Abstract
Background
Fruits from several species of the Rosaceae family are reported to cause allergic reactions in certain populations. The allergens identified belong to mainly four protein families: pathogenesis related 10 proteins, thaumatin-like proteins, lipid transfer proteins and profilins. These families of putative allergen genes in apple (Mal d 1 to 4) have been mapped on linkage maps and subsequent genetic study on allelic diversity and hypoallergenic traits has been carried out recently. In peach (Prunus persica), these allergen gene families are denoted as Pru p 1 to 4 and for almond (Prunus dulcis)Pru du 1 to 4. Genetic analysis using current molecular tools may be helpful to establish the cause of allergenicity differences observed among different peach cultivars. This study was to characterize putative peach allergen genes for their genomic sequences and linkage map positions, and to compare them with previously characterized homologous genes in apple (Malus domestica).
Results
Eight Pru p/du 1 genes were identified, four of which were new. All the Pru p/du 1 genes were mapped in a single bin on the top of linkage group 1 (G1). Five Pru p/du 2 genes were mapped on four different linkage groups, two very similar Pru p/du 2.01 genes (A and B) were on G3, Pru p/du 2.02 on G7,Pru p/du 2.03 on G8 and Pru p/du 2.04 on G1. There were differences in the intron and exon structure in these Pru p/du 2 genes and in their amino acid composition. Three Pru p/du 3 genes (3.01–3.03) containing an intron and a mini exon of 10 nt were mapped in a cluster on G6. Two Pru p/du 4 genes (Pru p/du 4.01 and 4.02) were located on G1 and G7, respectively. The Pru p/du 1 cluster on G1 aligned to the Mal d 1 clusters on LG16; Pru p/du 2.01A and B on G3 to Mal d 2.01A and B on LG9; the Pru p/du 3 cluster on G6 to Mal d 3.01 on LG12; Pru p/du 4.01 on G1 to Mal d 4.03 on LG2; and Pru p/du 4.02 on G7 to Mal d 4.02 on LG2.
Conclusion
A total of 18 putative peach/almond allergen genes have been mapped on five linkage groups. Their positions confirm the high macro-synteny between peach/almond and apple. The insight gained will help to identify key genes causing differences in allergenicity among different cultivars of peach and other Prunus species.
Background
In general, eating fruit is strongly recommended to improve health, but this can cause an allergic reaction in certain individuals. Most fruit crops in temperate and some subtropical zones belong to the Rosaceae family, mainly the subfamilies Maloideae and Prunoideae. Fruits from the Maloideae, like apple [1,2] and pear [3], and Prunoideae such as peach [4,5], sweet cherry [6,7], plum [8], apricot [9] and almond [10] have been reported to cause allergic reactions in Europe. The annual peach production in China has been around 6 million tons in recent years, and has lead to both an increase in peach consumption and more prevalent fruit allergy.
It is considered that general avoidance of fruit has a negative effect on health in allergic patients and also affects their quality of life. Therefore, the low allergic property of fruit is worth considering in new peach breeding programs. As cultivars of the same species differ in allergenicity [11], the selection and breeding of new, low-allergenic cultivars is becoming feasible by a multidisciplinary approach, such as in the EU-SAFE project [1], where doctors and plant geneticists identified the apple cultivar 'Santana' as low allergenic in the majority of Dutch patients with a history of allergy to apple [12]. A similar approach was initiated in peach in recent years through collaboration between China and Europe.
A variety of allergens from different fruits have been identified by means of experimental immunology and molecular biology, in particular by protein and gene identification and sequencing[13]. From the structural and biochemical point of view, fruit allergens identified belong to a limited number of protein families [13]. In the case of Rosaceae, the majority of allergens present in fresh fruit belong to four families: pathogenesis-related protein 10 (PR-10 protein, birch allergen Bet v 1 homologues), thaumatin-like proteins (TLP, PR-5 proteins), non-specific lipid transfer proteins (nsLTPs, PR-14 proteins) and profilins (PRF) [13]. Whole, unprocessed fruit containing PR-10 protein elicit allergy, while TLP, LTP and profilins are stable proteins because of intra-molecular disulfide bonds. These four allergen families in Prunus fruits are already on the international allergen list http://www.allergen.org. In peach they are denoted as Pru p 1 (PR10), Pru p 2 (TLP), Pru p 3 (LTP), and Pru p 4 (profilin), and in almond as Pru du 1, 2, 3, and 4, respectively. Due to the common structure and properties of each protein family over a wide range of plant species, genera and even families, allergy cross-reactivity has been frequently observed, for example between Pru p 1 and Mal d 1 proteins from peach and apple respectively with Bet v 1 proteins in birch pollen [14]. Peach allergy occurs more often in southern Europe, and two different peach allergy profiles have been observed in Spain. One of these is the oral allergy syndrome (OAS) caused by Pru p1 and Pru p 4, the other is the systemic gastrointestinal symptom caused by Pru p 3 [15]. In China both profiles have also been reported after eating fresh peach [16]. In addition, some contact with peach fruit hair or even with pollen during the blooming season caused adverse reactions (from some patients' claims).
Genomic characterization of the sequences and linkage map positions of the four apple allergen gene families were reported in 2005 [17-19], and subsequent associations between the genetic constitution and allergenicity of different cultivars gave the first indications on the isoallergens (genes) and protein variants (alleles) involved in low and high allergenic responses [20]. Comparison between apple (Malus domestica) and Prunus linkage maps suggests a high level of synteny of the linkage groups between these two genera [21]. For Prunus G1, the upper part contains several RFLP markers that aligned with apple LG13 and LG16 on which the majority of Mal d 1 genes are located, and the lower part aligned with LG8, where Mal d 4.03A is located. Prunus G3 aligns with apple LG9 where apple Mal d 2.01A/B and Mal d 4.01 were mapped. Therefore, some of the homologous allergen genes of Prunus fruit may be located at syntenic positions as compared with the apple genome.
In peach, differences in major allergen Pru p 1 (PR-10) and Pru p 3 (LTP) content among cultivars have been reported in recent years [11,22], which encouraged us to conduct research similar to that done in apple [17-19]. Advances in three aspects make the genomic research of Prunus allergen genes possible. First, a set of candidate genes is now available based on previous medical and molecular biology research [23-25]. Second, a large number of peach fruit ESTs are available in databases (mainly from researchers in Chile, Italy and USA), allowing us to find different members of the four putative allergen gene families. Third, a highly saturated genetic Prunus reference map T × E (based on the F2 progeny of 'Texas' almond and 'Earlygold' peach) and a simple bin-mapping strategy [26] provides an efficient and reliable way to map new genes.
Results
PCR-based cloning and individual clone sequencing with genomic DNA templates of the Prunus T × E F1 hybrid (MB1-73, the plant used for the generation of the T × E F2 population by selfing), produced 35 consensus sequences (GenBank: EU424239-EU424273) represent 18 different gene loci. Within a single gene locus, two putative alleles from peach and almond were distinguished through marker test and sequence comparison. The total length of each sequence and length of its open reading frames (ORFs), exon and intron are summarized in Table 1. Primers used for linkage mapping are presented in Table 2. Each gene family is described below in more detail.
Table 1.
Basic genomic features of four putative allergen genes identified from the MB1-73 plant (F1 hybrid between 'Texas' almond and 'Earlygold' peach)
| Genes1 | Total length2 | ORF | Exon 12 | Intron 12 | Exon 22 | Intron 22 | Exon 3 | GenBank accessions3 | Bin-map4 |
| Pru p/du 1.01 | 586 | 483 | 184 | 98 | 299 | EU424239, EU424240 | 1:14 | ||
| Pru p/du 1.02 | 772/762 | 483 | 184 | 233/223 | 299 | EU424241, EU424242 | 1:14 | ||
| Pru p/du 1.03 | 624 | 483 | 184 | 105/102 | 299 | EU424243, EU424244 | 1:14 | ||
| Pru p/du 1.04 | 615/622 | 480 | 184 | 124/131 | 296 | EU424245, EU424246 | 1:14 | ||
| Pru p/du 1.05 | 1503/1510 | 483 | 184 | 135 | 299 | EU424247, EU424248 | 1:14 | ||
| Pru p/du 1.06A | 1871/2096 | 483 | 184 | 105/109 | 299 | EU424251, EU424252 | 1:14 | ||
| Pru p/du 1.06B | 589 | 483 | 184 | 105 | 299 | EU424249, EU424250 | 1:14 | ||
| Pru p 1.06C | 1432 | 483 | 184 | 109 | 299 | EU424253 | 1:14 | ||
| Pru p/du 2.01A | 882/879 | 741 | 61 | 105/108 | 680 | EU424256, EU424257 | 3:37 | ||
| Pru p/du 2.01B | 892 | 741 | 61 | 118 | 680 | EU424258, EU424259 | 3:37 | ||
| Pru p/du 2.02 | 1373/1271 | 741/729 | 52/58 | 118/180 | 689/671 | EU424254, EU424255 | 7:25 | ||
| Pru p/du 2.03 | 1296/1290 | 783/834 | 61 | 160 | 722/773 | EU424260, EU424261 | 8:41 | ||
| Pru p/du 2.04 | 1799/1825 | 993 | 61 | 110 | 694 | 678/704 | 238 | EU424262, EU424263 | 1:50 |
| Pru p/du 3.01 | 645 | 354 | 344 | 193 | 10 | EU424264, EU424265 | 6:74 | ||
| Pru p/du 3.02 | 514/513 | 372 | 362 | 98/97 | 10 | EU424266, EU424267 | 6:74 | ||
| Pru p/du 3.03 | 528/527 | 351 | 341 | 104/103 | 10 | EU424268, EU424269 | 6:74 | ||
| Pru p/du 4.01 | 1041 | 396 | 123 | 391 | 138 | 210 | 135 | EU424270, EU424271 | 1:73 |
| Pru p/du 4.02 | 754 | 396 | 123 | 209 | 138 | 131 | 135 | EU424272, EU424273 | 7:56 |
1 p and du represent peach (P. persica) and almond (P. dulcis), respectively.
2 x/y indicates two different DNA lengths from 'Texas' and 'Earlygold' allele, the first is for 'Texas'.
3 Two accessions are from alleles of 'Texas' almond and 'Earlygold' peach respectively.
4 Bin map position is indicated by a first digit corresponding to the linkage group number followed by a colon and the distance in cM from the top of the linkage group to the last marker of this bin (i.e. 1:14 is linkage group 1 position 14 cM)
Table 2.
Markers developed and cloning and sequencing primers for mapping
| Gene | Primer sequence1 | Tm °C2 | Cycles3 | Allele length (nt)4 |
| Pru du 1.01 | For-ATGGGTGTCTTCACATATGAGAG Rev-CTGACAACATATATACTGTAACC |
58 | 35 | T-218 |
| Pru p 1.01 | For-TCATACAGCTACACCTTGTT Rev-CAATGAGCTTGAAGAGATTTG |
58 | 35 | E-205 |
| Pru p/du 1.02 | For-GTTGGAACCATCAAGAAGGAC GTTTCATGAGCATACAGAACTATC |
58 | 35 | T-188 E-179 |
| Pru p/du 1.03 | For-GGTGAAGGTTAGCTAGTTGA Rev-GTTTCCATCCCTGTGCTTCACAA |
58 | 35 | T-153 E-150 |
| Pru p/du 1.04 | For-CTTTGGTGAAGGTAAGTTTAGA Rev-GTTTCATACCTGTAAGTGCTACCTGC |
58 | 35 | T-157 E-164 |
| Pru p/du 1.05 | For-GCTTCCAACATCAAGTAATACCT Rev-GTTTGGGACTAGGTTGTCAGCA |
60 | 35 | T-179 E-182 |
| Pru p/du 1.06A | For-TAGTTATGAGTTGCTTGCAATGCT Rev-GAAAGTTCCAAAGTACATGTGC |
58 | 35 | 1871, sequencing |
| Pru du 1.06B | For-ATCCCCAAGATTGATCCCCTG Rev-GATTAGAATTTAAGAGCTTACTGT |
60/58 | 5/35 | T-182 |
| Pru p 1.06B | For-AAAGCCCTTGTTCTTGAAGCG Rev-GATTAGAATTTTAGAGCTTACTCC |
60/58 | 5/35 | E-212 |
| Pru p/du 1.06C | For-CTAATTAATAACATAAATCTTGAAACT Rev-TTGATCTCAACATCCCCTGC |
56/54 | 5/35 | E-272 |
| Pru du 2.01A | For-CTTAGCATTCAACCAGCCTC Rev-GATGAGGTAGTCAGGGCAAG |
60 | 35 | T-175 |
| Pru p 2.01A | For-CAATTGTCTCTGTAATCTGTTT Rev-ACGGAGCGTCCACAGAGTT |
60 | 35 | E-205 |
| Pru du 2.01B | For-TTGCCCTGCCAATGTTAACGC Rev-GCACTGGGTCTTAAAGAACTG |
60 | 35 | T-187 |
| Pru p 2.01B | For-CAATTGTCTCTGTAATCTTTAG Rev-GGTCTTGAAGAGCTTAGAGTGA |
60 | 35 | E-622 |
| Pru p/du 2.02 | For-AAGAATCCATCAACTGAATCC Rev-CTACTAGGGTCTTCATCATCGG |
56 | 40 | T-271 E-315 |
| Pru p/du 2.03 | For-ACATGCGCCTCTGCGGACTA Rev-CCATTTCTTGCACGATTCAA |
60 | 30 | T-237 E-229 |
| Pru p/du 2.04 | For-TCAGTGTCAGGCTGTCAGTG Rev-CTCCAGATTCAGAGAGGTGC |
60 | 35 | T-267 E-235 |
| Pru p/du 3.01 | For-CACCATGCATACCCTACGTG Rev-CGTGAGGAATCCCTAAGTGG |
60 | 35 | 530, sequencing |
| Pru p/du 3.02 | For-CTTACCCCAATGCAAATGCT Rev-CCACTGAAACCAACAGCAGA |
60 | 35 | 319, sequencing |
| Pru p/du 3.03 | For-AGGCAGCCCTTACTTGTCCT Rev-CCACATGCTTCTCCGTATCA |
60 | 35 | 413, sequencing |
| Pru p/du 4.01 | For-AGAAGAAATCAGAAGCAACG Rev1-CAAGATACAACCCAGTTGCG Rev2-CTTTGTCCCACCAAGGAATAGA |
55 | 35 | T-276 E-631 |
| Pru du 4.02 | For-AAGTACATGGTCATCCAAGCC Rev-GTCCCTGAAATTGAAGTGTC |
58 | 35 | T-188 |
| Pru p 4.02 | For-GGCAACCACCTCTCTCCT Rev-CAAGATACAACCCAGTTGCG |
55 | 35 | E-363 |
1Designed mismatching nucleotides are in bold and underlined, added pigtail bases in bold and italics.
2, Annealing temperature (Tm)
3Number of PCR cycles. In the case of two values, a touch-down PCR was performed in two steps: the first number of the "Tm" and "Cycles" column refers to the Tm and number of cycles, respectively, of the first step.
4T and E refer to parental source from 'Texas' almond and 'Earlygold' peach, respectively. Genes mapped by direct sequencing are indicated by 'sequencing'
Pru p/du 1 (PR-10) genes
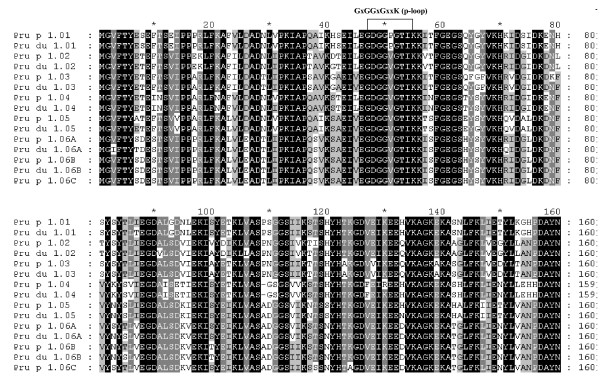
According to four Pru p 1 cDNA reference sequences derived from peach fruit in the GenBank database, we obtained 15 different PR-10 DNA sequences [GenBank: EU424239-EU424253] from MB1-73 by PCR cloning, and classified them into eight distinct gene members based on their DNA sequence identity less than 98%, of which four were new (Pru p/du 1.05 and Pru p/du 1.06A-C) (Table 1) and formed a new group distinct from the initial reference sequence [GenBank:DQ251187] (Pru p 1.01). All Pru p/du 1 genes had a single intron with variable size, (98–233 nt) and two exons of 184 nt and 299 nt each, except in the second exon in Pru p/du 1.04 that was 296 nt long. Alleles derived from almond cv 'Texas' and peach cv 'Earlygold' had different intron sizes in four Pru p/du 1 members (Table 1), used to map these genes in polyacrylamide gel electrophoresis (PAGE). DNA sequence divergence between allele from peach and almond is very low (1%–2.1%). Amino acid (aa) sequences from these genes were deduced from their coding DNA sequences (Figure 1). Most Pru p/du 1 had 160 aa, except for Pru p/du 1.04 with 159aa, with the common feature of a p-loop (GxGGxGxxK). Overall aa sequence identity among different isoallergen was 71.9%–98.8%, and three Pru p 1.06 members (A, B, C) have about 98% aa sequence identity (Figure 1). Segments of identical aa sequence were found in several locations, especially surrounding the p-loop. Pru p/du 1.01 had a proline (P) residue in the p-loop, which is similar to Mal d 1.01 and 02 and all Bet v 1, while the other Pru p/du 1 isoallergens had a valine (V) in this position. The predicted protein molecular weights were in the range of 17.1–17.5 kDa, and pI values ranged from 4.9 to 6.0. Comparing the aa composition between alleles from peach and almond of each gene in Figure 1, we found that there was only one difference for Pru p/du 1.01, 02, 03, 04 and 06B, while Pru p/du 1.05 and Pru p/du 1.06A differed by 2 and 5 aa, respectively. A phylogenetic tree of the deduced amino acids from peach cv. 'Earlygold' alleles and those in apple (mainly cv. 'Fiesta') (Figure 2A) showed that at least one Pru p 1 isoallergen member is present in each of the four subfamilies of apple Mal d 1 genes (PR-10) identified in a previous study [17]. Furthermore, we can see in the subfamily III branch that Pru p 1.05 and three highly similar Pru p 1.06 members (A-C) clustered with three Mal d 1.06 members (A-C), while Pru-1.04 clustered together with Mal d 1.07, one of the apple intronless genes in subfamily IV. All eight Pru p/du 1 genes mapped to bin 1:14, at the top of G1 (Figure 3A, Table 1).
Figure 1.
Alignment of amino acid sequences of Pru p/du 1 members identified in the Prunus 'Texas' almond × 'Earlygold' peach plant MB1-73. p, peach, du, almond. Four levels of identity are shown: (1) black, 100%; (2) grey, 80%; (3) light grey, 60%; and (4) white.
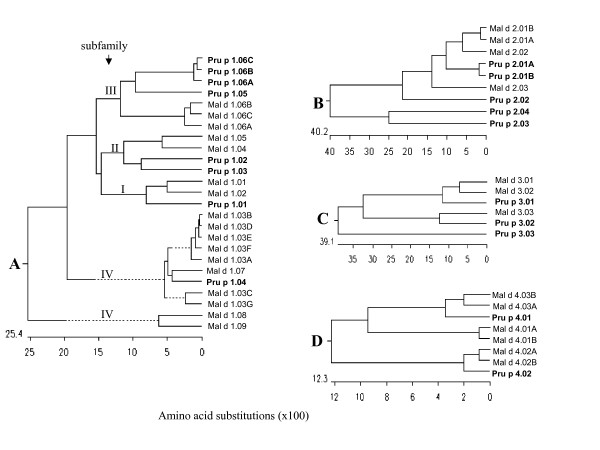
Figure 2.
Phylogenetic trees derived from the deduced PR-10, TLP, LTP and profilin (PRF) families in peach (Pru p 1–4) and apple (Mal d 1–4) based on our genomic sequences and ESTs (in bold) retrieved from the DNA NCBI Database (see their GenBank accession numbers below). A: PR-10 family. Peach: Pru p 1.01 [GenBank:EU424240], Pru p 1.02 [GenBank:EU424242], Pru p 1.03 [GenBank:EU424244], Pru p 1.04 [GenBank:EU424246], Pru p 1.05 [GenBank:EU424248], Pru p 1.06A [GenBank:EU424252], Pru p 1.06B [GenBank:EU424250], Pru p 1.06C [GenBank:EU424253]. Apple: Mal d 1.01 [GenBank:AY789238]; Mal d 1.02 [GenBank:AY789241]; Mal d 1.03A [GenBank:AY789263]; Mal d 1.03B [GenBank:AY789265]; Mal d 1.03C [GenBank:AY789266]: Mal d 1.03D [GenBank:AY789267], Mal d 1.03E [GenBank:AY789269], Mal d 1.03F [GenBank:AY789271], Mal d 1.03G [AY789274], Mal d 1.04 [GenBank:AY789244], Mal d 1.05 [GenBank:AY789246], Mal d 1.06A [GenBank:AY789249], Mal d 1.06B [GenBank:AY789251], Mal d 1.06C [GenBank:AY789254], Mal d 1.07 [GenBank:AY789257], Mal d 1.08 [GenBank:AY789260], Mal d 1.09 [GenBank:AY789262]. B: TLP family. Peach: Pru p 2.01A [GenBank:EU424257], Pru p 2.01B [GenBank:EU424259], Pru p 2.02 [GenBank:EU424255], Pru p 2.03 [GenBank:EU424261], Pru p 2.04 [GenBank:EU424263]. Apple: Mal d 2.01A [GenBank:AY792599], Mal d 2.01B [GenBank:AY792603]; Mal d 2.02 (consensus sequence derived from ESTs, [GenBank:CO9044477, CV082311, CO723595, CN445021] and Mal d 2.03 (consensus of 8 ESTs: [GenBank:CO866703, CO901275, CN495042, CV084040, CO866711, CN444138, CO866347, CN491711] [18]. C: LTP family: Peach: Pru p 3.01 [GenBank:EU424265], Pru p 3.02 [GenBank:EU424267], Pru p 3.03 [GenBank:EU424269]; Apple: Mal d 3.01 [GenBank:AY572501], Mal d 3.02 [GenBank:AY572517], Mal d 3.03 [GenBank:DY256246]. D: Profilin Family: Peach: Pru p 4.01 [GenBank:EU424271], Pru p 4.02 [GenBank:EU424273]; Apple: Mal d 4.01 [GenBank:AY792608], Mal d 4.02A [GenBank:AY792610], Mal d 4.02B (consensus of 3 ESTs, [GenBank:CN578860, CN992900, CV082623], Mal d 4.03A [GenBank:AY792616], Mal d 4.03B (consensus of 3 ESTs, [GenBank:CO722645, CO052497, CO418275].
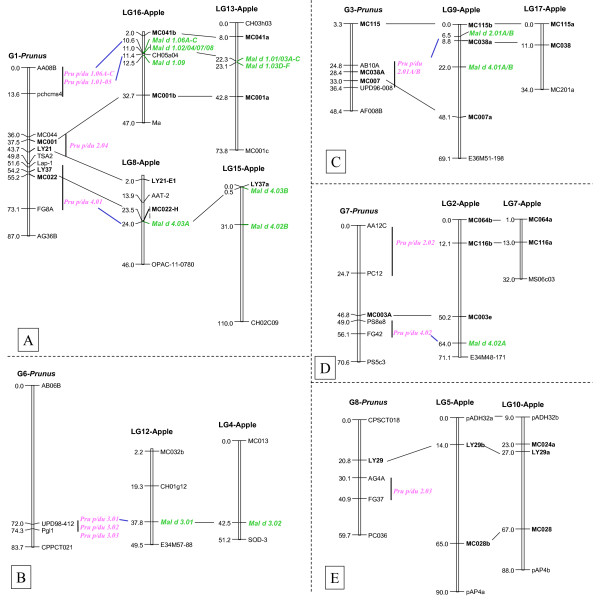
Figure 3.
Linkage map positions of the four allergen gene families in Prunus (T × E) reference map and alignment with those in apple (Malus domestica) reference map. Relevant linkage map information was derived from Prunus T × E updated maps (Arús et al.) and apple (Maliepaard et al., TAG, 1998, 97:60–73; Gao et al 2005, TAG 110:479–491; 111:174–183 and 1087–1097). An initial reference for alignment is based on Dirlewanger et al. (PNAS, 2004, 101:9891–9896). A bar indicates map position range of a bin. Homologous allergen genes and common RFLP probes are underlined to show their synteny.
Pru p/du 2 (TLP)
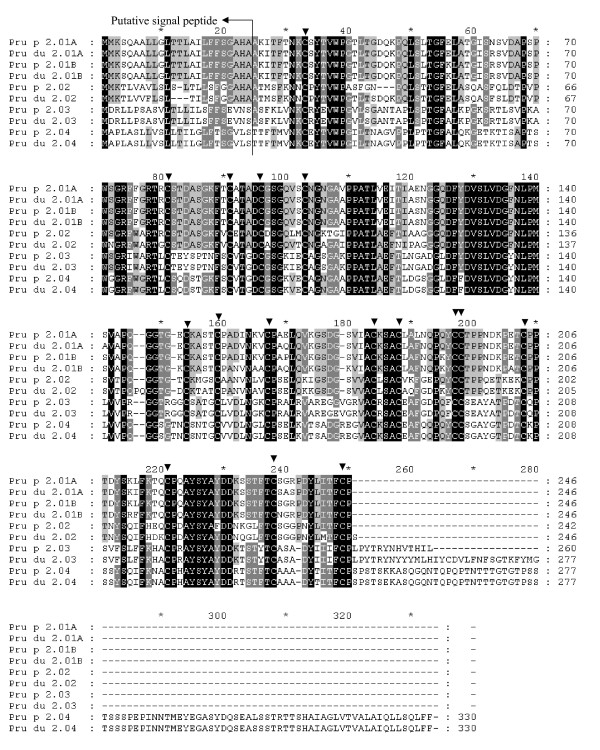
Analysis of the peach TLP sequences in the NCBI identified four members of this gene family at the start. A single primer pair based on [GenBank:AF362988] resulted in four very similar sequences assigned as two separate genes (Pru p/du 2.01A and 2.01B) with minor intron size differences. Genome walking to upstream was employed in Pru p/du 2.02 to search for more polymorphisms and downstream to Pru p/du 2.03, since its initial reference sequence was partial. Five members of Prunus TLPs were identified after analysis of the genomic sequences [GenBank: EU424254-EU424263]: Pru p/du 2.01A and -01B, Pru p/du 2.02, -03 and -04 (Table 1), of which Pru p/du 2.01B is new. The Pru p/du 2 members had distinct genomic structure (Table 1). Three of them had one intron separating a short (58 or 61 nt) and long (671–773 nt) exon, while one member (Pru p/du 2.04) had two introns. Comparing the deduced precursor of Prunus TLPs with the reference TLP amino acid sequences in the Rosaceae and other plants, we assigned the putative signal peptides to deduce the mature protein (Figure 4). Pru p/du 2.02 had a shorter signal peptide, 23 aa in peach and 21 aa in almond, while the other four members had a 24 aa signal peptide. They differed considerably in protein size, with Pru p/du 2.03 and Pru p/du 2.04 having longer peptides in the C-terminal than Pru p/du 2.01 and 02. Amino acid sequences identity from alleles from peach and almond for five Pru p/du 2 members are different, they are 84.4% for Pru p/du 2.02, about 96% for Pru p/du 2.01A and -01B, 98% for Pru p/du 2.03, and 99.4% for Pru p 2.04 p/du 2.04.
Figure 4.
Alignment of amino acid sequences of Pru p/du 2 members identified in the Prunus 'Texas' almond × 'Earlygold' peach hybrid MB1-73. Four levels of identity are shown: (1) black, 100%; (2) grey, 80%; (3) light grey, 60%; and (4) white. The signal peptides were deducted by comparison with other TLPs in Rosaceae fruits or other plants. 16 conserved cysteine residues are arrowed.
Based on the percentage amino acid identity among five Pru p 2 isoallergens, Pru p 2.01A, 01B and 2.02 were clustered with four apple Mal d 2 (01A, 01B, 02 and 03) (Figure 2B) proteins. Pru p 2.01A and 2.01B had 96.7% identity in their aa sequence, their closest homologous members being apple Mal d 2.01A and B. Pru p 2.03 and 2.04 are quite distant from 2.01 and 2.02 (Figure 2B) and no close counterparts were found in apple. The overall sequence identity level in amino acids among the four Pru p 2 proteins was around 40% (Figure 2B), but a long sequence region (DFYDVSLVDG [F/Y]NLPM) around position 130, and the typical 16 cysteine (C) to form 8 disulfide bonds, were conserved in all Pru p/du 2 members. Mature Pru p 2.01A, 02 and 04 proteins were acidic (pI around 4–5), whereas Pru p 2.01B and 3 were basic (pI 7.9–8.17). Mature Pru p 2.01 and 02 had a molecular weight of about 23 kDa. Mature Pru p 2.03 differed in length and weight with mature Pru du 2.03, with the molecular weight being 25.23 kDa and 27.46 kDa respectively. Mature Pru 2.04 had the highest number of aa (307) with a calculated molecular weight of 30.7 kDa. Unlike the Pru p 1 genes that were placed in a single bin, these five Pru p/du 2 members mapped on four linkage groups: Pru p/du 2.01 A and B on G3, -2.02 on G7, -2.03 on G8, and -2.04 on G1 (Figure 3A, C, D, E).
Pru p/du 3 (nsLTP)
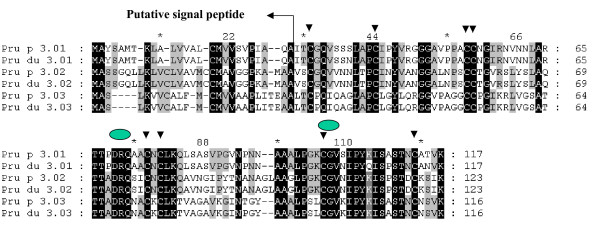
Four peach and almond nsLTP members were traced from the NCBI DNA database, three of them have complete coding sequences, the fourth (GenBank:BQ641139) is a partial almond cDNA sequence distant from the other three. Genomic cloning and sequencing was done with the first three members Pru p/du 3.01-03 [GenBank: EU424264-EU424269]. Pru p/du 3 gene sequences have two exons and one intron (Table 1). The size of the second exon was conserved and extremely short (10 nt), while the first was variable (344, 362 and 341 nt). Sequences from peach and almond LTPs are highly conserved in exon and intron size, with only one base pair length difference in introns of Pru p/du 3.02 and 3.03 (Table 1).
The deduced pre-mature Pru p 3 proteins had different signal peptide lengths of 26 aa, 30 aa and 25 aa for Pru p/du 3.01–03, respectively (Figure 5). In peach and almond (E and T), Pru p/du 3.01 alleles had identical signal amino acids, but their mature proteins differed in 5 aa, of which 4 are at the C-terminal end. Pru p/du 3.02 had identical aa, and Pru p/du 3.03, only one amino acid difference P/T at position 88. The three Prunus LTPs isoallergens differed greatly in their signal peptide region, with about 40–80% sequence identity (Figure 2C), their alignments were according to the eight conserved cysteine residues (Figure 5). All mature Pru p/du 3 were alkaline (pI = 8.5–9.22) and their molecular weight was around 9 kDa. Pru p 3.01 grouped closely with apple Mal d 3.01 and 02 (Figure 2C), and Pru p 3.02 with the putative Mal d 3.03 (e-cloning analysis assigned). Pru p 3.03 is distant from Pru p 3.01 and -02, and shares about 50% aa identity with the others. Interestingly, all three Pru p 3 genes mapped on bin 6:74 (Figure 3B). One of them, Pru p 3.03 (as Ltp2), had previously been mapped using the whole TxE population and was located at 72.5 cM from the top of G6 [19].
Figure 5.
Alignment of amino acid sequences of LTPs members identified in the Prunus 'Texas' almond × 'Earlygold' peach hybrid MB1-73. Eight conserved cysteine residues are arrowed. The lipid-binding motifs are shown by green oval.
Pru p/du 4 (profilin, PRF)
Genomic sequences for two profilin genes Pru p/du 4.01 and Pru p/du 4.02 were obtained based on their cDNA data [GenBank: EU424270-EU424273]. Both genes have three exons with conserved length, 123, 138 and 135 nt, in agreement with that found in most plant species. DNA polymorphisms in Pru p/du 4.01 occurred mainly in the first intron, but in Pru p/du 4.02 they were in the first and second exon. These two genes gave the same protein sequences for peach and almond (Figure 6), although a few synonymous polymorphisms existed at the gDNA level. Molecular weights were 14.06 and 14.19 kDa for Pru p 4.01 and 4.02, respectively, and both were acidic (calculated pI 4.5 and 4.6). Compared with the six known Mal d 4 isoallergens, Pru p 4.01 was similar to Mal d 4.03A and B (93.1% and 93.9% identity), while Pru p 4.02 was similar to Mal d 4.02A and B (95.4% and 96.9% identity) (Figure 2D). Pru p 4.01 was mapped on G1 and Pru p 4.02 on G7 (Figure 3A and 3D).
Figure 6.
Alignment of amino acid sequences of two profilin members from peach and almond.
Discussion
This study provides basic genomic sequences and map positions of four allergen families in Prunus persica (peach) and Prunus dulcis (almond). This information is a prerequisite for unravelling the genetic basis of allergenic characteristics of Prunus fruits, and is likely to be similar for other Prunus species due to the high level of synteny of the species within this genus [21].
Gene families and relevance of individual genes in allergenicity
Gene family is becoming a prevailing topic in genomic research. It is thought that the pressure to conserve protein sequence and structure is associated with an increase in copy number of certain genes during evolution [27]. Although food allergens belong to a limited number of protein families [28], each protein family in a single species can be putatively encoded (experimentally demonstrated in some cases) by multiple genes in specific tissues, thus making the in vivo allergic reaction very complex. There is a need to identify genes implicated in the allergic reaction from the many putative allergen genes existing. Searching the updated Arabidopsis thaliana DNA sequence database, we found 29 putative Pru p 1-homologous genes scattered over the five chromosomes (Table 3). More than half of these (17/29) are located on chromosome 1 in four clusters, each with 3 to 7 genes. Nineteen TLP genes are present on four chromosomes. LTP genes form a large gene family with 98 members, more or less equally distributed over all five chromosomes, and profilin is a small gene family with 5 members mapping to three chromosomes. It is probable that, in peach and apple, only a small fraction of PR-10, TLP and LTP gene members are relevant to allergenic trait.
Table 3.
Members of four gene families related to allergenicity, pathogenesis related 10 proteins (PR-10), thaumatin-like proteins (TLP), lipid transfer proteins (LTP) and profilin (PRF) on the five Arabidopsis thaliana chromosomes traced from the GenBank database
| Gene | Chrom-1 | Chrom-2 | Chrom-3 | Chrom-4 | Chrom-5 | Total |
| PR-10 | 17 | 3 | 2 | 3 | 4 | 29 |
| TLP | 8 | 2 | 6 | 3 | 19 | |
| LTP | 19 | 14 | 16 | 25 | 24 | 98 |
| PRF | 2 | 2 | 1 | 5 |
A limited number of putative allergen genes may be expressed in consumed fruits, their expression profiles at mRNA and protein level are essential to allergenicity. Evidence of cDNA present in the ripe fruit is not sufficient. The need to qualify and quantify their individual or combined allergenicity is a major challenge for further investigation, where the application of new experimental and computational tools in genomics and proteomics will be crucial. Results in this area are starting to emerge and integrate, such as differential expression of LTP 1 (Pru p 3.01) and LTP2 (Pru p 3.02) [29], the expression studies of Pru p 3.01 (Pp-LTP1 in two peach genotypes [23], where major differences were detected in expression at mRNA and protein accumulation levels between them, and the research on IgE binding capacity [30], where specific IgE-binding protein regions of Pru p 3 were found for two types of allergy (OAS and systemic) by the mimotope mapping technique. Of four Pru p 2 isoallergen genes, a clone PpAz44 [GenBank: AF362988] matching to Pru p 2.01 was abundantly expressed in ripe peach fruit, while clone PpAz8 [GenBank:AF362987] (Pru p 2.02) was undetected by northern blot analysis [24]. Pru p 2.03 and Pru 2.04 were identified from the fruit ESTs, the latter may be in large amount of transcript, since 29 ESTs can be assembled into one consensus gene. A very low level of Pru p 4 (profilin) was has been identified as pollen-associated allergen, and two isoallergen genes were isolated from fruit tissue [25]. Different Pru p 1 genes expression was not reported previously, our preliminary expression study was performed by means of real-time PCR for six Pru p 1 isoallergen genes on full ripe fruits of two cultivars. The result showed that the most abundantly expressed member is Pru p 1.01, followed by Pru p 1.06, then very low expression level for Pru p 1.04 and -1.05, while Pru p 1.02 and -03 were hardly detected (data not shown).
In apple, a very recent comprehensive gene expression of four classes of putative allergens in apple fruit has gained insight of the genetic and environmental factors affecting the allergenic potential of apples [31]. They showed that some putative members are statistically different in transcript accumulation in ripe fruit, while others were not detectable. This was a very nice reference to think about comparative transcription and protein expression for all set of peach putative allergen genes among diverse peach cultivars.
We found that the putative genes encoding the major allergens reported in peach fruit, Pru p 1 and Pru p 3 [15], were located on two linkage groups, G1 and G6. Based on their bin map positions, we can use SSR markers tightly linked or flanking these two major Prunus allergen genes for a diversity survey of a large number of genotypes to search for potential candidates for further sequencing of the allergen genes. Clinical evaluation of allergenicity for different peach cultivars is needed for association analysis to find low allergenic alleles.
Comparative mapping between Prunus and Malus using the sequence and position of allergen genes
Dirlewanger et al. [21] compared the Prunus and apple genomes using a limited number of common markers. They found that three Prunus linkage groups (G1, G3 and G4) were syntenic to seven apple linkage groups, G1 to LG13 and LG16 at the top and LG8 at the bottom, G3 to LG9 and LG17, and G4 to LG5 and LG10. Information is also available on the correspondence between homoeologous linkage groups within the apple genome obtained initially with RFLPs by Maliepaard et al. [32], to which various allergen genes and SSRs were recently added by Gao et al. [17-19] and Celton et al. [33], respectively. With the availability of genomic sequences and the linkage map positions of the four allergen-families in apple (26 apple allergen genes on eight linkage groups) [17-19] and peach (18 genes on 5 linkage groups, this study), we have additional information useful for updating the synteny between these two species.
Three of the eight Prunus bins where we located putative allergen genes corresponded to known syntenic and colinear regions of the apple genome (Figure 3): the Pru p/du 1 on G1, corresponding to regions with clusters of homologous genes (Mal d 1) of apple on LG13 and LG16, Pru p/du 4.01 on the lower part of G1 and Mal d 4.03 on LG8, and Pru p/du 2.01A/B on G3 and Mal d 2.01A/B on LG9. Three more syntenic regions were detected in this study: Pru p/du 4.02 on G7 and the region of LG2 where Mal d 4.02 maps, Pru p/du 3.01-03 on G6 corresponding to Mal d 3.01 on LG12 and Mal d 3.02 on LG4; and Pru p/du 4.01 which, in addition to the region of LG8 already mentioned, is syntenic with the upper part of LG15 where Mal d 4.03B maps. No counterparts were found to the Pru p/du 2.02, -03, and -04 genes, which were mapped on G7, G8 and G1, respectively, on the apple genome. The positions of these three genes in the Prunus genome indicate syntenic regions of apple where additional Mal d 2 genes may be located. Because Mal d 1 on apple LG 13 and LG16 are clustered in a region of about 2 cM, so we can deduce our mapped Pru p 1 genes are also in a cluster.
There are two hypotheses on the origin of the two genomes that generated the apple amphydiploid genome (2n = 34). One proposes that these were from members of two Rosaceae subfamilies, one from the Amygdaloideae (thus closer to Prunus with x = 8) and another from the Spiraeoideae (x = 9) [34]. The other hypothesis proposes that both ancestral genomes had a basic chromosome number of x = 9 at least one being Spiraeoideae (x = 9) [35]. Combining map position and sequence comparison, the cluster of Pru p 1 genes mapped on Prunus G1 is more similar to that of the Mal d 1 genes of LG16 than to LG13: a) Mal d 1 isoallergen genes were classified into four subfamilies and members of sub-families were distributed asymmetrically to LG 13 and LG16 (Figures 2, 3), i.e. LG13 contains Mal d 1 members from subfamilies I and IV, but LG16 contains representative members of the four subfamilies; b) Pru p 1 genes fall into the four subfamilies as defined in apple, and had a higher deduced amino acid sequence similarity to the Mal d 1 members on LG16 (Figure 2 and data of percentage amino acid identity in the Results section). These results favor the two primitive genomes for apple and one is closer to Prunus than the other.
Bin mapping feasibility and effectiveness
The bin mapping approach provides a fast and cheap way to map new genes [26] that has worked very well in our hands. Once we cloned and sequenced the two putative alleles for each gene from MB1-73, we either created an allele-specific marker and genotyped the eight plants of the bin set, or cloned the fragments from the individuals of the bin set and sequenced them to determine their genotypes. One of the advantages of using the T × E population for bin mapping is that, due to its inter-specific nature, polymorphism is very high [26]. The disadvantage of the bin mapping approach is that there is a loss of precision in marker location (8–25 cM), but this can be solved for markers of particular interest by running the whole population to establish a more precise position or narrow down a region where closely related genes or common RFLP markers in apple has been fine mapped. For example, we estimate Pru p/du 1 genes (in bin 1:14) are in a cluster and likely around 13 cM from the top based on location of Mal d 1 genes and anchor marker MC001 (Figure 3A). A very large number of ESTs from peach fruit are on public databases. This information is essential to find candidate genes to be mapped, and their positions compared to that of major genes or quantitative trait loci of interest mapped using phenotypic data. Bin mapping, as in this case, can facilitate the positioning of these candidate genes. Exchange of DNA and genotype data of the T × E bin set in the peach research community is helping candidate gene mapping, with genetic analysis and breeding applications following on.
Conclusion
Eighteen putative Prunus allergen genes were located on five of the eight linkage groups of this genus. Genomic characterization of Prunus allergens provided further evidence for synteny between Prunus and Malus. This information will be directed towards identifying the key genes or isoforms causing the differences in allergenicity among different cultivars by additional gene and protein expression and genetic association studies. These results are a first step towards the understanding of allergenicity caused by peach fruit and for the development of new cultivars with enhanced health properties and subject to specific recommendations on fruit consumption.
Methods
Plant materials
PCR cloning and sequencing for targeting allergen genes to identify different gene members and alleles was mainly on the 'Texas' (T) almond and 'Earlygold' (E) peach hybrid (MB1-73) used to generate the reference mapping population (T × E) and it is now used by the international Prunus community. Therefore, two putative alleles, one from 'Texas' (T) and another from 'Earlygold' (E), will be simultaneously amplified with a pair of primers based on a reference sequence. For T and E allele-specific marker tests, DNA from T and E were used as controls. Eight plants were used for the bin set (the set of plants used for bin mapping): the grandparent E, the parent MB1-73, and six plants from the F2 progeny (# 5, 12, 23, 30, 34, 83) [26]. DNA samples were isolated and provided by IRTA using standard protocols [36].
PCR primers, cloning and sequencing
Four families of putative allergens from peach and almond, PR-10 (Bet v 1 homologous gene), TLPs, LTPs and profilins (PRF), were searched in the GenBank, most of them were derived from peach fruit tissues. The sequences obtained were assembled using Seqman program (DNAstar, Lasergene 6) to identify different gene members for each family. DNA sequences were assumed to belong to different gene members when they displayed less than 98% identity. This resulted in four members for Pru p1 (PR10) and Pru p 2 (TLP), three for Pru p 3 (LTP) and two members for Pru p 4 (profilin). Primer pairs were designed using the software program PRIMER DESIGNER v. 2.0 (Scientific and Educational Software, Cary, N.C.) based on these sequences. Preferably, the forward and reverse primers covered the whole region of the gene encoding sequences (Table 4). Detailed PCR cloning and sequencing procedures have been described previously [19], here we mention only the key points and changes. First, the melting temperature for amplification was optimized by gradient PCR with hybrid (MB1-73) DNA, then PCR was performed in two steps with Pfu polymerase (Stratagene, La Jolla, Calif.) and Taq (Takara Biotechnology, Dalian, China) using an Eppendorf Masterycler. The amplified fragments were purified by Qiaquick Gel Extraction Kit (Qiagen, Germany), ligated into the pGEM-T easy vector (Promega, Madison, Wis.) and used to transform DH5α competent cells (Takara Biotechnology, Dalian, China) according to the manufacturer's protocol. For each purified fragment, 6–8 white colonies were selected for plasmid isolation and sequencing by Invitrogen (Shanghai, China) using an ABI 3730 (Applied Biosystems, Foster City, Calif.) platform. If there are more than two distinct sequences with initial one primer pair, additional colonies were sequenced to explore new gene members. All primers were synthesised by Invitrogen (Shanghai, China).
Table 4.
PCR primer pairs used for genomic cloning of the different peach/almond allergen genes and for (bin) mapping the genes
| Primers group | Reference Sequence 1 | Primer name | Primer sequence (5' – 3') | Tm °C/cycles | Product | Gene | |
| Pfu | Taq | (nt) | |||||
| 1 | DQ251187 | PpPr10f PpPr10r |
ACCATGGGTGTCTTCACATA AATTTAGTTGTAGGCATCGG |
56/30 | 58/2 | 586/623 | Pru p/du 1.01/05 |
| 2 | DY646736 | Pp1-2f Pp1-2r |
GTTTGTCCTTTAAACTCTCC GATTTAGTTGTAGGCATCGG |
53/30 | 55/2 | 762/765 | Pru p/du 1.02 |
| 3 | DY647300 | Pp1-3f Pp1-3r |
CTTTGATCAGTTTCCCAAGT TTAGTTGTAGGCATCAGGGT |
53/30 | 55/2 | 624 | Pru p/du 1.03 |
| 4 | DY653062 | Pp1-4f1 Pp1-4r |
TTTACAGAATCATGGGTGTG TTAGTTGTAGGCATCATGGT |
53/30 | 55/2 | 615/622 | Pru p/du 1.04 |
| 5 | GW1 | Pp1-1A-GSP1 Pp1-1A-GSP2 |
AACCTTCACCAAAGCTAGTCTTCTTGATG GTCAGCATCCAGAACAAGGGCTTTGAACA |
1100 | Pru p/du 1.05 /6A/C |
||
| 6 | GW2 | P1-1A3-GSP1 P1-1A3-GSP2 |
GGTGTCTTCACATACTCAGACGAGTC CCACCTCAGTCATCCCCCCACCAAGA |
1546 | Pru p/du 1.06A/C | ||
| 7 | P1-1-6 PpPr10r |
CACCTCCAGTGCTCATAGC See above |
57/30 | 59/2 | 720 | Pru p/du 1.05 | |
| 8 | P1-7f P1-7r |
ATGGGTGTCTTCACATACTCAG TTTAGTTGTAGGCATCTGGGT |
57/30 | 59/2 | 593 | Pru p/du 1.06A/B | |
| 9 | P1-8f P1-8r2 |
TAGTTATGAGTTGCTTGCAATGCT GGGCATGCTTGCTTTGGTAACTC |
58/30 | 60/2 | 1346 | Pru p/du 1.06C | |
| 10 | P1-8f P1-9r |
See above GAAAGTTCCAAAGTACATGTGC |
58/30 | 60/2 | 1908 | Pru p/du 1.06A | |
| 11 | AF362988 | PpTL2f PpTL2r |
ACAGCAAGCCAATTAAGACA GCTTATGGGCAGAATGTGATGAG |
56/30 | 58/2 | 879/892 | Pru p/du 2.01A/B |
| 12 | AF362987 | PpTL1f PpTL1r |
ATGATGAAGACCCTAGGAGCAG GTCGAGAGTCCTATCTTTAT |
56/30 | 58/2 | 974 | Pru p/du 2.02 |
| 13 | GW3 | PTLP1-GSP1 PTLP1-GSP2 |
GATAATTGAGGATTTCCAAAGGAGGCTG AGGAGGGTTAAGCTGAGGCTGAGGACTG |
500/1500 | Pru p/du 2.02 | ||
| 14 |
BF717226, DW349103 |
PpTLP3f PpTLP3r |
AAGTAAAGTTCTTTGGCGTC CACGTGGCACCACAAGCATC |
53/30 | 55/2 | 659 | Pru p/du 2.03 |
| 15 | GW4 | PruTLP3-GSP1 PruTLP3-GSP2 |
AGTACTCACCCACCAACTTCTCCTGCGT GCTACTTTAGCCGAGTTCACTCTCAACG |
800/1000 | Pru p/du 2.03 | ||
| 16 | Consensus of 29 ESTs |
PpTLP4f PpTLP4r |
ACTTGTCTGAACTTATGGCTCC GAAGTTAGAAGAAGAGCTGCG |
58/30 | 60/2 | 1800/1825 | Pru p/du 2.04 |
| 17 |
AY620230 X96714 |
PpLTP1f1 PpLTP1r |
ATCATAGTCAAGAGAGATGG CCCTAAGTGGATCACATAGC |
52/30 | 54/2 | 645 | Pru p/du 3.01 |
| 18 |
AY093699 X96716 |
PpLTP2f PpLTP2r |
TATCAGCTTTACTTACGACG CTGGCTTCCACAGAAACCTC |
58/30 | 60/2 | 513/514 | Pru p/du 3.02 |
| 19 | X96715 | PpLTP3f PpLTP3r |
CCCAAGCGAAAGAAACACTA CATCTCATATCATCCTTCCA |
58/30 | 60/2 | 527/528 | Pru p/du 3.03 |
| 20 |
AJ491881 AY081852 |
Pp4.01f Pp4.01r |
AGAAGAAATCAGAAGCAACG AAATAGTCACTCGGAGCAAT |
51/30 | 53/2 | 1041 | Pru p/du 4.01 |
| 21 | AJ491882 | Pp4.02f Pp4.02r1 |
CAGCAACAACAACAAAGATG TCTAGAGACCCTGCTCAACC |
53/30 | 55/2 | 754 | Pru p/du 4.02 |
Genome walking
Genome walking was applied to gDNA of MB1-73 using the Universal Genome Walker kit (Clontech, Palo Alto Calif.) for gene sequences that were not cover the ORF or to avoid the mistakes induced by primer sequence, or to explore polymorphisms in the flanking regions. Four libraries were constructed using Dra I, EcoR V, Pvu II and Stu I enzymes to digest 2.5 μg of gDNA from MB1-73. Adaptors were ligated to the digested DNA fragments. Two gene specific primers (GSP, Table 4) and two adaptors were used for nest PCR. The product from two of the four libraries was excised from the gel and subsequently purified, ligated, transformed and sequenced as described in the above section.
Sequence analysis
DNA sequences were analysed and single nucleotide polymorphisms (SNPs) were identified using the SEQMAN program (Lasergene 6, Madison, Wis.). Intron sizes were deduced by comparing the genome sequences with cDNA (ESTs) sequences or by putative conserved splicing patterns in plants. The phylogenetic tree was created and percentage sequence identity calculated using Clustal W by Megalign (Lasergene 6). The GenDoc program http://www.psc.edu/biomed/genedoc was used for multiple DNA and amino acid sequence alignment.
Allele-specific markers and bin mapping approach
Three types of molecular markers were used: 1) two dominant single nucleotide amplification polymorphism (SNAP) markers [18] representing T and E respectively, 2) co-dominant markers based on length differences due to Indels and variation in the number of simple sequence repeats (SSRs) and 3) SNP markers based on direct sequencing of the PCR products from 8 bin samples. Markers 1) and 2) were used for the mapping of Pru p 1, 2, and 4 genes and 3) for Pru p 3 genes.
With SNAP markers, the annealing temperature is usually critical. It was optimised by gradient PCR using gDNAs of 'Texas' almond and 'Earlygold' peach (the two parents of MB1-73) as contrasting templates, and then applied to genomic DNA of 8 bin samples. Indels and SSR markers were revealed by 6% PAGE and silver staining. For direct sequencing, after PCR amplification (GoTaq, Promega), an aliquot of the PCR reaction was loaded on an agarose gel to check for specific amplification. If the PCR produced a single fragment, the remaining PCR reaction was purified using Roche High Pure PCR Product purification kit (Germany). An aliquot of 0.5 μl to 5 μl of purified PCR was used as template for sequencing with forward and/or reverse primers. SNPs and/or indels were detected using Sequencher software, version 4.7.
For mapping, we followed the strategy of "selective" or "bin" mapping [26,37] where, based on the data of a detailed map already constructed (in this case the TxE map), a few plants are selected having a high number of recombination breakpoints and an approximately even distribution of these breakpoints throughout the genome. The joint genotype of these plants for a new marker defines a single map position, allowing its mapping with less time and cost, and with a moderate loss of precision compared with the use of the whole population. For Prunus, a set of only eight plants: one of the parents ('Earlygold') the F1 hybrid (MB1-37) and six plants (# 5, 12, 23, 30, 34 and 83) from the TxE progeny, was selected by Howad et al. [26] allowing to establish the position of any marker to 67 fragments of the genome ("bins") of an average size of 7.8 cM. The identification of each bin is unequivocal except for two pairs (2:45/3:04 and 5:41/8:30), which have an identical genotype and require the genotype of an additional F2 plant (#27) to separate them. By comparing the genotype appeared in H and E, we were able to distinguish T allele from that of E. Marker tests were performed on these eight individuals of the bin set that were genotyped as homozygous for the 'Texas' allele (noted as A), homozygous for the 'Earlygold' allele (B) or heterozygous as MB1-73 (H). The resulting segregation pattern was compared to the defined data for each bin, and the matching bin was identified [26].
Nomenclature of the putative allergen sequences
We followed the systematic allergen nomenclature [38] for the four different protein classes and encoding genes. Generally, allergens are designated by the first three letters of the name of the genus, followed by a space and the first one or two letters of the species name. These letters are usually followed by a space and an Arabic number according to the order in which the allergens were identified, whereby the same number is used to designate homologous allergens of related species. Additional numbers and letters can be added to distinguish different isoallergens and variants [19]. Accordingly, PR-10, TLP, LTP and PRF proteins are denoted as Pru p 1 to 4 in peach and Pru du 1 to 4 in almond. Amino acid sequences with more than 5% dissimilarity usually belong to a different isoallergen and are distinguished by a two-digit code. If there are several genes within isoallergen group (on the DNA sequence identity, usually more than 95%), they are further distinguished by adding a capital letter after their isoallergen name, such as Pru p 2.01A and Pru p 2.01B. This procedure has been employed in apple [17] and birch [39]. The plant material in this study is an interspecies hybrid between peach and almond; we assigned two alleles of the same gene locus by its original species source, from Prunus persica as Pru p and from Prunus dulci as Pru du. Therefore, Pru p/du refers to a gene in interspecies hybrid parent MB1-37.
Authors' contributions
ZSG, KSC, EVW, PA and SDW initiated this study; ZSG, LC and WH designed the experiment. LC, SMZ, LJS and ZSG performed the gene cloning, sequence analysis and mapping of Pru p/du 1, Pru p/du 2 and Pru p/du 4. EI and WH did the bin mapping of Pru p/du 3; ZSG, LC, EVW, PA and KSC drafted the paper.
Acknowledgments
Acknowledgements
This study has been carried out with financial support from the China Ministry of Education (107064 and 1108), Natural Science Foundation of China (30771496), Science and Technology Department of Zhejiang Province (2007C22068) and the 111 project (B06014). It was supported in part by the Commission of the European Communities for the project ISAFRUIT (Contract FP6-FOOD-CT-2006-016279). Prof. Xu Changjie and Dr. Zhang Bo are acknowledged for their advice and assistance on the lab work, and Dr. Luud Gilissen for his critical reading of this manuscript.
Contributor Information
Lin Chen, Email: d05chenlin@zju.edu.cn.
Shuiming Zhang, Email: zhangshuiming@zju.edu.cn.
Eudald Illa, Email: eudald.illa@irta.es.
Lijuan Song, Email: slj2006215.slij@yahoo.com.cn.
Shandong Wu, Email: wushandong@sohu.com.
Werner Howad, Email: werner.howad@irta.es.
Pere Arús, Email: pere.arus@irta.es.
Eric van de Weg, Email: eric.vandeweg@wur.nl.
Kunsong Chen, Email: akun@zju.edu.cn.
Zhongshan Gao, Email: gaozhongshan@zju.edu.cn.
References
- Hoffmann-Sommergruber K, the SAFE consortium The SAFE project: 'plant food allergies: field to table strategies for reducing their incidence in Europe' an EC-funded study. Allergy. 2005;60:436–442. doi: 10.1111/j.1398-9995.2005.00809.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-Rivas M, Bolhaar S, Gonzalez-Mancebo E, Asero R, van Leeuwen A, Bohle B, Ebner C, Rigby N, Sancho AI, Miles S, Zuidmeer L, Knulst A, Breiteneder H, Mills C, Hoffmann-Sommergruber K, van Ree R. Apple allergy across Europe: How allergen sensitization profiles determine the clinical expression of allergies to plant foods. Journal of Allergy and Clinical Immunology. 2006;118:481–488. doi: 10.1016/j.jaci.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Karamloo F, Scheurer S, Wangorsch A, May S, Haustein D, Vieths S. Pyr c 1, the major allergen from pear (Pyrus communis), is a new member of the Bet v 1 allergen family. J Chromatogr B Biomed Sci Appl. 2001;756:281–293. doi: 10.1016/S0378-4347(01)00087-1. [DOI] [PubMed] [Google Scholar]
- Fernandez-Rivas M, van Ree R, Cuevas M. Allergy to Rosaceae fruits without related pollinosis. Journal of Allergy and Clinical Immunology. 1997;100:728–733. doi: 10.1016/S0091-6749(97)70265-3. [DOI] [PubMed] [Google Scholar]
- Pastorello EA, Farioli L, Pravettoni V, Ortolani C, Ispano M, Monza M, Baroglio C, Scibola E, Ansaloni R, Incorvaia C, Conti A. The major allergen of peach (Prunus persica) is a lipid transfer protein. Journal of Allergy and Clinical Immunology. 1999;103:520–526. doi: 10.1016/S0091-6749(99)70480-X. [DOI] [PubMed] [Google Scholar]
- Scheurer S, Metzner K, Haustein D, Vieths S. Molecular cloning, expression and characterization of Pru a 1, the major cherry allergen. Molecular Immunology. 1997;34:619–629. doi: 10.1016/S0161-5890(97)00072-2. [DOI] [PubMed] [Google Scholar]
- Scheurer S, Lauer I, Foetisch K, Moncin MSM, Retzek M, Hartz C, Enrique E, Lidholm J, Cistero-Bahima A, Vieths S. Strong allergenicity of Pru av 3, the lipid transfer protein from cherry, is related to high stability against thermal processing and digestion. Journal of Allergy and Clinical Immunology. 2004;114:900–907. doi: 10.1016/j.jaci.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Pastorello EA, Farioli L, Pravettoni V, Giuffrida MG, Ortolani C, Fortunato D, Trambaioli C, Scibola E, Calamari AM, Robino AM, Conti A. Characterization of the major allergen of plum as a lipid transfer protein. J Chromatogr B Biomed Sci Appl. 2001;756:95–103. doi: 10.1016/S0378-4347(01)00074-3. [DOI] [PubMed] [Google Scholar]
- Pastorello EA, Purello D' Ambrosio F, Pravettoni V, Farioli L, Giuffrida G, Monza M, Ansaloni R, Fortunato D, Scibola E, Rivolta F, Incorvaia C, Bengtsson A, Conti A, Ortolani C. Evidence for a lipid transfer protein as the major allergen of apricot. Journal of Allergy and Clinical Immunology. 2000;105:371–377. doi: 10.1016/S0091-6749(00)90090-3. [DOI] [PubMed] [Google Scholar]
- Tawde P, Venkatesh YP, Wang F, Teuber SS, Sathe SK, Roux KH. Cloning and characterization of profilin (Pru du 4), a cross-reactive almond (Prunus dulcis) allergen. Journal of Allergy and Clinical Immunology. 2006;118:915–922. doi: 10.1016/j.jaci.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Brenna OV, Pastorello EA, Farioli L, Pravettoni V, Pompei C. Presence of allergenic proteins in different peach (Prunus persica) cultivars and dependence of their content on fruit ripening. Journal of Agricultural and Food Chemistry. 2004;52:7997–8000. doi: 10.1021/jf0491052. [DOI] [PubMed] [Google Scholar]
- Kootstra HS, Vlleg-Boerstra BJ, Dubois AEJ. Assessment of the reduced allergenic properties of the Santana apple. Ann Allergy Asthma Immunol. 2007;99:522–525. doi: 10.1016/S1081-1206(10)60381-X. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Ebner C. Molecular and biochemical classification of plant-derived food allergens. Journal of Allergy and Clinical Immunology. 2000;106:27–36. doi: 10.1067/mai.2000.106929. [DOI] [PubMed] [Google Scholar]
- Bohle B. The impact of pollen-related food allergens on pollen allergy. Allergy. 2007;62:3–10. doi: 10.1111/j.1398-9995.2006.01258.x. [DOI] [PubMed] [Google Scholar]
- Gamboa PM, Caceres O, Antepara I, Sanchez-Monge R, Ahrazem O, Salcedo G, Barber D, Lombardero M, Sanz ML. Two different profiles of peach allergy in the north of Spain. Allergy. 2007;62:408–414. doi: 10.1111/j.1398-9995.2006.01284.x. [DOI] [PubMed] [Google Scholar]
- Wen ZM, Ye ST. A report of 50 patients with artemisia pollenosis and plant food allergy. National Medical Journal of China. 2002;82:626–629. (in Chinese with English abstract) [PubMed] [Google Scholar]
- Gao ZS, Weg WE van de, Schaart JG, Schouten HJ, Tran DH, Kodde LP, Meer IM van der, Geest AHM van der, Kodde J, Breiteneder H, Hoffmann-Sommergruber H, Bosch D, Gilissen LJWJ. Genomic cloning and linkage mapping of the Mal d 1 (PR-10) gene family in apple (Malus domestica) Theoretical and Applied Genetics. 2005;111:171–183. doi: 10.1007/s00122-005-2018-4. [DOI] [PubMed] [Google Scholar]
- Gao ZS, Weg WE van de, Schaart JG, van Arkel G, Breiteneder H, Hoffmann-Sommergruber K, Gilissen LJWJ. Genomic characterization and linkage mapping of the apple allergen genes Mal d 2 (thaumatin-like protein) and Mal d 4 (profilin) Theoretical and Applied Genetics. 2005;111:1087–1097. doi: 10.1007/s00122-005-0034-z. [DOI] [PubMed] [Google Scholar]
- Gao ZS, Weg WE van de, Schaart JG, Meer IM van der, Kodde L, Laimer M, Breiteneder H, Hoffmann-Sommergruber K, Gilissen LJWJ. Linkage map positions and allelic diversity of two Mal d 3 (non-specific lipid transfer protein) genes in the cultivated apple (Malus domestica) Theoretical and Applied Genetics. 2005;110:479–491. doi: 10.1007/s00122-004-1856-9. [DOI] [PubMed] [Google Scholar]
- Gao ZS, Weg WE Van de, Matos CI, Arens P, Bolhaar STHP, Knulst AC, Li YH, Hoffmann-Sommergruber , Gilissen LJWJ. Assessment of allelic diversity in intron-containing Mal d 1 genes and their association to apple allergenicity. BMC Plant Biology. 2008;8:116. doi: 10.1186/1471-2229-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldere F, Cosson P, Howad W, Arus P. Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9891–9896. doi: 10.1073/pnas.0307937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrazem O, Jimeno L, Lopez-Torrejon G, Herrero M, Espada JL, Sanchez-Monge R, Duffort O, Barber D, Salcedo G. Assessing allergen levels in peach and nectarine cultivars. Ann Allergy Asthma Immunol. 2007;99:42–47. doi: 10.1016/S1081-1206(10)60619-9. [DOI] [PubMed] [Google Scholar]
- Botton A, Vegro M, De Franceschi F, Ramina A, Gemignani C, Marcer G, Pasini G, Tonutti P. Different expression of Pp-LTP1 and accumulation of Pru p 3 in fruits of two Prunus persica L. Batsch genotypes. Plant Science. 2006;171:106–113. doi: 10.1016/j.plantsci.2006.02.016. [DOI] [Google Scholar]
- Ruperti B, Cattivelli L, Pagni S, Ramina A. Ethylene-responsive genes are differentially regulated during abscission, organ senescence and wounding in peach (Prunus persica) Journal of Experimental Botany. 2002;53:429–437. doi: 10.1093/jexbot/53.368.429. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Perez R, Fernandez-Rivas M, Gonzalez-Mancebo E, Sanchez-Monge R, Diaz-Perales A, Salcedo G. Peach profilin: cloning, heterologous expression and cross-reactivity with Bet v 2. Allergy. 2003;58:653–640. doi: 10.1034/j.1398-9995.2003.00198.x. [DOI] [PubMed] [Google Scholar]
- Howad W, Yamamoto T, Dirlewanger E, Testolin R, Cosson P, Cipriani G, Monforte AJ, Georgi L, Abbott AG, Arus P. Mapping with a few plants: Using selective mapping for microsatellite saturation of the Prunus reference map. Genetics. 2005;171:1305–1309. doi: 10.1534/genetics.105.043661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatters RG, Boykin LM, Lapointe SL, Hunter WB, Weathersbee AA. Phylogenetic and structural relationships of the PR5 gene family reveal an ancient multigene family conserved in plants and select animal taxa. Journal of Molecular Evolution. 2006;63:12–29. doi: 10.1007/s00239-005-0053-z. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Mills C. Structural bioinformatic approaches to understand cross-reactivity. Molecular Nutrition & Food Research. 2006;50:628–632. doi: 10.1002/mnfr.200500274. [DOI] [PubMed] [Google Scholar]
- Botton A, Begheldo M, Rasori A, Bonghi C, Tonutti P. Differential expression of two lipid transfer protein genes in reproductive organs of peach (Prunus persica L. Batsch) Plant Science. 2002;163:993–1000. doi: 10.1016/S0168-9452(02)00271-6. [DOI] [Google Scholar]
- Pacios LF, Tordesillas L, Cuesta-Herranz J, Compes E, Sanchez-Monge R, Palacin A, Salcedo G, Diaz-Perales A. Mimotope mapping as a complementary strategy to define allergen IgE-epitopes: Peach Pru p 3 allergen as a model. Molecular Immunology. 2008;45:2269–2276. doi: 10.1016/j.molimm.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Botton A, Lezzer P, Dorigoni A, Barcaccia G, Ruperti B, Ramina A. Genetic and environmental factors affecting allergen-related gene expression in apple fruit (Malus domestica L. Borkh) Journal of Agricultural and Food Chemistry. 2008;56:6707–6716. doi: 10.1021/jf800813d. [DOI] [PubMed] [Google Scholar]
- Maliepaard C, Alston FH, Van Arkel G, Brown LM, Chevreau E, Dunemann F, Evans KM, Gardiner S, Guilford P, Van Heusden AW, Janse J, Laurens F, Lynn JR, Manganaris AG, Den Nijs APM, Periam N, Rikkerink E, Roche P, Ryder C, Sansavini S, Schmidt H, Tartarini S, Verhaegh JJ, Vrielink-Van Ginkel M, King GJ. Aligning male and female linkage maps of apple (Malus pumila Mill.) using multi-allelic markers. Theoretical and Applied Genetics. 1998;97:60–73. [Google Scholar]
- Celton JM, Tustin S, Chagné D, Gardiner S. Construction of a dense genetic linkage map for apple rootstocks using SSRs developed from Malus ESTs and Pyrus genomic sequences. Tree Genetics and Genomes. 2008.
- Arús P, Yamamoto T, Dirlewanger E, Abbott AG. Synteny in the Rosaceae. In: Jules Janick, editor. Plant Breeding Reviews. Vol. 27. 2005. pp. 175–211. [Google Scholar]
- Phipps JB, Robertson KR, Rohrer JR. Origins and evolution of subfam. Maloideae (Rosaceae) Systematic Botany. 1991;16:303–332. doi: 10.2307/2419283. [DOI] [Google Scholar]
- Viruel MA, Messeguer R, de Vicente MC, Garcia-Mas J, Puigdomenech P, Vargas F, Arus P. A linkage map with RFLP and isozyme markers for almond. Theoretical and Applied Genetics. 1995;91:964–971. doi: 10.1007/BF00223907. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Shmoys DB, Durrett RT, Tanksley SD. Selective mapping: a strategy for optimizing the construction of high-density linkage maps. Genetics. 2000;155:407–420. doi: 10.1093/genetics/155.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TP, Hoffman D, Lowenstein H, Marsh DG, Platts TAE, Thomas W. Allergen nomenclature. Journal of Allergy and Clinical Immunology. 1995;96:5–14. doi: 10.1016/S0091-6749(95)70027-7. [DOI] [PubMed] [Google Scholar]
- Schenk MF, Gilissen L, Esselink GD, Smulders MJM. Seven different genes encode a diverse mixture of isoforms of Bet v I, the major birch pollen allergen. BMC Genomics. 2006;7:168. doi: 10.1186/1471-2164-7-168. [DOI] [PMC free article] [PubMed] [Google Scholar]