Abstract
Cells of Flavobacterium johnsoniae move rapidly over surfaces by gliding motility. The mechanism of this form of motility is not known. Six genes (gldA, gldB, gldD, gldF, gldG, and ftsX) that are required for gliding have been described. Tn4351 mutagenesis was used to identify another gene, gldH, which is required for cell movement. GldH mutants formed nonspreading colonies, and individual cells lacked the cell movements and ability to propel latex spheres along their surfaces that are characteristic of wild-type cells. gldH mutants also failed to digest chitin and were resistant to bacteriophages that infect wild-type cells. Introduction of pMM293, which carries wild-type gldH, restored to the gldH mutants colony spreading, cell motility, the ability to move latex spheres, phage sensitivity, and the ability to digest chitin. gldH encodes a predicted 141-amino-acid protein that localized to the membrane fraction. Labeling studies with [3H]palmitate demonstrated that GldH is a lipoprotein. GldB and GldD, which were previously described, also appear to be lipoproteins. GldH does not exhibit significant amino acid similarity to proteins of known function in the databases. Putative homologs of gldH of unknown function are found in motile (Cytophaga hutchinsonii) and apparently nonmotile (Bacteroides thetaiotaomicron, Bacteroides fragilis, Tannerella forsythensis, Porphyromonas gingivalis, and Prevotella intermedia) members of the Cytophaga-Flavobacterium-Bacteroides group.
Flavobacterium johnsoniae, like many other members of the Cytophaga-Flavobacterium-Bacteroides (CFB) group, moves rapidly over surfaces in a process called gliding motility (22). Individual cells of F. johnsoniae glide at speeds of up to 10 μm/s (27). Cells also adsorb latex spheres and propel these rapidly around the cell in multiple paths (21, 28). Several models have been proposed to explain the cell movement of F. johnsoniae and related bacteria, but the mechanism remains unknown (22).
Genetic techniques have been developed for F. johnsoniae, and several genes that are required for gliding have been identified (1, 13-15, 23). gldA, gldF, and gldG are thought to encode components of an ATP-binding cassette (ABC) transporter that is required for motility. The exact role of this transporter in cell movement is not known. gldB and gldD encode additional membrane proteins that are required for cell movement. GldB and GldD do not exhibit similarity to proteins of known function, and their roles in gliding remain to be determined. ftsX is required for both cell division and gliding motility. The inability of ftsX mutants to glide may be an indirect effect of the defects in cell division. Here we describe the identification of gldH, a lipoprotein that is required for F. johnsoniae gliding motility.
MATERIALS AND METHODS
Bacterial and bacteriophage strains, plasmids, and growth conditions.
F. johnsoniae UW101 (ATCC 17061) was the wild-type strain used in these studies, and all mutants were derived from this strain. gld mutants used in this study which were previously characterized included the gldA mutant CJ101-288 (1), the gldB mutant CJ569 (Tn4351 insert Ω569) (15), the gldD mutant CJ282 (14), the gldF mutant CJ787 (13), and the gldG mutant CJ776 (13). Fifty nonmotile mutants of F. johnsoniae were obtained from J. Pate and were previously described (5, 15, 47). The bacteriophages active against F. johnsoniae that were used in this study were φCj1, φCj13, φCj23, φCj28, φCj29, φCj42, φCj48, and φCj54 (5, 29, 47). The Escherichia coli strains used were DH5αMCR (GibcoBRL Life Technologies), S17-1 (39), BW19851 (24), and NovaBlue (Novagen Inc., Madison, Wis.). E. coli strains were grown in Luria-Bertani medium (36) at 37°C, and F. johnsoniae strains were grown in Casitone-yeast extract (CYE) medium at 30°C, as previously described (23). To observe colony spreading, F. johnsoniae was grown on PY2 agar medium at 25°C (1). For radiolabeling experiments, cells were grown in SDY medium. SDY medium was identical to the defined medium SD (4) except that yeast extract was added to 0.1 g/liter to increase the growth rate. Antibiotics were used at the following concentrations when needed: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; erythromycin, 100 μg/ml; kanamycin, 30 μg/ml; and tetracycline, 20 μg/ml. The plasmids and primers used in this study are listed in Table 1.
TABLE 1.
Plasmids and primers used in this study
| Plasmid or primer | Description and/or sequencea | Source or reference |
|---|---|---|
| Plasmids | ||
| pBC SK(+) | ColE1; Cmr | Stratagene |
| pSTBlue-1 | ColE1 ori; Apr | Novagen |
| PT7Blue | ColE1 ori; Apr | Novagen |
| PHP45ΩKan | Plasmid carrying Kmr cassette with transcriptional terminators at each end; Apr Kmr | 8 |
| pMAL-c2 | MalE fusion protein expression vector; Apr | New England Biolabs |
| pCP23 | E. coli-F. johnsoniae shuttle plasmid; Apr (Tcr) | 1 |
| pSA21 | gldA in pCP23; Apr (Tcr) | 1 |
| pDH233 | gldBC in pCP23; Apr (Tcr) | 15 |
| pMM213 | gldD in pCP23; Apr Kmr (Tcr) | 14 |
| pMK314 | gldFG in pCP29; Apr (Cfr Emr) | 13 |
| pMK301 | 5.5-kb HindIII fragment of CJ778 chromosomal DNA spanning the site of insertion of the Tn4351 insertion cloned into pBC SK(+) | This study |
| pMM219 | 6.1-kb SacI fragment spanning gldH and adjacent genes cloned into pBC SK(+); Cmr | This study |
| pMM224 | 6.1-kb SacI fragment spanning gldH and adjacent genes cloned into pCP23; Apr (Tcr) | This study |
| pMM225 | 6.1-kb SacI fragment spanning gldH and adjacent genes cloned into pCP23 (opposite orientation of pMM224); Apr (Tcr) | This study |
| pMM286 | 933-bp gldH PCR product cloned into pSTBlue-1; Apr | This study |
| pMM293 | 933-bp gldH fragment of pMM286 cloned into pCP23; Apr (Tcr) | This study |
| pMM298 | pMM293 with the Kmr cassette from pHP45Ωkan inserted upstream of gldH; Apr Kmr (Tcr) | This study |
| pMM299 | Identical to pMM298 except that the Kmr cassette is inserted in the opposite orientation; Apr Kmr (Tcr) | This study |
| pJB200 | gldH expression plasmid, 430-bp gldH fragment in pMal-c2; Apr | This study |
| pTB39 | Recombinant gldB-his in pCP23; expresses GldB with a carboxy-terminal His tag; Apr (Tcr) | This study |
| pTB41 | Recombinant gldH-his in pCP23; expresses GldH with a carboxy-terminal His tag; Apr (Tcr) | This study |
| Primers | ||
| T7 promoter primer | 5′ CGTAATACGACTCACTATAGGGC 3′ | |
| 24 | 5′ CAGGGATGTATATTTGCAG 3′; primer used during construction of pTB39. | |
| 340 | 5′ GACTTGGATACCTCACGCC 3′; primer in IS4351; used for inverse PCR amplification of Tn4351 insertion junction sites and for DNA sequencing | |
| 341 | 5′ TTGGAAATTTTCTGGGAGG 3′; primer in ermF of Tn4351; used for inverse PCR amplification of Tn4351 insertion junction sites and for DNA sequencing | |
| 425 | 5′ TTGAATTTCAGGGAGACGG 3′; primer used during preparation of fjo17 probe for Northern blotting | |
| 429 | 5′ GAACCGAAAAGTCCCCAAG 3′; primer used for construction of gldH-containing plasmids pMM286 and pMM293 | |
| 432 | 5′ ATGCAATGGGGCAGGAAAG 3′; primer used for construction of gldH-containing plasmids pMM286 and pMM293 | |
| 439 | 5′ GCAGCCATAAAGCTTTTTCTAATCTTTTTG 3′; primer used for construction of gldH expression plasmid pJB200 | |
| 452 | 5′ TCTAAAGCCTCAACTCCG 3′; primer used during preparation of gldH probe for Northern blotting | |
| 468 | 5′ ATACTTCTTCTGCAGTGCGATAAAAAAAG 3′; primer used for construction of gldH expression plasmid pJB200 | |
| 512 | 5′ CAAATCTGTTGGAAGTGCC 3′; primer used during preparation of gldH probe for Northern blotting | |
| 558 | 5′ CACGTTTCGATCAGCCGA 3′; primer used during construction of pTB41 | |
| 569 | 5′ TTATTAATGATGATGATGATGATGATGATGTTTCTTAGGTTTATATTTTGACTTTTCG 3′; primer used during construction of pTB39 | |
| 571 | 5′ TTATTAATGATGATGATGATGATGATGATGATCTTTTTGTTCTATTCTAAAACCTACG 3′; primer used during construction of pTB41 | |
| 594 | 5′ GTTTTTGGCAGACAGCATCACC 3′; primer used during preparation of fjo17 probe for Northern blotting |
Antibiotic resistance phenotypes: ampicillin, Apr; chloramphenicol, Cmr; erythromycin, Emr; kanamycin, Kmr; tetracycline, Tcr. Unless indicated otherwise, antibiotic resistance phenotypes are those expressed in E. coli. Antibiotic resistance phenotypes listed in parentheses are those expressed in F. johnsoniae but not in E. coli.
Tn4351 mutagenesis of F. johnsoniae and identification of sites of insertion in gldH.
Tn4351 was introduced into wild-type F. johnsoniae, and nonmotile mutants were isolated as described previously (15). The sites of transposon insertion in the nonmotile mutants CJ778, CJ779, CJ925, and CJ1043 were determined by cloning the Tn4351-disrupted genes essentially as previously described (18) and by inverse PCR (13, 26).
Nucleic acid sequencing.
Nucleic acid sequencing was performed by the dideoxy nucleotide procedure with an automated sequencing system (Applied Biosystems). Sequences were analyzed with MacVector and AssemblyLign software (Accelrys, San Diego, Calif.), and comparisons to database sequences were made by using the BLAST (2) and FASTA (30) algorithms. Predictions regarding cellular localization were made by using PSORT (http://PSORT.NIBB.AC.JP/) (25).
Cloning of gldH and surrounding DNA from wild-type F. johnsoniae.
A library of wild-type F. johnsoniae DNA was constructed in LambdaGEM-11 essentially as previously described (18). Lambda clones containing the regions of interest were detected by hybridization with radiolabeled DNA prepared by using the 5.5-kb HindIII fragment of pMK301 and the Prime-a-Gene labeling kit (Promega, Madison, Wis.). DNA from one of the lambda clones was isolated, and a 6.1-kb SacI fragment, which contained gldH and adjacent genes, was subcloned into pBC SK(+) to generate pMM219. For complementation of the gldH mutants, the 6.1-kb SacI fragment from pMM219 was cloned into the shuttle vector pCP23 to generate pMM224 and pMM225. PCR amplification was used to obtain a product containing gldH and 303 bp of upstream DNA. This 933-bp product was amplified from chromosomal DNA by using primers 432 and 429 and cloned into the EcoRV site of pSTBlue-1 to generate pMM286. The gldH-containing DNA was transferred from pMM286 to pCP23 as a PstI-XbaI fragment to generate pMM293. pMM298 was generated by inserting the 2.1-kb BamHI fragment of pHP45Ωkan, which carries a kanamycin resistance cassette with transcription termination signals on each end, into the SmaI site of pMM293. pMM299 was identical to pMM298 except that the kanamycin resistance cassette was inserted in the opposite orientation. For complementation analyses, plasmids were introduced into the F. johnsoniae mutants by conjugation or electroporation as previously described (15, 23).
Modified versions of gldB and gldH which encode proteins containing eight histidine residues at their C termini were constructed to allow rapid isolation of the recombinant gld proteins from F. johnsoniae cell extracts. gldB was amplified by using primers 24 and 569, and gldH was amplified by using primers 558 and 571. Elongase (Life Technologies) was used for PCR amplifications, and the products were cloned into the SmaI site of pCP23. The resulting plasmids (pTB39 [gldB-his] and pTB41 [gldH-his]) were sequenced to ensure that mutations were not introduced during amplification.
Microscopic observations of cell movement.
Wild-type and mutant cells of F. johnsoniae were examined for movement over glass and agar surfaces and for their ability to propel polystyrene latex spheres by phase-contrast microscopy as previously described (14, 15), except that cells grown to late exponential phase in CYE broth were examined both directly in their growth medium and after harvesting by centrifugation and suspension in 10 mM Tris (pH 7.5), since it was determined that this treatment often resulted in increased binding of latex spheres, increased sphere movement, and increased gliding motility on glass surfaces.
Measurements of phage sensitivity.
Sensitivity to F. johnsoniae phages was determined essentially as previously described by spotting 2.5 μl of phage lysates (6 × 107 PFU/ml) onto lawns of cells in CYE overlay agar (15). The plates were incubated for 24 h at 25°C to observe lysis.
Chitin utilization.
Chitin powder (practical grade from crab shells; Sigma Chemical Co., St. Louis, Mo.) was prepared as a 2% slurry essentially as described previously (31). Three milliliters of 2% chitin was added on top of solid PY2 agar medium in a 9-cm-diameter petri dish, or 7.2 ml of 2% chitin was added on top of PY2 agar in 14-cm-diameter dishes, and the plates were allowed to dry. Wild-type or mutant strains of F. johnsoniae were grown overnight in CYE broth with appropriate antibiotics. Cells from 1 ml of each stationary-phase culture were collected by centrifugation at 7,000 × g for 3 min, washed with 1 ml of a buffer consisting of 10 mM Tris and 8 mM CaCl2 (pH 7.3) (TC buffer), and suspended in 0.5 ml of TC buffer. Two microliters of this suspension (approximately 4 × 107 cells) was spotted on PY2-chitin agar. Plates were incubated at 25°C in a moist chamber.
RNA analysis.
Total RNA was isolated from overnight cultures of F. johnsoniae UW101 by cold phenol extraction (37), and Northern blotting was performed essentially as described previously (36). Probes were prepared by using the DIG RNA labeling kit (Roche Diagnostics Corp., Indianapolis, Ind.). An internal fragment of fjo17 was amplified by using primers 425 and 594, and an internal fragment of gldH was amplified by using primers 452 and 512. Products were separated by agarose gel electrophoresis and ligated into pT7Blue. A second amplification with the T7 primer and either primer 425 (for fjo17) or 512 (for gldH) resulted in products that allowed in vitro transcription to produce the digoxigenin-labeled RNA probes.
Protein expression and antibody production.
A 430-bp fragment coding for the C-terminal 141 amino acids of GldH was amplified by using Taq polymerase and primers 468 and 439 and cloned into pSTBlue-1. The gldH fragment was isolated as a BamHI-HindIII fragment and ligated into pMal-c2 to produce pJB200. Expression of the fusion protein was induced in DH5αMCR by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside. The fusion protein was purified by binding to amylose resin (New England Biolabs, Beverly, Mass.) and was eluted with 10 mM maltose. To generate antibodies, recombinant GldH (100 μg) was mixed with 600 μl of Freund's incomplete adjuvant and injected into female New Zealand White rabbits. The animals were boosted with 50 μg of protein in Freund's incomplete adjuvant every 4 weeks until test bleeds showed suitable reactivity with proteins in Western blot assays. Anti-GldH antiserum was affinity purified essentially as previously described (15).
Cell fractionation and Western blot analyses.
F. johnsoniae cells were disrupted with a French press and fractionated into soluble and membrane fractions as described previously (13). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blot analyses were performed as previously described (15). Antigens were detected by using anti-GldH antiserum, goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate, and Opti-4CN (Bio-Rad, Hercules, Calif.). Protein concentrations were measured with the bicinchoninic acid reagent (Pierce, Rockford, Ill.).
In vivo radiolabeling with [3H]palmitate or [3H]glutamate and detection of labeled proteins.
Cells of F. johnsoniae were labeled with [9,10-3H]palmitate (48 Ci/mmol; Perkin-Elmer Life Sciences, Boston, Mass.) to determine whether GldB, GldD, and GldH were lipoproteins. In parallel experiments, l-[3,4-3H]glutamic acid (51 Ci/mmol; Perkin-Elmer Life Sciences) was used to label essentially all cell proteins. Cells were grown in SDY broth containing appropriate antibiotics at 30°C. When the cell density reached approximately 4 × 108 cells/ml (50 Klett units), [3H]palmitate or [3H] glutamic acid was added to a final concentration of 50 μCi/ml. Incubation was continued for 3 h, by which time the culture had reached approximately 109 cells/ml. Cells were collected by centrifugation and washed four times with TC buffer, and cell pellets were stored at −80°C until they were processed. For analysis of total cell extracts for lipoproteins, cell pellets from 1 ml of culture were solubilized for 3 min at 85°C in 60 μl of a protein loading buffer which consisted of 2% SDS, 10% glycerol, 20 mM dithiothreitol, 0.02% bromophenol blue, and 63 mM Tris (pH 6.8). Protein samples (20 μl/lane) were separated by SDS-PAGE, and labeled bands were detected by autoradiography. For purification of His-tagged lipoproteins, cell pellets from 3 ml of culture were suspended in 100 μl of B-per reagent (Pierce) to which was added 25 μg of lysozyme, 20 μg of DNase, 0.6 mM phenylmethylsulfonyl fluoride, and 1× Halt EDTA-free protease inhibitor cocktail (Pierce). After incubation for 15 min at 24°C, solubilization was completed by adding 11 μl of 10% Triton X-100 and vortexing. One hundred microliters of 50% Ni-nitrilotriacetic acid (Ni-NTA) His-Bind resin (Novagen) in Ni-NTA bind buffer (300 mM NaCl, 1% Triton X-100, 10 mM imidazole, 50 mM Na2HPO4-NaH2PO4 [pH 8.0]) was added, and extracts were incubated with gentle mixing for 30 min at 24°C. The resin was sedimented by centrifugation and washed five times with 1 ml of Ni-NTA wash buffer (300 mM NaCl, 1% Triton X-100, 20 mM imidazole, 50 mM Na2HPO4-NaH2PO4 [pH 8.0]) and once with 300 mM NaCl-1% Triton X-100-10 mM Na2HPO4-NaH2PO4 (pH 8.0). Proteins were eluted with two 100-μl aliquots of 1% SDS-150 mM NaCl-250 mM imidazole-100 mM NaH2PO4 (pH 4.5). The eluted fractions were pooled and neutralized with 3 μl of 1 M Tris (pH 9.1). Samples were prepared for electrophoresis in loading buffer as described above, and equal volumes were separated by SDS-PAGE. The gels were fixed in isopropanol-water-acetic acid (25:65:10) for 60 min, soaked in Amplify (Amersham Biosciences, Piscataway, N.J.) for 60 min, dried under vacuum at 65°C, and exposed to Hyperfilm MP autoradiography film (Amersham Biosciences). Radiolabeled GldD was isolated by immunoprecipitation, since we were unable to construct a stable C-terminal histidine-tagged version of GldD. For immunoprecipitation, cells were lysed as described above, except that 5 mM EDTA was included in the lysis buffer. After lysis, 10 μl of affinity-purified antiserum against GldD was added, and the samples were incubated for 1 h at 25°C. One hundred microliters of a 50% slurry of protein A-agarose (Novagen) was added, and incubation was continued for an additional 30 min at 25°C. Proteins bound to protein A-agarose were collected by centrifugation; washed four times with a buffer consisting of 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, and 50 mM Tris (pH 7.5); and eluted with protein loading buffer by incubation at 85°C for 3 min.
Genetic nomenclature.
Genes required for gliding motility were given the designation gld followed by a letter. Open reading frames (ORFs) that exhibited strong sequence similarity to genes with known function were named after the corresponding genes. ORFs of unknown function that did not exhibit strong similarity to previously described genes were given the provisional designation fjo (for F. johnsoniae ORF) followed by a number.
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the GenBank database under accession number AF527791.
RESULTS
Tn4351 mutagenesis and identification of gldH.
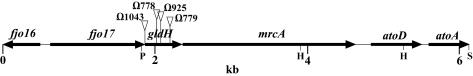
F. johnsoniae was mutagenized with Tn4351, approximately 20,000 erythromycin-resistant transconjugants were screened, and 80 nonmotile mutants were isolated. These mutants formed nonspreading colonies on PY2 agar and lacked the single-cell movements that are characteristic of wild-type cells. Twenty-five of the mutants had defects in cell division resulting in filamentous cells similar to those of ftsX mutants (18). These mutants were not considered further in this study. Southern blot analyses revealed that another 17 mutants had multiple Tn4351 insertions. The remaining 38 mutants were analyzed further. To identify the regions disrupted by Tn4351 in each of the mutants, the DNA flanking the sites of insertion was cloned or amplified as described in Materials and Methods. Sequence analysis revealed that 21 of the mutants had independent insertions in genes that have been previously characterized. Three of the mutants had insertions in gldA (1), 12 had insertions in gldB (15), 2 had insertions in gldF (13), and 4 had insertions in gldG (13). Four of the remaining nonmotile mutants (CJ778, CJ779, CJ925, and CJ1043) contained insertions within a gene that we refer to as gldH (Fig. 1).
FIG. 1.
Map of the gldH region of F. johnsoniae. The sites of the Tn4351 insertions in CJ778, CJ779, CJ925, and CJ1043 are indicated by triangles. Restriction sites: P, PstI; H, HindIII; S, SacI.
The gldH coding region is 477 nucleotides in length. Its primary product contains 159 amino acids and has a predicted molecular mass of 18.1 kDa. The amino-terminal sequence (MRIKNSGILLLAAILLFSCDKKRV) contains a hydrophobic stretch terminated by a cysteine (underlined), which is characteristic of lipoproteins. Lipoproteins undergo a series of modifications that typically result in cleavage of the signal peptide, leaving the cysteine at the amino terminus covalently attached to a fatty acid via an amide bond and to diacyl glycerol through a thioether bond (12). The lipid modifications result in localization to either the cytoplasmic membrane or the outer membrane, depending on the particular protein. GldH does not exhibit sequence similarity to proteins with known function, but it does display similarity to the Bacteroides thetaiotaomicron BT3818 gene product (29% amino acid identity over 160 amino acids) (50), to the putative Cytophaga hutchinsonii GldH (CH2431 gene product [GenBank accession no. ZP_00119033]; 27% identity over 159 amino acids), and to related proteins from Bacteroides fragilis, Tannerella forsythensis, Porphyromonas gingivalis, and Prevotella intermedia. These bacteria are all members of the CFB branch of the bacterial phylogenetic tree. C. hutchinsonii exhibits gliding motility, but the other organisms are apparently nonmotile. All of the GldH homologs listed above display similar N-terminal sequences, suggesting that they may all be lipoproteins.
fjo17 lies upstream of gldH and overlaps the N-terminal coding region by 17 nucleotides. The predicted product of fjo17 does not exhibit amino acid sequence similarity to any proteins of known function in the databases, but it does exhibit similarity to a number of hypothetical proteins, such as the B. thetaiotaomicron BT3819 gene product (50), the C. hutchinsonii CH2430 gene product (GenBank accession no. ZP_00119032), Bacillus subtilis YaaT (20), and Treponema pallidum Tpl (9). B. thetaiotaomicron BT3819 and C. hutchinsonii CH2430 lie just upstream of their respective gldH homologs, indicating a conserved gene order among these members of the CFB group. fjo16 lies 434 bp upstream of the putative fjo17 start codon and is oriented in the opposite direction. fjo16 does not exhibit obvious similarity to sequences in the databases. The predicted mrcA start codon lies 11 nucleotides downstream of the gldH stop codon. The mrcA gene product is similar in sequence to E. coli penicillin binding protein 1A (PBP 1a) (26% identity over 768 amino acids) (3) and to predicted homologs of PBP 1a from other bacteria, such as the B. thetaiotaomicron BT0743 gene product (46% identity over 740 amino acids) (50). E. coli PBP 1a is involved in peptidoglycan synthesis (44). atoD and atoA lie downstream of mrcA. The atoD start codon lies 168 nucleotides downstream of the mrcA stop codon. AtoD and AtoA are similar in sequence to the AtoD and AtoA proteins, respectively, from E. coli (3) and to related proteins from other bacteria. In E. coli these proteins are subunits of acetoacetate coenzyme A transferase (16). Mutations in fjo16, fjo17, mrcA, atoD, or atoA have not been isolated, and we do not know whether these genes are required for gliding motility.
Complementation of gldH mutants.
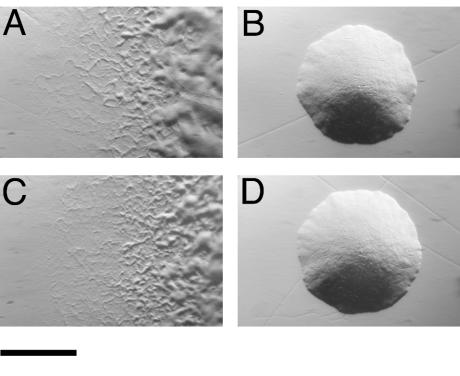
Introduction of pMM224 or pMM225, which carry fjo17, gldH, mrcA, and atoD, into CJ778, CJ779, CJ925, and CJ1043 resulted in complementation of each mutant (data not shown). The resulting colonies spread over agar, and cells exhibited rapid gliding motility. pMM293, which carries just gldH with 303 bp of upstream DNA and 150 bp of downstream DNA, also restored motility to each of the mutants (Fig. 2 and data not shown). Wild-type cells of F. johnsoniae and related bacteria bind latex spheres and propel them along their surfaces (21, 28). Cells of the gldH mutant CJ1043 failed to propel latex spheres. Introduction of pMM293 into CJ1043 restored the ability to propel spheres, in addition to restoring gliding motility.
FIG. 2.
Photomicrographs of F. johnsoniae colonies. Colonies were grown for 2 days at 25°C on PY2 agar medium. Photomicrographs were taken with a Kodak DC290 digital camera mounted on an Olympus IMT-2 inverted microscope. Bar, 1 mm. (A) Wild-type F. johnsoniae. (B) gldH mutant CJ1043. (C) CJ1043 complemented with pMM293, which carries gldH. (D) CJ1043 carrying pMM298.
Pate and colleagues isolated 50 spontaneous and chemically induced nonmotile mutants of F. johnsoniae (5, 47). The mutations responsible for the motility defects of 16 of these mutants have previously been identified (1, 13-15). pMM224 was introduced into the remaining 34 mutants to determine whether any have defects in gldH or in neighboring genes. pMM224 restored motility to two of the mutants (UW102-52 and UW102-64). Colonies of the complemented mutants spread over agar, and cells exhibited wild-type motility in wet mounts. pMM293, which contains just gldH, also restored motility to these two mutants. The fact that pMM224, which contains fjo17, mrcA, and atoD in addition to gldH, did not complement any additional mutants suggests that fjo17, mrcA, and atoD may not be required for gliding. The exact sites of the mutations in UW102-52 and UW102-64 were determined by amplification and sequencing of the gldH region. UW102-52 had an A inserted at position 266 of gldH (numbered from the A of the gldH start codon), while UW102-64 had a large insertion at position 213 of gldH.
gldH appears to be cotranscribed with fjo17.
To determine whether the 933-bp fragment in pMM293 carries the gldH promoter or whether gldH expression was driven by a promoter from the vector, the kanamycin resistance omega fragment (8), which has transcription terminators at each end, was inserted 303 bp upstream of the gldH start codon to block transcriptional readthrough from the vector. The resulting plasmids, pMM298 and pMM299, were identical except that they contained the omega fragment inserted in opposite orientations. pMM298 and pMM299 failed to complement any of the mutants (Fig. 2D and data not shown). These results suggest that gldH expression from pMM293 was driven by a promoter within the vector. The apparent absence of a promoter in the 303-bp fragment upstream of gldH suggests that gldH is normally cotranscribed with fjo17. Northern blot analysis was used to determine the sizes of the gldH and fjo17 transcripts produced by wild-type cells (Fig. 3). A probe constructed internal to fjo17 and one constructed internal to gldH each hybridized to a band of approximately 2.8 kb, providing support for the suggestion that fjo17 and gldH are cotranscribed. The region upstream of gldH in pMM293 is derived from the Flavobacterium psychrophilum plasmid pCP1 (1, 23) (GenBank accession number AY277637). gldH in pMM293 lies downstream of the ORF1 promoter and appears to be expressed from this promoter.
FIG. 3.

Northern blot analysis of F. johnsoniae gldH. Wild-type RNA was separated on an agarose gel, transferred to a nylon membrane, and probed with digoxigenin-labeled RNA internal to fjo17 (lane 1) or to gldH (lane 2). Numbers correspond to the sizes of RNA molecular size markers.
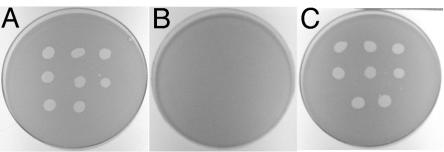
Bacteriophage resistance of gldH mutants.
It has previously been reported that many nonmotile mutants of F. johnsoniae are resistant to infection by a number of F. johnsoniae bacteriophages (46). The reason for this pleiotropy is not known. The sensitivity of F. johnsoniae strains UW101, CJ1043, and CJ1190 (CJ1043 carrying pMM293) to the F. johnsoniae bacteriophages φCj1, φCj13, φCj23, φCj28, φCj29, φCj42, φCj48 and φCj54 was tested (Fig. 4). F. johnsoniae UW101 was readily lysed by these phages, whereas the gldH mutant CJ1043 was resistant to infection by each of them. Introduction of wild-type gldH on pMM293 into CJ1043 restored sensitivity to each of these phages in addition to restoring gliding motility.
FIG. 4.
Effect of mutation in gldH on bacteriophage resistance. Bacteriophages (2.5 μl of lysates containing approximately 6 × 107 phage/ml) were spotted onto lawns of cells in CYE overlay agar. The plates were incubated at 25°C for 24 h to observe lysis. Phages were spotted in the following order from left to right: top row, φCj1, φCj13, and φCj23; middle row, φCj28, φCj29, and φCj42; bottom row, φCj48 and φCj54. (A) Wild-type F. johnsoniae. (B) gldH mutant CJ1043. (C) CJ1043 complemented with pMM293, which carries gldH. The diameter of the petri dish is 9 cm.
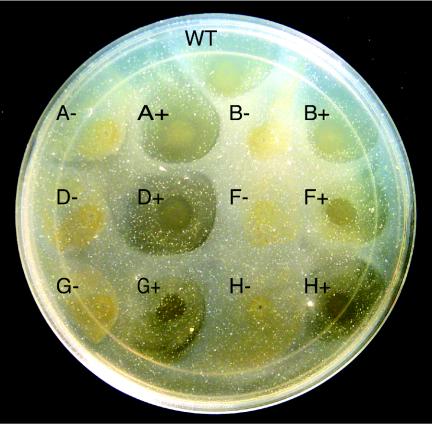
Mutations in gldH eliminate the ability to utilize chitin.
Wild-type cells of F. johnsoniae digest chitin (42). Chang et al. demonstrated that many nonmotile mutants fail to utilize chitin (5). The mutations in these strains were not well defined, and genetic techniques to allow complementation were not available. In this study the ability of wild-type F. johnsoniae, nonmotile mutants, and complemented mutants to digest chitin was analyzed (Fig. 5). Previous studies of chitin digestion by F. johnsoniae typically involved suspending the insoluble polymer in a thin layer of agar and spotting cells on the surface (5). In preliminary experiments we determined that under these conditions chitin was hydrolyzed slowly by our wild-type strain, and there was no noticeable clearing until after 10 days of incubation at 25°C (data not shown). In contrast, when a suspension of chitin was added to the surface of PY2 agar medium and allowed to dry, it was readily attacked by wild-type cells that were spotted directly on the chitin layer (Fig. 5, spot WT), and clearing zones were visible after 2 days of incubation at 25°C (data not shown). Cells of the gldH mutant CJ1043 failed to digest chitin (Fig. 5, spot H−). Complementation with pMM293 restored the ability to digest chitin, in addition to restoring gliding motility (Fig. 5, spot H+). Strains with mutations in other genes that are required for gliding motility, such as gldA (CJ101-288), gldB (CJ569), gldD (CJ282), gldF (CJ787), and gldG (CJ776), also failed to digest chitin (Fig. 5). Complementation of each mutant by introduction of the appropriate plasmid (pSA21 for gldA, pDH233 for gldB, pMM213 for gldD, pMK314 for gldF, and pMK314 for gldG) restored the ability to digest chitin in addition to restoring gliding motility. The clearing zones exhibited by wild-type cells or complemented mutants did not appear to extend beyond the edge of the cell growth. The inability of cells to rapidly digest chitin embedded in agar and the lack of a clearing zone extending beyond the region of cell growth suggest that direct contact with chitin was necessary for efficient utilization.
FIG. 5.
Mutations in gld genes disrupt chitin utilization. Approximately 4 × 107 cells of wild-type and mutant strains were spotted on PY2-chitin agar and incubated at 25°C for 10 days. Abbreviations: WT, wild-type F. johnsoniae; A−, gldA mutant CJ101-288; A+, CJ101-288 complemented with pSA21; B−, gldB mutant CJ569; B+, CJ569 complemented with pDH233; D−, gldD mutant CJ282; D+, CJ282 complemented with pMM213; F−, gldF mutant CJ787; F+, CJ787 complemented with pMK314; G−, gldG mutant CJ776; G+, CJ776 complemented with pMK314; H−, gldH mutant CJ1043; H+, CJ1043 complemented with pMM293. The diameter of the petri dish is 14 cm.
Immunodetection of GldH.
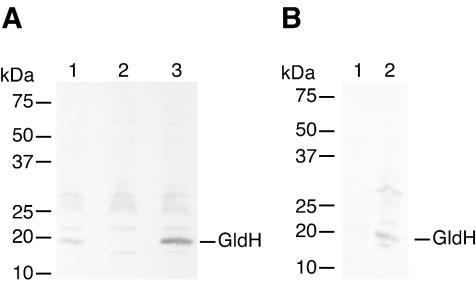
Antisera against GldH were used to detect GldH in cell extracts. GldH, which migrated with an apparent molecular mass of approximately 20 kDa, was detected in extracts of wild-type cells (Fig. 6A, lane 1) but was absent from extracts of the gldH mutant CJ1043 (Fig. 6A, lane 2). Introduction of gldH on pMM293 restored production of GldH (Fig. 6A, lane 3). pMM293 has a copy number of approximately 10 and resulted in overexpression of gldH in F. johnsoniae (compare lanes 1 and 3 of Fig. 6A). GldH was found primarily in membrane fractions of wild-type cells (Fig. 6B). Strains with mutations in other gld genes were used to determine whether these mutations affected GldH protein levels. The level of GldH protein produced by wild-type cells was approximately the same as that produced by strains with mutations in gldA (CJ101-288), gldB (CJ569), gldD (CJ282), and gldFG (CJ787) (data not shown). Western blots developed with antibodies against GldA, GldB, GldD, and GldG were used to determine the effect of a gldH mutation on the level of each of these Gld proteins. The gldH mutants CJ778 and CJ1043 produced approximately the same amounts of each of these Gld proteins as did wild-type cells (data not shown).
FIG. 6.
Immunodetection and localization of GldH. (A) Cell extracts were examined for GldH by Western blot analysis. Lane 1, wild-type F. johnsoniae. Lane 2, CJ1043 (Tn4351 insertion in gldH). Lane 3, CJ1043 with pMM293, which carries gldH. (B) Cell fractions were examined for GldH by Western blot analysis. Lane 1, soluble fraction. Lane 2, membrane fraction.
GldB, GldD, and GldH are minor lipoproteins of F. johnsoniae.
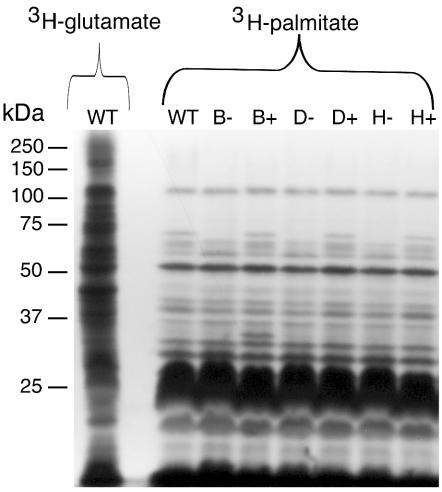
Sequence analysis of the predicted F. johnsoniae GldH revealed a hydrophobic amino-terminal region followed by a cysteine, suggesting that GldH may be a lipoprotein. Inspection of the amino terminal sequences of GldB (MKMYRFVVVLCLFFLSCDQKTKVE) and GldD (MLKKILSVTAILLTLTVVSCKDDV) suggested that these may also be lipoproteins. To detect lipoproteins of F. johnsoniae, cells were labeled with [3H]palmitate as described in Materials and Methods. In a parallel experiment, cells were labeled with [3H]glutamate to label essentially all proteins. A limited subset of total cell proteins of wild-type cells were labeled with [3H]palmitate (Fig. 7) indicating that label from palmitate was not rapidly incorporated into amino acids. The labeled proteins included several abundant lipoproteins that migrated at approximately 25 kDa and a large number of less abundant lipoproteins. Digestion of extracts with proteinase K before separation by SDS-PAGE eliminated all of the labeled bands with apparent molecular masses of greater than 15 kDa, indicating that these were lipoproteins rather than other lipid-containing molecules (data not shown). Digestion of extracts with lysozyme prior to electrophoresis did not alter the pattern of labeled bands, indicating that the detectable lipoproteins were not covalently attached to peptidoglycan (data not shown). Cells with mutations in gldB, gldD, or gldH exhibited lipoprotein profiles that were similar to those of wild-type cells except that a band of approximately 70 kDa was absent or reduced in intensity in each case (Fig. 7, lanes B−, D−, and H−). GldB (approximately 34 kDa), GldD (approximately 22 kDa), and GldH (approximately 20 kDa) are each much smaller than 70 kDa, and Western blot analyses demonstrated that the 70-kDa band was not composed of any of these proteins (data not shown). Complementation of CJ569, CJ282, and CJ1043 with pDH233 (which carries gldB), pMM213 (which carries gldD), and pMM293 (which carries gldH), respectively, restored the level of the 70-kDa lipoprotein to approximately wild-type levels (Fig. 7, lanes B+, D+, and H+). Cells carrying wild-type gldB on pDH233 overproduced a lipoprotein which migrated at approximately 34 kDa (Fig. 7, lane B+), which is close to the previously observed size of GldB (15). pDH233 has a copy number of about 10 and has been shown to overexpress GldB about 10-fold (15). Overexpression of GldD from pMM213 (Fig. 7, lane D+) or of GldH from pMM293 (Fig. 7, lane H+) did not result in a detectable increase in the intensity of any additional band. These plasmids overexpress their respective proteins (14; this study), but these proteins may have been masked by the presence of abundant lipoproteins of similar size.
FIG. 7.
Lipoproteins of F. johnsoniae. Cells of F. johnsoniae were labeled with either [3H]glutamate (to label nearly all proteins) or [3H]palmitate (to label lipoproteins). Proteins were separated by SDS-PAGE and detected by autoradiography. The first lane contains extracts of wild-type cells labeled with [3H]glutamate, whereas the remaining lanes contain extracts of wild-type or mutant cells labeled with [3H]palmitate. Lanes: WT, wild-type F. johnsoniae; B−, gldB mutant CJ569; B+, CJ569 complemented with pDH233; D−, gldD mutant CJ282; D+, CJ282 complemented with pMM213; H−, gldH mutant CJ1043; H+, CJ1043 complemented with pMM293.
To verify that GldB was labeled by [3H]palmitate and to determine whether GldH was labeled, F. johnsoniae strains expressing recombinant GldB or GldH proteins carrying eight histidine residues at their carboxy termini were utilized. Plasmids expressing GldB-His or GldH-His were introduced into F. johnsoniae, and the recombinant proteins were each functional, since they restored motility to the appropriate gld mutants (data not shown). Cells were incubated with [3H]palmitate or [3H]glutamate and extracted, and proteins were isolated by using Ni-NTA His-Bind resin (Novagen) as described in Materials and Methods. The eluted proteins were separated by SDS-PAGE, and radiolabeled proteins were detected by autoradiography. GldB-His and GldH-His were each labeled by [3H]palmitate, suggesting that they are both lipoproteins (Fig. 8A, lanes 1 and 2). Incubation with [3H]palmitate resulted in more intense labeling of the Gld proteins than did incubation with [3H]glutamate (Fig. 8A, lanes 3 and 4), despite the fact that label from [3H]glutamate was more readily incorporated into most proteins (Fig. 7). This suggests that labeling was not the result of metabolism of [3H]palmatate into amino acids before incorporation into GldB-His and GldH-His. Many proteins in addition to GldB-His and GldH-His were detected when extracts from cells labeled with [3H]glutamate were analyzed (Fig. 8A, lanes 3 and 4). These proteins presumably interacted with the Ni-NTA His-Bind resin or with the Gld-His proteins. These proteins were not detected when cells were incubated with [3H]palmitate (Fig. 8A, lanes 1 and 2), presumably because they are not lipoproteins and only lipoproteins incorporated significant amounts of [3H]palmitate.
FIG. 8.
Radiolabeling of GldH-His, GldB-His, and GldD with [3H]palmitate. Cells of F. johnsoniae were labeled with [3H]palmitate or [3H]glutamate. Proteins were isolated by precipitation with Ni-NTA His-Bind resin (A) or by immunoprecipitation (B), separated by SDS-PAGE, and detected by autoradiography. (A) Labeling of His-tagged Gld proteins. Lane 1, gldH mutant CJ1043 expressing GldH-His from pTB41, labeled with [3H]palmitate. Lane 2, gldB mutant CJ569 expressing GldB-His from pTB39, labeled with [3H]palmitate. Lane 3, CJ1043 expressing GldH-His from pTB41, labeled with [3H]glutamate. Lane 4, CJ569 expressing GldB-His from pTB39, labeled with [3H]glutamate. (B) Labeling of GldD. Lane 1, Wild-type F. johnsoniae labeled with [3H]palmitate. Lane 2, gldD mutant CJ282 labeled with [3H]palmitate. Lane 3, CJ282 complemented with pMM213, labeled with [3H]palmitate. Lane 4, wild-type F. johnsoniae labeled with [3H]glutamate. Lane 5, gldD mutant CJ282 labeled with [3H]glutamate. Lane 6, CJ282 complemented with pMM213, labeled with [3H]glutamate.
A plasmid that should express GldD-His in F. johnsoniae was also constructed. GldD was expressed, as determined by Western blot analysis with antisera against GldD and by complementation of a gldD mutant, but repeated attempts to isolate the protein by Ni affinity chromatography or to detect it by using His-Tag monoclonal antibody (Novagen) were unsuccessful. This suggested that the recombinant GldD lacked the C-terminal His tag and raised the possibility that native GldD undergoes carboxy-terminal processing. To determine whether GldD is a lipoprotein, wild-type cells, cells of the gldD mutant CJ282, and cells of CJ282 complemented with pMM213 were labeled with [3H]palmitate as described in Materials and Methods. A labeled protein of approximately 15 kDa was immunoprecipitated from extracts of wild-type cells but was absent from extracts of the gldD mutant (Fig. 8B, lanes 1 and 2). Complementation of the gldD mutant with pMM213, which carries gldD, restored the 15-kDa lipoprotein (Fig. 8B, lane 3). GldD was previously observed to migrate with an apparent molecular mass of approximately 22 kDa (14). The immunoprecipitation procedure used in this study involved incubation of extracts at 25°C, which may have resulted in proteolysis of the full-length protein, producing the 15-kDa species. In any event, the presence of labeled bands in extracts of wild-type and complemented cells and the absence of the labeled band in extracts of gldD mutant cells suggest that GldD, like GldB and GldH, is a lipoprotein.
DISCUSSION
The mechanism responsible for F. johnsoniae gliding motility is unknown. Six genes (gldA, gldB, gldD, gldF, gldG, and ftsX) that are required for gliding have been described (1, 13-15, 18). GldA, GldF, and GldG are thought to constitute an ABC transporter that is required for cell movement. The functions of the other proteins in gliding are not known. The results presented in this paper identify another gene, gldH, which is required for gliding. Transposon insertions in gldH eliminated gliding motility, and introduction of a wild-type copy of gldH on a plasmid restored motility to the mutants.
GldH does not exhibit sequence similarity to proteins of known function, but it is similar to hypothetical proteins from bacteria belonging to the CFB group whose genomes have been sequenced. C. hutchinsonii, which has a GldH homolog, exhibits gliding motility. The other bacteria that have GldH homologs (B. thetaiotaomicron, B. fragilis, T. forsythensis, P. gingivalis, and P. intermedia) are not known to glide. GldH may perform functions unrelated to gliding motility in these bacteria. The requirement of F. johnsoniae GldH for chitin utilization may indicate that the homologs from members of the Bacteroides branch function in polysaccharide utilization, but other functions are also possible. The function of F. johnsoniae fjo17, which lies upstream of and appears to be cotranscribed with gldH, is unknown. The gene order, fjo17-gldH, is conserved in the distantly related bacteria F. johnsoniae, C. hutchinsonii, and B. thetaiotaomicron, which may indicate that the products of these genes have related functions. Analysis of the complete B. thetaiotaomicron genome resulted in the identification of genes that exhibit sequence similarity to F. johnsoniae gldA and gldB in addition to gldH, but genes with obvious similarity to gldD, gldF, and gldG were not identified. T. forsythensis, for which an incomplete genome sequence is available, has apparent homologs to gldA, gldB, gldD, gldF, and gldH but not gldG. From these preliminary analyses it is clear that apparently nonmotile members of the CFB group have genes that are similar to many of the F. johnsoniae gld genes.
Homologs to GldH have not been found outside the CFB group, but gliding motility is found in many bacteria that do not belong to this group. It has recently become clear that there are multiple mechanisms for gliding. Myxococcus xanthus, for example, has two apparently independent mechanisms for movement, i.e., S (social) and A (adventurous) motility. S motility relies on type IV pilus extension and retraction (43, 49), while A motility does not appear to require pili (17) and may depend on polysaccharide secretion (45). Gliding of F. johnsoniae does not appear to be closely related to that of M. xanthus (22). F. johnsoniae moves much more rapidly than M. xanthus and exhibits behaviors such as sphere movement and rotary movements that are not characteristic for M. xanthus. A number of genes that are required for the M. xanthus motility systems or for F. johnsoniae gliding have been identified, but there is little similarity between the known motility genes of these two bacteria.
Sequence analysis suggested that GldH might be a lipoprotein, since it has a hydrophobic N-terminal sequence followed by a cysteine. Inspection of previously described Gld proteins suggested that GldB and GldD might also be lipoproteins. Labeling of cells with [3H]palmitate provided evidence that GldB, GldD, and GldH are lipoproteins. F. johnsoniae produced many additional lipoproteins. One of these is a 70-kDa lipoprotein that is absent or decreased in abundance in cells with mutations in gldB, gldD, or gldH. The 70-kDa lipoprotein may be a component of the motility apparatus that requires GldB, GldD, and GldH for expression or stability. We recently identified a gene, gldJ, which is required for gliding and appears to encode a lipoprotein of approximately this size (T. F. Braun and M. J. McBride, unpublished data). We do not know why such a large fraction of the identified Gld proteins are lipoproteins. The Gld proteins may assemble to form a multiprotein complex in the cell envelope. Anchoring of the appropriate Gld lipoproteins in the cytoplasmic membrane or outer membrane may facilitate formation of such a complex.
The properties of gldH mutants were similar to those of previously studied gld mutants (1, 13-15). In addition to showing complete loss of cell movement and colony spreading, cells of gldH mutants, like those of other gld mutants, failed to propel latex spheres and were resistant to a variety of bacteriophages that infect wild-type cells. Wild-type cells digest chitin, and it has previously been reported that some nonmotile mutants are unable to digest this polysaccharide (5). We determined that disruption of gldA, gldB, gldD, gldF, gldG, or gldH eliminated chitin digestion in addition to gliding motility. The connection between cell movement, phage resistance, and ability to digest chitin is not known, although it is easy to imagine that alterations to the cell surface could affect each process. Bacteriophage infection and chitin utilization each require transport across the cell envelope. Bacteriophage infection requires the export of receptor molecules to the cell surface and the entry of phage nucleic acid into the cell. Chitin utilization requires the transport of chitinase and chitin binding proteins to their sites of action and the transport of breakdown products of chitin into the cell. GldA, GldF, and GldG are required for gliding motility, bacteriophage sensitivity, and chitin utilization and are thought to be components of an ABC transporter. GldA is similar in sequence to Sinorhizobium meliloti NodI (33% identity over 299 amino acids), which is thought to be involved in export of the lipochitin oligosaccharide Nod factor (40, 41). GldA is also similar to components of transporters that transport peptide antibiotics or that are involved in maturation of nitrous oxide reductase, so we cannot conclude that GldA is involved in transport of a compound that is chemically or structurally similar to chitin. Nevertheless, the connection between chitin transport-utilization and gliding is difficult to ignore.
Sugars and polysaccharides have previously been implicated in colony spreading by F. johnsoniae. Mutants with a deficiency in the ability to produce a high-molecular-weight cell surface polysaccharide maintained some ability to glide but formed colonies that spread much less than wild-type colonies (10). Addition of some sugars, including N-acetylglucosamine and glucosamine, severely inhibited colony spreading by wild-type cells but had little if any effect on the ability of cells to glide on glass microscope slides (11, 48). The basis for the defective colony spreading of mutants deficient in cell surface polysaccharides or of wild-type cells exposed to certain sugars is not known.
Many members of the CFB group are proficient at polymer utilization (19, 32, 35, 50). B. thetaiotaomicron utilizes starch and other polysaccharides by a mechanism that may be common to other polymer-utilizing members of the CFB group (35, 38). B. thetaiotaomicron binds the polysaccharide starch on the cell surface. Much of the hydrolysis into smaller units is thought to occur during or after transit of the outer membrane. We suggest a model for CFB gliding motility that could have evolved by modifying such a polysaccharide transport system. In this model, export and uptake of macromolecules at different sites form “conveyor belts” along the cell surface which propel cells. The belts may be individual polymer molecules or fibers composed of multiple molecules, and they could be composed of polysaccharide, protein, or other macromolecules. Export alone could not account for the types of cell surface sphere movements that are observed. Latex spheres move along the cell surface, but they are not propelled away from the cell as would be expected if polymer extrusion propelled cells (21, 28, 34). Propulsion by simple polymer extrusion also could not account for the ability of individual cells to glide rapidly for extended periods of time in the absence of exogenous nutrients (6).
Several cell behaviors may be difficult to account for by the macromolecule export-import model and require further discussion. (i) How would export and import of a macromolecule cause the rotary movements that are sometimes observed when cells attach to a surface by one pole (28)? If the transporters had a rotary component, attachment to these structures at a cell pole could result in cell rotation. Cells attached to the substratum along the length of the cell would be unlikely to rotate, since they would have multiple points of contact that could prevent rotation around any one site. (ii) Several investigators have reported the movement of latex spheres along the length of a cell, around the tip, and back down the other side (21, 28). How can such movements be reconciled with the proposed mechanism? Apparent continuous movement around a cell pole might be explained if the polymers were threaded around the pole, perhaps by attachment to cell surface polymer binding proteins, or if the cell had many short conveyor belts and latex spheres were passed between belts to navigate the pole.
A number of questions remain unanswered. Do the putative exporters and importers of an individual belt coordinate their activities? Does the cell know whether the cargo of an individual transporter is moving toward the head or the tail of the cell? How are multiple conveyor belts coordinated to allow directional cell movement? Proton motive force has been demonstrated to be required for gliding of F. johnsoniae and related bacteria (6, 7, 28, 33). Is proton motive force linked to cell movement through the functioning of a transporter or by some other mechanism? While our data have led us to propose the model described above, other mechanisms put forward to explain gliding have not been ruled out. Lapidus and Berg (21) proposed that outer membrane components are driven along tracks by periplasmic and cytoplasmic membrane proteins that obtain energy from the proton motive force. In this model, a transporter that translocates molecules across the cell envelope may function in assembly or modification of the motility machinery. Further analysis of gldH and of other gld genes and proteins will help to determine the actual mechanism of F. johnsoniae gliding motility.
Acknowledgments
This research was supported by grants from the National Science Foundation (MCB-9727825 and MCB-0130967) and by a Shaw Scientist Award to M.J.M. from The Milwaukee Foundation.
DNA sequencing was performed by the Automated DNA Sequencing Facility of the University of Wisconsin-Milwaukee Department of Biological Sciences. Preliminary sequence data for C. hutchinsonii were obtained from the DOE Joint Genome Institute at http://jgi.doe.gov/. We thank M. Kempf for construction of pMK301 and D. Hunnicutt for assistance in raising anti-GldH antisera.
REFERENCES
- 1.Agarwal, S., D. W. Hunnicutt, and M. J. McBride. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. USA 94:12139-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 4.Chang, L. Y. E., and J. L. Pate. 1981. Nutritional requirements of Cytophaga johnsonae and some of its auxotrophic mutants. Curr. Microbiol. 5:235-240. [Google Scholar]
- 5.Chang, L. Y. E., J. L. Pate, and R. J. Betzig. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 159:26-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duxbury, T., B. A. Humphrey, and K. C. Marshall. 1980. Continuous observations of bacterial gliding motility in a dialysis microchamber: the effects of inhibitors. Arch. Microbiol. 124:169-175. [Google Scholar]
- 7.Dzink-Fox, J. L., E. R. Leadbetter, and W. Godchaux, III. 1997. Acetate acts as a protonophore and differentially affects bead movement and cell migration of the gliding bacterium Cytophaga johnsonae (Flavobacterium johnsoniae). Microbiology 143:3693-3701. [DOI] [PubMed] [Google Scholar]
- 8.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 9.Fraser, C., S. Norris, G. Weinstock, O. White, G. Sutton, R. Dodson, M. Gwinn, E. Hickey, R. Clayton, K. Ketchum, E. Sodergren, J. Hardham, M. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. Cotton, C. Fuji, S. Garland, B. Hatch, K. Horst, K. Roberts, M. Sandusky, J. Weidman, H. Smith, and J. Venter. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 10.Godchaux, W., III, M. A. Lynes, and E. R. Leadbetter. 1991. Defects in gliding motility in mutants of Cytophaga johnsonae lacking a high-molecular-weight cell surface polysaccharide. J. Bacteriol. 173:7607-7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorski, L., W. Godchaux III, and E. R. Leadbetter. 1993. Structural specificity of sugars that inhibit gliding motility of Cytophaga johnsonae. Arch. Microbiol. 160:121-125. [Google Scholar]
- 12.Hayashi, S., and H. C. Wu. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22:451-471. [DOI] [PubMed] [Google Scholar]
- 13.Hunnicutt, D. W., M. J. Kempf, and M. J. McBride. 2002. Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J. Bacteriol. 184:2370-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunnicutt, D. W., and M. J. McBride. 2001. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes gldD and gldE. J. Bacteriol. 183:4167-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunnicutt, D. W., and M. J. McBride. 2000. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes, gldB and gldC. J. Bacteriol. 182:911-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins, L., and W. Nunn. 1987. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J. Bacteriol. 169:42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kempf, M. J., and M. J. McBride. 2000. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J. Bacteriol. 182:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 20.Kunst, F., N. Ogasawara, I. Moszer, A. Albertini, G. Alloni, V. Azevedo, M. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. Brignell, S. Bron, S. Brouillet, C. Bruschi, B. Caldwell, V. Capuano, N. Carter, S. Choi, J. Codani, I. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 21.Lapidus, I. R., and H. C. Berg. 1982. Gliding motility of Cytophaga sp. strain U67. J. Bacteriol. 151:384-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 23.McBride, M. J., and M. J. Kempf. 1996. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 178:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metcalf, W. W., W. Jiang, L. L. Daniels, S.-K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 25.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in gram-negative bacteria. Protein Struct. Funct. Genet. 11:95-110. [DOI] [PubMed] [Google Scholar]
- 26.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pate, J. L. 1988. Gliding motility. Can. J. Microbiol. 34:459-465. [Google Scholar]
- 28.Pate, J. L., and L.-Y. E. Chang. 1979. Evidence that gliding motility in prokaryotic cells is driven by rotary assemblies in the cell envelopes. Curr. Microbiol. 2:59-64. [Google Scholar]
- 29.Pate, J. L., S. J. Petzold, and L.-Y. E. Chang. 1979. Phages for the gliding bacterium Cytophaga johnsonae that infect only motile cells. Curr. Microbiol. 2:257-262. [Google Scholar]
- 30.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 31.Reichenbach, H. 1992. The genus Lysobacter, p. 3256-3275. In A. Balows, H. Truper, M. Dworkin, W. Harder, and K. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 32.Reichenbach, H. 1992. The order Cytophagales, p. 3631-3675. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. M. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 33.Ridgway, H. F. 1977. Source of energy for gliding motility in Flexibacter polymorphus: effects of metabolic and respiratory inhibitors on gliding movement. J. Bacteriol. 131:544-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ridgway, H. F., and R. A. Lewin. 1988. Characterization of gliding motility in Flexibacter polymorphus. Cell Motil. Cytoskel. 11:46-63. [DOI] [PubMed] [Google Scholar]
- 35.Salyers, A. A., A. Reeves, and J. D'Elia. 1996. Solving the problem of how to eat something as big as yourself: diverse bacterial strategies for degrading polysaccharides. J. Indust. Microbiol. 17:470-476. [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Sawers, G., and A. Bock. 1989. Novel transcriptional control of the pyruvate formate-lyase gene: upstream regulatory sequences and multiple promoters regulate anaerobic expression. J. Bacteriol. 171:2485-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shipman, J. A., J. E. Berleman, and A. A. Salyers. 2000. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J. Bacteriol. 182:5365-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 2:784-791. [Google Scholar]
- 40.Spaink, H. P. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54:257-288. [DOI] [PubMed] [Google Scholar]
- 41.Spaink, H. P., A. H. M. Wijfjes, and B. J. J. Lugtenberg. 1995. Rhizobium NodI and NodJ proteins play a role in the efficiency of secretion of lipochitin oligosaccharides. J. Bacteriol. 177:6276-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanier, R. Y. 1947. Studies on non-fruiting myxobacteria. I. Cytophaga johnsonae, n. sp., a chitin-decomposing myxobacterium. J. Bacteriol. 53:297-315. [PMC free article] [PubMed] [Google Scholar]
- 43.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143-1146. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, H., Y. Nishimura, and Y. Hirota. 1978. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc. Natl. Acad. Sci. USA 75:664-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolgemuth, C., E. Hoiczyk, D. Kaiser, and G. Oster. 2002. How myxobacteria glide. Curr. Biol. 12:369-377. [DOI] [PubMed] [Google Scholar]
- 46.Wolkin, R. H., and J. L. Pate. 1986. Phage adsorption and cell adherence are motility-dependent characteristics of the gliding bacterium Cytophaga johnsonae. J. Gen. Microbiol. 132:355-367. [Google Scholar]
- 47.Wolkin, R. H., and J. L. Pate. 1985. Selection for nonadherent or nonhydrophobic mutants co-selects for nonspreading mutants of Cytophaga johnsonae and other gliding bacteria. J. Gen. Microbiol. 131:737-750. [Google Scholar]
- 48.Wolkin, R. H., and J. L. Pate. 1984. Translocation of motile cells of the gliding bacterium Cytophaga johnsonae depends on a surface component that may be modified by sugars. J. Gen. Microbiol. 130:2651-2669. [Google Scholar]
- 49.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547-558. [DOI] [PubMed] [Google Scholar]
- 50.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]