified Stm1 as a high-copy suppressor of the temperature-sensitive growth defect of the pat1Δ strain. Stm1 has been shown to associate with ribosomes (Van Dyke et al. 2004, 2006) and was initially identified as a suppressor of Tom1, which has a role in the export of messenger RNAs from the nucleus (Utsugi et al. 1995). In this study we show that Stm1 has genetic interactions with Pat1 and Dhh1, affects the accumulation of Dhh1 in P-bodies, and can affect the decay of a subclass of yeast mRNAs. Taken together, this identifies Stm1 as a component of the decapping machinery that also interacts with the translation machinery.
MATERIALS AND METHODS
Yeast strains, growth conditions, and plasmids:
The genotypes of all strains used in this study are listed in Table1. Strains were grown in either standard yeast extract/peptone medium (YP) or synthetic medium (SC) supplemented with appropriate amino acids and 2% dextrose. Strains were grown at 30° unless otherwise stated. For overexpression studies, strains were grown in YP or SC media supplemented with appropriate amino acids, 2% galactose, and 0.5% sucrose. All plasmids and oligonucleotides used in the study are listed in Table 2 and Table 3, respectively.
TABLE 1.
Yeast strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| yRP2065 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Coller and Parker (2005) |
| yRP2066 | MATa, his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 dhh1∷NEO | Coller and Parker (2005) |
| yRP2323 | MATα, his3Δ1 leu2Δ0 ura3Δ0 dhh1∷NEO, pat1∷HIS3 | Gift from J. Coller |
| yRP1437 | MATa, his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 stm1∷NEO | Winzeler et al. (1999)a |
| yRP840 | MATaleu2–3,112 trp1 ura3-52 his4-539 cup1∷LEU2/PGK1pG/MFA2pG | Hatfield et al. (1996) |
| yRP1372 | MATaleu2–3,112 trp1 ura3-52 his4-539 cup1:LEU2/PGK1pG/MFA2pG pat1∷LEU2 | Tharun et al. (2000) |
Purchased from Open Biosystems.
TABLE 2.
Plasmids used in this study
| Name | Description | Reference |
|---|---|---|
| pRP948 | Stm1 under the control of its endogenous promoter on a 2μ URA3 vector | This study |
| pRP1360 | Stm1 overexpression using GAL10 promoter on a 2μ URA3 vector | Gelperin et al. (2005)a |
| pRP1728 | Stm1 overexpression using GAL10 promoter on a 2μ LEU2 vector | This study |
| pRP1361 | Overexpression of Dhh1 using GAL10 promoter on a 2μ URA3 vector | Gelperin et al. (2005)a |
| pRP485 | MFA2 mRNA with a poly(G) tract under GAL10 promoter on a CEN URA3 vector | Muhlrad and Parker (1992) |
| pRP469 | PGK1 mRNA with a poly(G) tract under GAL10 promoter on a CEN URA3 vector | Decker and Parker (1993) |
| pRP1189 | EDC1 mRNA with a poly(G) tract under GAL10 promoter on a CEN TRP1 URA3 vector | Muhlrad and Parker (2005) |
| pRP1151 | GFP-tagged Dhh1 on a CEN LEU2 vector | Coller et al. (2001) |
| pRP1574 | mCherry-tagged Edc3 on CEN URA3 vector | Buchan et al. (2008) |
| pRP1007 | COX17 mRNA under GAL10 promoter on a 2μ LEU2 vector (pG74/ST30) | Olivas and Parker (2000) |
Purchased from Open Biosystems.
TABLE 3.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| oRP140 | ATATTGATTAGATCAGGAATTCC |
| oRP141 | AATTGATCTATCGAGGAATTCC |
| oRP100 | GTCTAGCCGCGAGGAAGG |
| oRP1211 | AATTGCTTTGGATGACCAGATCC |
| oRP1427 | GGTTGTCGGCAGACTGTCAG |
RNA analysis:
All RNA analyses were performed as described in Muhlrad and Parker (1992). For half-life measurements, cells were grown to mid-log phase containing 2% galactose. Transcription was repressed by the addition of media containing 4% dextrose. Aliquots were collected over a brief course of time and frozen. Total RNA was extracted as described in Caponigro et al. (1993) and analyzed by running 20 μg of total RNA on 1.25% formaldehyde agarose gels. All Northern analyses were performed using radiolabeled oligonucleotide probes directed against MFA2pG (oRP140), PGK1pG (oRP141), COX17 (oRP1427), and EDC1pG (oRP1121). Loading corrections were done using oRP100, an oligonucleotide probe directed against SCR1 RNA, a stable RNA polymerase III transcript. Half-lives were determined by quantitation of blots using a Molecular Dynamics phosphorimager (Sunnyvale, CA).
Microscopy:
For the analysis of P-bodies under logarithmic growth conditions, cultures were grown to an OD600 of 0.3–0.4 in the appropriate media. Cells grown in YP media were harvested and washed with SC supplemented with appropriate amino acids and 2% dextrose and observed under the microscope. Cells grown in SC media were directly harvested, resuspended in a smaller volume of the same media, and observed under the microscope.
For the analysis of P-bodies following glucose depletion, cells grown to mid log phase were washed with SC supplemented with appropriate amino acids (no sugar), resuspended in the same media, and incubated in a flask in a shaking water bath for 15 min. The cells were then harvested and observed under the microscope.
For the analysis of P-bodies at high cell densities, the cultures were allowed to grow overnight to an OD600 of 1.0 or for 2 days to an OD600 of >3.0. The cells were harvested and observed under the microscope. All microscopy was done on a deconvolution microscope (Deltavision RT, Applied Precision) using an objective (UPlan Sapo ×100 1.4 NA; Olympus). Images were collected using software (softWoRx) as 512 × 512-pixel files with a camera (CoolSNAP HQ; Photometrics) using 1 × 1 binning. Images are Z series that have been adjusted to the same contrast range with ImageJ software.
Western analysis:
Western analysis of proteins was conducted by preparing whole-cell extracts from the appropriate strains. Protein concentration was determined by Bio-Rad protein assay and equal amounts of total protein were loaded on the gel. Protein-A-tagged Dhh1 and Stm1 proteins were detected using peroxidase antiperoxidase antibody (DAKO). GFP-tagged Dhh1 and Stm1 proteins were detected using anti-GFP antibody (Covance).
RESULTS
Stm1 is a high-copy suppressor of pat1Δ temperature sensitivity:
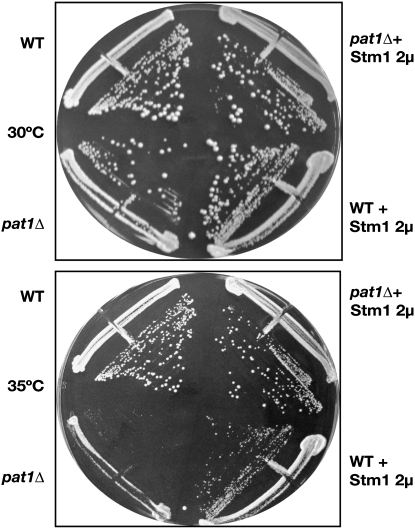
To identify additional proteins involved in translation repression and/or mRNA decay, we screened for high-copy suppressors of the temperature-sensitive phenotype of PAT1 loss-of-function alleles using a PAT1 nonsense allele termed mrt1-3 (Hatfield et al. 1996; Tharun et al. 2000). A mrt1-3 strain was transformed with a 2μ URA3 high-copy yeast genomic library, and >10,000 transformants were screened for colonies able to grow at the nonpermissive temperature (35°). The plasmids from yeast capable of growth at nonpermissive temperature were rescued, and the yeast genomic sequences within the plasmids were identified by DNA sequence analysis. The analysis of overlapping plasmid inserts suggested that STM1 was a high-copy suppressor of the mrt1-3 temperature-sensitive phenotype, which was confirmed by subcloning only STM1 on a high-copy plasmid and showing that it could suppress the temperature sensitivity of both the mrt1-3 allele and a complete pat1Δ (data not shown and Figure 1). We also observed the RCN2 gene as a weak high-copy suppressor of the mrt1-3 and pat1Δ temperature sensitivity (data not shown). However, since the RCN2 suppression phenotype was weak, it was not pursued. These results identified STM1 as a high-copy suppressor of the temperature sensitivity of loss-of-function alleles in PAT1.
Figure 1.—
Stm1 is a high-copy suppressor of pat1Δts. Stm1 was overexpressed using a 2μ overexpression plasmid in wild-type (yRP840) and pat1Δ (yRP1372) strain backgrounds and the growth was monitored under the conditions mentioned. Overexpression of Stm1 allowed the pat1Δ strains to survive at nonpermissive temperatures.
Stm1 has genetic interactions with Dhh1 and Pat1:
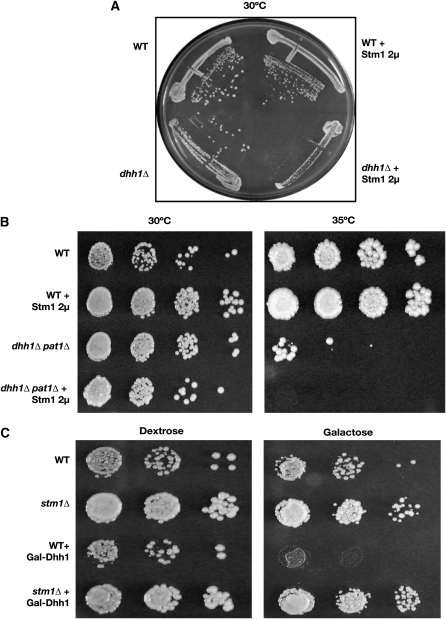
To examine possible mechanisms by which Stm1 suppressed pat1Δ, we examined how overexpression of Stm1 from a 2μ plasmid affected the growth of a dhh1Δ strain, which is also temperature sensitive. Surprisingly, we observed that overexpression of Stm1 impaired the growth of the dhh1Δ strain as compared to a dhh1Δ strain carrying the empty vector (Figure 2A). In contrast, 2μ overexpression of Stm1 in a wild-type strain had little or no effect on growth rate (Figure 2A). This observation demonstrated that Stm1 affects the cells' requirement for Dhh1 function and suggested that Stm1 and Dhh1 might functionally interact.
Figure 2.—
Stm1 has genetic interactions with the general translation repressors Dhh1 and Pat1. Stm1 was overexpressed using a 2μ plasmid in wild type (yRP2065) and dhh1Δ (yRP2066) (A) and dhh1Δ pat1Δ (yRP2323) (B) double-deletion strain backgrounds. Growth was monitored over a period of 3 days. Overexpression of Stm1 has a deleterious effect on growth in dhh1Δ and dhh1Δ pat1Δ strain backgrounds. (C) Dhh1 was overexpressed using GAL10 promoter in wild-type (yRP2065) and stm1Δ (yRP1437) strains. Growth was monitored over a period of 3 days. Deletion of STM1 prevented the overexpression lethality of Dhh1.
The different effects of Stm1 overexpression on the pat1Δ and dhh1Δ strains could be explained in two ways. First, overexpression of Stm1 might suppress the pat1Δ by promoting the function of Dhh1 in translation repression and mRNA decay. In this model, the overexpression of Stm1 in a dhh1Δ might be detrimental by committing mRNAs to a Dhh1-mediated event that would be lacking in a dhh1Δ strain. An alternative possibility is that Stm1 functions independently of Dhh1 and Pat1 in a third pathway that compensates for the loss of Pat1. To test these two possibilities, we overexpressed Stm1 (using a 2μ plasmid) in a dhh1Δ pat1Δ double deletion. If Stm1 functions independently of both Pat1 and Dhh1, overexpression of Stm1 in the double mutant should improve its growth at high temperatures. Alternatively, if Stm1 suppresses the growth defects of the pat1Δ by increasing the function of Dhh1, one predicts that overexpression of Stm1 should be detrimental to the growth of dhh1Δ pat1Δ. We observed that the dhh1Δ pat1Δ strain grows poorly at the restrictive temperature, but overexpressing STM1 made it considerably worse (Figure 2B). We interpret this result to suggest that Stm1 suppresses the pat1Δ growth defect by promoting the function of Dhh1.
The above results suggested that Stm1 overexpression enhances the function of Dhh1. If Stm1 functions to increase Dhh1 activity in some manner, this would predict that a deletion of STM1 reduces the function of Dhh1. To test this possibility, we took advantage of previous work showing that overexpression of Dhh1 under control of the GAL10 promoter inhibits cell growth (Coller and Parker 2005). Given this, we asked if deletion of STM1 had an effect on the overexpression lethality of the Dhh1.
We observed that deletion of STM1 prevents the growth inhibition by overexpression of Dhh1 (Figure 2C). Moreover, as judged by Western analysis the stm1Δ strain showed similar levels of GAL-Dhh1 expression as compared to the wild-type strain (data not shown). Taken together, these observations argue that Stm1 promotes Dhh1 function.
Stm1 affects, but is not required for, the accumulation of Dhh1 in P-bodies:
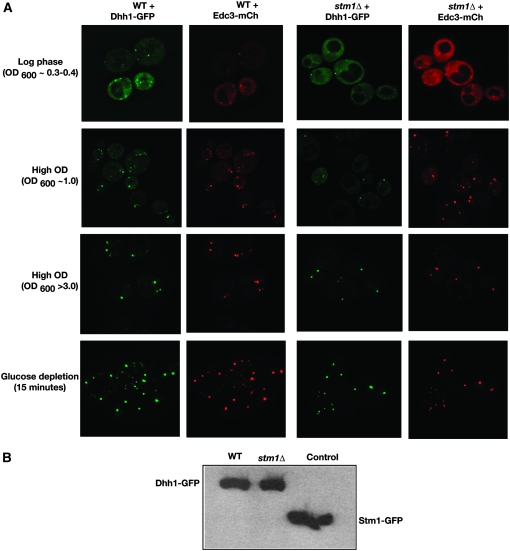
The above results suggested that Stm1 acts to enhance the function of Dhh1. Dhh1 is thought to function in translation repression and P-body formation by first decreasing translation initiation and then by accumulating with the translationally repressed mRNA in a P-body (Coller and Parker 2005). A prediction of Stm1 enhancing Dhh1 function is that strains lacking Stm1 might show a deficit in Dhh1 accumulation in P-bodies. To test this possibility, we examined the formation of P-bodies in stm1Δ strains and their ability to accumulate a GFP-tagged version of Dhh1. For this experiment, we cotransformed a GFP-tagged plasmid of Dhh1 and a mCherry-tagged plasmid of another P-body marker, Edc3, into wild-type and stm1Δ strains. The strains were examined for P-body formation during glucose depletion, which is a stress condition that leads to rapid assembly of P-bodies (Teixeira et al. 2005), and at higher cell density where P-bodies are enlarged (Teixeira et al. 2005).
We first examined P-body formation at higher cell densities where P-bodies enlarge over time from the small P-bodies observed in mid-log phase cultures (Teixeira et al. 2005). We observed that in wild-type strains, P-bodies as judged by the accumulation of Dhh1 and Edc3 were clearly visible when the cells reached an OD600 of 1.0 (Figure 3A). In stm1Δ strains, P-bodies formed at similar cell densities as judged by the accumulation of Edc3 in these foci (Figure 3A), but these P-bodies contained reduced amounts of Dhh1 as compared to wild-type cells (Figure 3A). To be sure that these differences in P-body formation were not due to changes in the levels of Dhh1, we showed by Western analysis that the protein levels of Dhh1-GFP were comparable between the wild-type and the stm1Δ strains (Figure 3B). This indicates that Stm1 can affect the accumulation of Dhh1 in P-bodies.
Figure 3.—
Stm1 promotes the accumulation of Dhh1 in P-bodies. (A) Wild-type (yRP2065) and stm1Δ (yRP1437) strains were cotransformed with GFP-tagged plasmid of Dhh1 and mCherry-tagged plasmid of Edc3, and the localization of these proteins was observed by microscopy under glucose depletion or high cell densities (see materials and methods). Deletion of STM1 affects the accumulation of Dhh1 in P-bodies under high cell densities. (B) Whole-cell extracts were prepared from wild-type (yRP2065) and stm1Δ (yRP1437) strains carrying Dhh1-GFP plasmid and Western analysis was performed using anti-GFP antibody (Covance). Equal amounts of total protein were loaded on the gel.
Two observations indicate that, although Stm1 affects Dhh1 accumulation in P-bodies, Stm1 is not absolutely required for Dhh1 accumulation in P-bodies. First, we observed that when cells reached higher cell densities (OD600 > 3.0 after 2 days of growth), Dhh1-GFP accumulated in P-bodies, even in the stm1Δ strain (Figure 3A). Moreover, during acute glucose deprivation, where translation decreases to <5% of normal in a few minutes and mRNA accumulates in P-bodies (Ashe et al. 2000; Teixeira et al. 2005), Dhh1-GFP accumulated in P-bodies in both wild-type and stm1Δ strains (Figure 3A). Taken together, these observations indicate that Stm1 promotes, but is not required for, the accumulation of Dhh1 in P-bodies and is consistent with Stm1 acting upstream of Dhh1 to promote its function.
Stm1 enhances the degradation of the EDC1 and COX17 mRNAs:
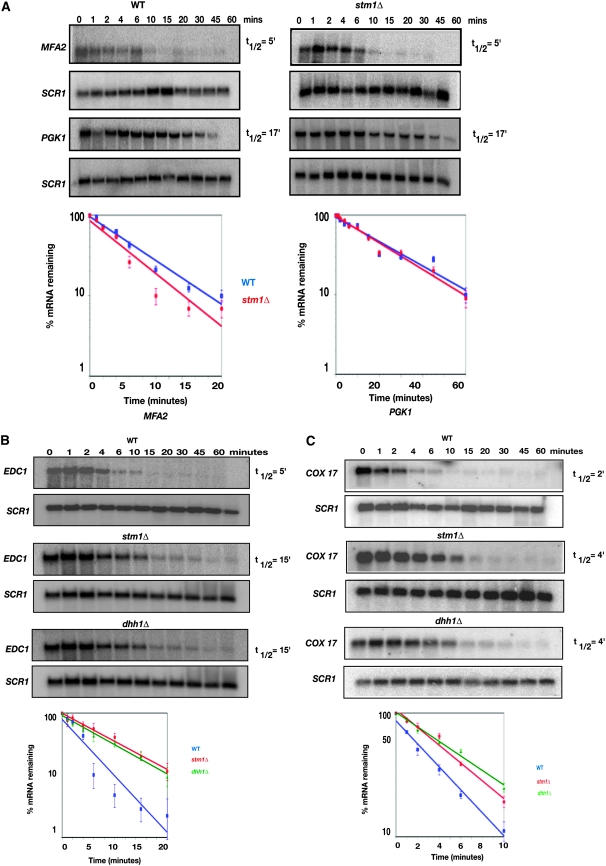
The above results document genetic and cell biological interactions between Stm1 and Dhh1, suggesting that Stm1 enhances the function of Dhh1. Since Dhh1 functions in both translation repression and mRNA decay, it suggested that Stm1 might affect the decay of some mRNAs. To determine if Stm1 affects mRNA degradation, we examined the effect of stm1Δ on the decay of specific mRNA reporters. The specific reporters used were the PGK1pG, MFA2pG, COX17, and EDC1pG mRNAs under the control of the GAL10 promoter, which allows transcriptional shutoff by the addition of glucose (Decker and Parker 1993). The PGK1pG and MFA2pG transcripts are canonical reporters that represent general mRNA decay, while COX17 and EDC1pG mRNAs are transcripts that are more sensitive to the loss of DHH1 (Cheng et al. 2005; Muhlrad and Parker 2005). These experiments resulted in the following observations.
First, we observed that the PGK1pG and MFA2pG mRNAs showed similar decay rates in the wild-type and stm1Δ strains (Figure 4A). In contrast, the dhh1Δ strain or pat1Δ strains show significant changes in the decay rates of these mRNAs by affecting their rates of decay (Tharun et al. 2000; Coller et al. 2001). These observations indicate that Stm1 is not normally required for decay at least of the MFA2 and PGK1 transcripts.
Figure 4.—
Stm1 deletion stabilizes EDC1 and COX17 mRNAs. (A) Decay of the reporters MFA2pG and PGK1pG following transcriptional repression in wild-type (yRP2065) and stm1Δ (yRP1437) strains. MFA2pG and PGK1pG were detected using oligonucleotide probes oRP140 and oRP141, respectively. SCR1 was detected using oRP100. (B) The decay of the EDC1pG reporter following transcriptional repression in wild-type (yRP2065), stm1Δ (yRP1437), and dhh1Δ (yRP2066) strains. Oligonucleotide probes oRP1211 and oRP100 were used to detect EDC1 and SCR1 transcripts, respectively. (C) The decay of the COX17 mRNA following transcriptional repression in wild-type (yRP2065), stm1Δ (yRP1437), and dhh1Δ (yRP2066) strains. Oligonucleotide probes oRP1427 and oRP100 were used to detect COX17 and SCR1 transcripts, respectively. (A–C) Time points represent minutes after transcriptional repression. SCR1 was used as a loading control. Each decay experiment was done in triplicate. A graphical representation of the data is shown by plotting the percentage of mRNA remaining as a function of time. Best-fit lines were determined by exponential curve fitting using the graphing software Deltagraph. Error bars denote the calculated standard deviation.
A second, and important, observation was that the half-life of the EDC1 transcript in a stm1Δ strain was prolonged (t1/2 = 15 min) as compared to a wild-type strain (t1/2 = 5 min) (Figure 4B) and was similar to the observed decay rate seen in a dhh1Δ strain (t1/2 = 15 min). Overexpression of Stm1 did not alter the half-life of the EDC1 transcript (data not shown). Similarly, the COX17 mRNA was approximately twofold more stable in the stm1Δ and dhh1Δ strains as compared to wild-type cells (Figure 4C). The three- to fourfold stabilization of EDC1 mRNA and twofold stabilization of COX17 mRNA observed in a stm1Δ strain provide direct evidence that Stm1 has a role in mRNA decay, although its function may primarily be limited to specific mRNAs.
Stm1 overexpression does not restore mRNA decay in a pat1Δ strain:
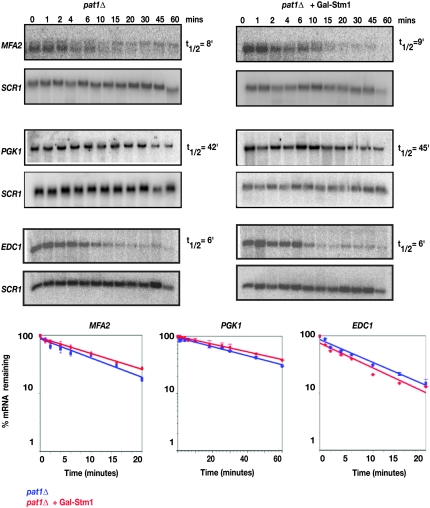
The above results indicated that Stm1 is a high-copy suppressor of the temperature sensitivity of pat1Δ and appeared to act by promoting Dhh1 function. Given this, one possibility is that Stm1 overexpression would suppress the decay defect in the pat1Δ strain by enhancing Dhh1 function. To examine this possibility, we compared the decay rates of MFA2pG, PGK1pG, and EDC1pG mRNAs between pat1Δ strains and pat1Δ strains overexpressing Stm1.
We observed that overexpression of Stm1 did not restore the defect in mRNA degradation seen in the pat1Δ strain for some mRNAs. Specifically, strains lacking PAT1 show increased stability of the MFA2pG and PGK1pG transcripts, while the EDC1 mRNA is not affected by pat1Δ, which is consistent with this mRNA being predominantly dependent on Dhh1 for its degradation (Figure 5). When Stm1 was overexpressed in a pat1Δ strain, the half-lives of MFA2pG (t1/2 = 9 min), PGK1pG (t1/2 = 45 min), and EDC1pG (t1/2 = 6 min) were not significantly altered. This argues that the suppression of the temperature sensitivity of pat1Δ, brought about by Stm1, is not due to a general stimulation of mRNA decay.
Figure 5.—
Overexpression of Stm1 does not affect the half-life of mRNAs in a pat1Δ strain. The decay of MFA2pG, PGK1pG, and EDC1pG reporters following transcriptional repression in the pat1Δ (yRP1372) strain overexpressing Stm1 or empty vector under a GAL10 promoter is shown. Time points indicated represent minutes after transcriptional repression. SCR1 was used as a loading control. MFA2pG, PGK1pG, and EDC1pG transcripts were detected using oligonucleotide probes oRP140, oRP141, and oRP1211, respectively, while SCR1 was detected using oRP100. A graphical representation of the data is shown by plotting the percentage of mRNA remaining as a function of time. Best-fit lines were determined by exponential curve fitting using the graphing software Deltagraph. Experiments were done in triplicate and error bars denote the standard deviation.
Strains lacking Pat1 also show a defect in translation (Wyers et al. 2000), and this may account for the temperature sensitivity of pat1Δ. To examine whether Stm1 overexpression was suppressing the defect in translation in the pat1Δ strains, we measured the incorporation of 35S-labeled amino acids into protein with and without Stm1 overexpression. We observed that pat1Δ strains showed a defect in amino acid incorporation (65% of wild type at 37°), which was partially suppressed by Stm1 overexpression (up to 90% of wild type). In contrast, overexpression of Stm1 in wild-type strains had no effect on amino acid incorporation. This suggests that at least part of the suppression of pat1Δ temperature sensitivity is due to Stm1 overexpression correcting a defect in translation rates. However, we cannot rule out that Stm1 overexpression may also suppress a defect in mRNA degradation for a subset of mRNAs.
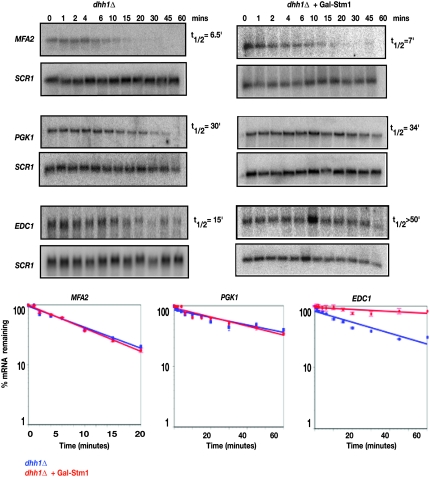
Stm1 overexpression inhibits mRNA degradation in a dhh1Δ strain:
One surprising genetic interaction was that the overexpression of Stm1 in a dhh1Δ affected its growth adversely (Figure 2A). This suggested that Stm1 performs a function that both enhances the activity of Dhh1 and increases the cellular need for Dhh1 function. A prediction of this model is that overexpression of Stm1 might also increase the role of Dhh1 in the degradation of mRNAs. To examine this possibility, we compared the decay rates of the MFA2pG, PGK1pG, and EDC1pG mRNAs between dhh1Δ strains and a dhh1Δ overexpressing Stm1.
We observed that overexpression of Stm1 strongly inhibited the degradation of the EDC1 mRNA in the dhh1Δ strain where the half-life went from ∼15 min to ≥50 min (Figure 6). In contrast, overexpression of Stm1 in the dhh1Δ strain did not affect the decay rates of the MFA2pG or PGK1pG mRNA. We interpret these observations to suggest that overexpression of Stm1 commits an mRNA to a Dhh1-specific pathway of decay. Moreover, the different effects on MFA2pG and EDC1pG suggest that Stm1 might affect only a subset of mRNAs that includes EDC1.
Figure 6.—
Overexpression of Stm1 prolongs the half-life of EDC1 mRNA in a dhh1Δ strain. The decay of MFA2pG, PGK1pG, and EDC1pG reporters following transcriptional repression in dhh1Δ (yRP2066) strain overexpressing Stm1 or empty vector under a galactose promoter is shown. Time points indicated represent minutes after transcriptional repression. SCR1 was used as a loading control. MFA2pG, PGK1pG, and EDC1pG transcripts were detected using oligonucleotide probes oRP140, oRP141, and oRP1211, respectively, while SCR1 was detected using oRP100. A graphical representation of the data is shown by plotting the percentage of mRNA remaining as a function of time. Best-fit lines were determined by exponential curve fitting using the graphing software Deltagraph. Experiments were done in triplicate and error bars denote the standard deviation.
DISCUSSION
Stm1 is a modulator of translation repression and/or mRNA decay:
Several lines of evidence argue that Stm1 affects the process of targeting an mRNA for degradation. First, we show that Stm1 is a high-copy suppressor of the temperature-sensitive phenotype of the pat1Δ strain (Figure 1). Second, we observe that stm1Δ strains show defects in the decay of the EDC1 and COX17 mRNAs, but not in MFA2 or PGK1 mRNAs (Figure 4). This demonstrates that Stm1 can affect decay of at least a subset of mRNAs. In addition, previous work has shown that overexpression of Stm1 partially suppresses a pop2Δ strain (Hata et al. 1998) and suppresses the synthetic lethality of a pop2Δ pab1–rrm2Δ combination (Ohn et al. 2007). Taken together, we suggest that Stm1 can influence the process of targeting an mRNA for degradation. Moreover, since translation repression and targeting an mRNA for degradation are often coupled, these results imply that Stm1 may also function in the process of translation repression.
Stm1 promotes the function of Dhh1 in translation repression and mRNA decay:
Several observations argue that Stm1 acts to enhance the function of Dhh1 in promoting translation repression and mRNA decay. First, the high-copy suppression by Stm1 of the temperature-sensitive phenotype of pat1Δ strains requires Dhh1 (Figure 1 and Figure 2B). Second, overexpression of Stm1 in a dhh1Δ strain affects growth adversely (Figure 2A); suggesting that Stm1 function creates a condition that requires Dhh1 for resolution. Consistent with this proposal, we observed that overexpression of Stm1 in dhh1Δ strains further inhibited the rates of decay of EDC1 mRNA in these conditions (Figure 6). Third, we observed that in a stm1Δ strain, Dhh1-GFP is less efficiently recruited to P-bodies as yeast cells reach high cell densities (Figure 3A). Finally, stm1Δ strains are resistant to inhibition of cell growth due to Dhh1 overexpression (Figure 2C). Taken together, the simplest interpretation of these observations is that Stm1 function stimulates Dhh1 function.
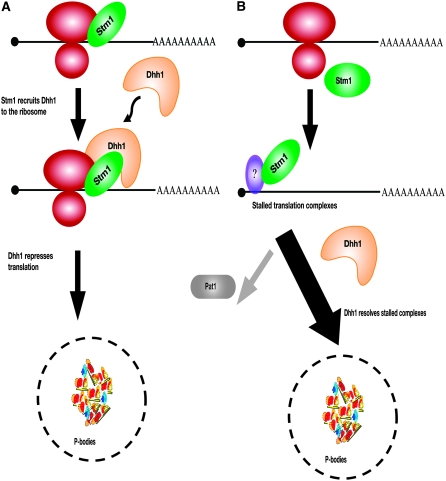
Possible mechanisms of Stm1 function:
An unresolved issue is the specific mechanism by which Stm1 promotes Dhh1 function. Stm1 has previously been shown to interact with yeast ribosomes (Van Dyke et al. 2004, 2006). This suggests two general models for how Stm1 could function (Figure 7). In one model, Stm1 recruits Dhh1 through physical interactions to translating ribosomes and by doing so leads to Dhh1 triggering translation repression. One limitation of this model is that it would not provide an explanation for why overexpression of Stm1 in the absence of Dhh1 is growth inhibitory. An alternate model is that Stm1 binds to the ribosome and inhibits a specific step of its function. This would create a stalled translation complex that would then be disassembled/resolved by Dhh1 promoting the mRNAs repression. The latter model is appealing since it would explain why Stm1 overexpression in the absence of Dhh1 is detrimental. Translation complexes would be predicted to be stalled in unproductive states that are poorly resolved without Dhh1 function. Consistent with this model, it is known that Dhh1 function in targeting an mRNA for decapping requires that ribosomes are able to load on the mRNA (Coller and Parker 2005). In either case, on the basis of its reported interactions with ribosomes, Stm1 could provide a necessary link between the translation machinery and Dhh1, serving as a link to repression and mRNA degradation. Unraveling this process will be important in understanding the regulation of translation and decay of mRNA.
Figure 7.—
Possible models for the function of Stm1. (A) Stm1 physically recruits Dhh1 to the translating ribosomes, and this allows Dhh1 to repress translation. (B) Stm1 stalls ribosomes at some stage of translation and represses translation, and this triggers the function of Dhh1.
Acknowledgments
We thank all members of the Parker laboratory, especially Wehai He, Guy Pilkington, Tracy Nissan, Denise Muhlrad, and Carolyn Decker for helpful discussions, technical assistance, and critical review of the manuscript. Thanks are also due to Jeff Coller for the gift of the yeast strain YRP2323. This work was supported by funds from the National Institutes of Health (grant R37 GM45443) and the Howard Hughes Medical Institute.
References
- Ashe, M. P., S. K. De Long and A. B. Sachs, 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman, C. A., and R. Parker, 1994. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 269 9687–9692. [PubMed] [Google Scholar]
- Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield et al., 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382 642–646. [DOI] [PubMed] [Google Scholar]
- Buchan, J. R., D. Muhlrad and R. Parker, 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G., D. Muhlrad and R. Parker, 1993. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 13 5141–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., J. Coller, R. Parker and H. Song, 2005. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA 11 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J., and R. Parker, 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73 861–890. [DOI] [PubMed] [Google Scholar]
- Coller, J., and R. Parker, 2005. General translational repression by activators of mRNA decapping. Cell 122 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J. M., M. Tucker, U. Sheth, M. A. Valencia-Sanchez and R. Parker, 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C. J., and R. Parker, 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7 1632–1643. [DOI] [PubMed] [Google Scholar]
- Dunckley, T., and R. Parker, 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio, A., I. Behm-Ansmant and E. Izaurralde, 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8 9–22. [DOI] [PubMed] [Google Scholar]
- Garneau, N. L., J. Wilusz and C. J. Wilusz, 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8 113–126. [DOI] [PubMed] [Google Scholar]
- Gelperin, D. M., M. A. White, M. L. Wilkinson, Y. Kon, L. A. Kung et al., 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19 2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata, H., H. Mitsui, H. Liu, Y. Bai, C. L. Denis et al., 1998. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield, L., C. A. Beelman, A. Stevens and R. Parker, 1996. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C. L., and A. Stevens, 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur, T., and R. Parker, 1999. The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context of the translational start codon. RNA 5 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., and R. Parker, 1992. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 6 2100–2111. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., and R. Parker, 2005. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 24 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., C. J. Decker and R. Parker, 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 8 855–866. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., C. J. Decker and R. Parker, 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn, T., Y. C. Chiang, D. J. Lee, G. Yao, C. Zhang et al., 2007. CAF1 plays an important role in mRNA deadenylation separate from its contact to CCR4. Nucleic Acids Res. 35 3002–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas, W., and R. Parker, 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., and U. Sheth, 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25 635–646. [DOI] [PubMed] [Google Scholar]
- Parker, R., and H. Song, 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11 121–127. [DOI] [PubMed] [Google Scholar]
- Schwartz, D. C., and R. Parker, 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D. C., and R. Parker, 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 20 7933–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U., and R. Parker, 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu, A. B., M. F. Wilkinson and A. van Hoof, 2008. Messenger RNA regulation: to translate or to degrade. EMBO J. 27 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, D., U. Sheth, M. A. Valencia-Sanchez, M. Brengues and R. Parker, 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun, S., W. He, A. E. Mayes, P. Lennertz, J. D. Beggs et al., 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404 515–518. [DOI] [PubMed] [Google Scholar]
- Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis et al., 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104 377–386. [DOI] [PubMed] [Google Scholar]
- Utsugi, T., A. Toh-e and Y. Kikuchi, 1995. A high dose of the STM1 gene suppresses the temperature sensitivity of the tom1 and htr1 mutants in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1263 285–288. [DOI] [PubMed] [Google Scholar]
- Van Dyke, M. W., L. D. Nelson, R. G. Weilbaecher and D. V. Mehta, 2004. Stm1p, a G4 quadruplex and purine motif triplex nucleic acid-binding protein, interacts with ribosomes and subtelomeric Y′ DNA in Saccharomyces cerevisiae. J. Biol. Chem. 279 24323–24333. [DOI] [PubMed] [Google Scholar]
- Van Dyke, N., J. Baby and M. W. Van Dyke, 2006. Stm1p, a ribosome-associated protein, is important for protein synthesis in Saccharomyces cerevisiae under nutritional stress conditions. J. Mol. Biol. 358 1023–1031. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901–906. [DOI] [PubMed] [Google Scholar]
- Wyers, F., M. Minet, M. E. Dufour, L. T. A. Vo and F. Lacroute, 2000. Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast. Mol. Cell. Biol. 20 3538–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]