Abstract
Background
The peptide hormone ghrelin has many important physiological and pathophysiological roles, including the stimulation of growth hormone (GH) release, appetite regulation, gut motility and proliferation of cancer cells. We previously identified a gene on the opposite strand of the ghrelin gene, ghrelinOS (GHRLOS), which spans the promoter and untranslated regions of the ghrelin gene (GHRL). Here we further characterise GHRLOS.
Results
We have described GHRLOS mRNA isoforms that extend over 1.4 kb of the promoter region and 106 nucleotides of exon 4 of the ghrelin gene, GHRL. These GHRLOS transcripts initiate 4.8 kb downstream of the terminal exon 4 of GHRL and are present in the 3' untranslated exon of the adjacent gene TATDN2 (TatD DNase domain containing 2). Interestingly, we have also identified a putative non-coding TATDN2-GHRLOS chimaeric transcript, indicating that GHRLOS RNA biogenesis is extremely complex. Moreover, we have discovered that the 3' region of GHRLOS is also antisense, in a tail-to-tail fashion to a novel terminal exon of the neighbouring SEC13 gene, which is important in protein transport. Sequence analyses revealed that GHRLOS is riddled with stop codons, and that there is little nucleotide and amino-acid sequence conservation of the GHRLOS gene between vertebrates. The gene spans 44 kb on 3p25.3, is extensively spliced and harbours multiple variable exons. We have also investigated the expression of GHRLOS and found evidence of differential tissue expression. It is highly expressed in tissues which are emerging as major sites of non-coding RNA expression (the thymus, brain, and testis), as well as in the ovary and uterus. In contrast, very low levels were found in the stomach where sense, GHRL derived RNAs are highly expressed.
Conclusion
GHRLOS RNA transcripts display several distinctive features of non-coding (ncRNA) genes, including 5' capping, polyadenylation, extensive splicing and short open reading frames. The gene is also non-conserved, with differential and tissue-restricted expression. The overlapping genomic arrangement of GHRLOS with the ghrelin gene indicates that it is likely to have interesting regulatory and functional roles in the ghrelin axis.
Background
Ghrelin, a hormone with many physiological and pathophysiological roles, was initially described as the endogenous ligand for the growth hormone secretagogue receptor (GHSR 1a), through which it stimulates the release of growth hormone from the anterior pituitary [1]. Ghrelin is primarily produced in the stomach and plays a key role in regulating appetite, gut motility and energy balance [2-6]. Ghrelin is also an autocrine factor in a number of tissues, as it regulates insulin release and has therapeutic potential for inflammatory diseases, heart disease, cancer cachexia, diabetes mellitus and obesity [7]. Despite the importance of ghrelin in a range of physiological systems and pathophysiological conditions, little is known about the regulation of ghrelin synthesis and secretion. We previously identified a gene on the opposite strand of the ghrelin gene, ghrelinOS (GHRLOS), which spans the promoter and untranslated regions of the ghrelin gene (GHRL) [8]. However, the genomic structure, expression pattern and potential function of GHRLOS remains to be investigated. It is not known whether GHRLOS RNA species with open reading frames exist, or whether GHRLOS is a non-coding RNA gene.
There is strong support for the hypothesis that antisense transcripts provide a widespread and important mechanism for the regulation of the human genome [9,10]. Our understanding of the genome is currently undergoing a paradigm shift, as a previously hidden and complex layer of antisense and non-coding RNAs is emerging, which controls gene transcription and translation through a diverse range of mechanisms [11]. Much phenotypic diversity between humans and other species is likely to be due to regulation by RNA [12].
In this study, we examined the genomic structure and organisation of GHRLOS. We have found that GHRLOS spans approximately 44 kb of genomic DNA and transcribes long, 5' capped, polyadenlyated RNA species that are extensively spliced and differentially expressed. High levels of GHRLOS expression occur in the emerging non-coding RNA tissues, the brain, testis and thymus. We have also examined GHRLOS RNA species in silico, revealing that GHRLOS is a candidate non-coding RNA gene. These data provide a strong basis for further functional studies to determine whether GHRLOS plays a role in the regulation of ghrelin gene expression.
Results
Characterisation of GHRLOS start sites and alternative splicing
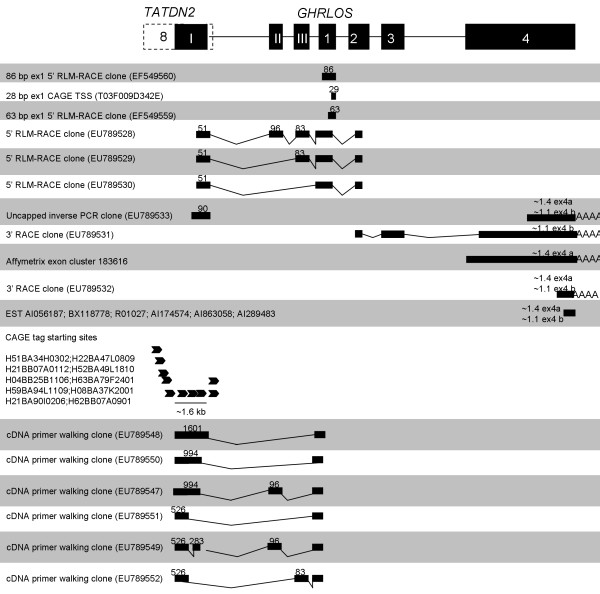
An initial aim of this study was to characterise GHRLOS in a range of tissues. As we have previously demonstrated the expression of GHRLOS mRNA transcripts in the human stomach [8], we performed 5' RLM-RACE (RNA ligase mediated rapid amplification of cDNA ends) on this tissue. Unexpectedly, we identified a number of new exons (exon I-III) [GenBank:EU789528, EU789529, and EU789530] that are 2.5 to 4.8 kb upstream of the previously reported GHRLOS transcription start sites [8] in exon 4* (4*a-c). To simplify the numbering of GHRLOS exons, the previously reported [8] exons 4*, 2**, 2* and -1* have been renamed exon 1 to 4, respectively; while the exons upstream of the reported start sites in exon 1 are denoted by Roman numerals (I-III). Importantly, in the GHRLOS variants demonstrated via 5' RLM-RACE, exon 1 is extended, with a 106 nt region (exon 1d, hereafter termed exon 1) overlapping exon 4 of the ghrelin gene (see [Additional file 1]). The novel first exon, exon I, is 51 bp in size (Fig. 1) and overlaps the 3' untranslated region of the adjacent gene, TATDN2 (TatD DNase domain containing 2, also known as hypothetical protein KIAA0218) [13]. This gene initiates on the same DNA strand as GHRLOS, approximately 32,000 base pairs upstream of the 51 bp exon I of GHRLOS.
Figure 1.
Mapping of GHRLOS transcript 5' and 3' ends. Relative positions of the 5' and 3' ends of GHRLOS transcripts were derived from RACE, inverse PCR and cDNA primer walking RT-PCR experiments, as well as from publically available data from EST databases, Affymetrix exon arrays and CAGE tags. TATDN2 exons are shown as white boxes, GHRLOS exons as black boxes, introns as horizontal lines. Size (bp) of selected exons are shown above each exon. All known 3' ends of GHRLOS transcripts contain the same polyadenylation signal. Poly(A) tails are represented by AAAA. Cap-analysis gene expression (CAGE) tags are indicated by arrows, and CAGE-aided cDNA primer walking amplicons are displayed below the arrows. Not drawn to scale.
In order to determine the polyadenylation site(s) of GHRLOS transcripts, 3' RACE and inverse PCR were conducted using normal stomach and prostate tissues, the RWPE-1 prostate cell line and the PC3 prostate cancer cell line (which is derived from prostate cancer cells that have metastasised to the bone). We obtained 3' RACE clones from the PC3 prostate cancer cell line, which demonstrated a 1.4 kb exon 4a and a 1.1 kb exon 4b, with the putative polyadenylation signal AAATTA [GenBank:EU789531, and EU789532] (Fig. 1). The 3' RACE data matches a number of Expressed Sequence Tags (ESTs) from subchondral bone [GenBank:BM991802], foetal liver [GenBank:AI056187], pooled liver and spleen [GenBank:BX118778; R01027], Wilms' tumour (a paediatric cancer of the kidney) [GenBank:AI174574], pooled brain cancers [GenBank:AI863058] and high grade serous carcinoma of the uterus [GenBank:AI289483]. Moreover, we performed uncapped inverse PCR, and sequencing of amplicons from the PC3 cell line revealed a 90 bp exon I and exon 4 sequence with a 3' end differing by less than 20 base pairs when compared to the 3' RACE and reported ESTs [GenBank:EU789533]. Interestingly, the putative 3' end of exon 4 corresponds to publicly available Affymetrix Human Exon 1.0 ST array data (Exon Cluster ID 183613). Furthermore, all of the amplicons that we obtained were followed by a stretch of adenosines at the 3' ends that are not present in the genomic sequence. We, therefore, concluded that we had reached a genuine 3' end of GHRLOS.
Complex pattern of GHRLOS variant exon expression
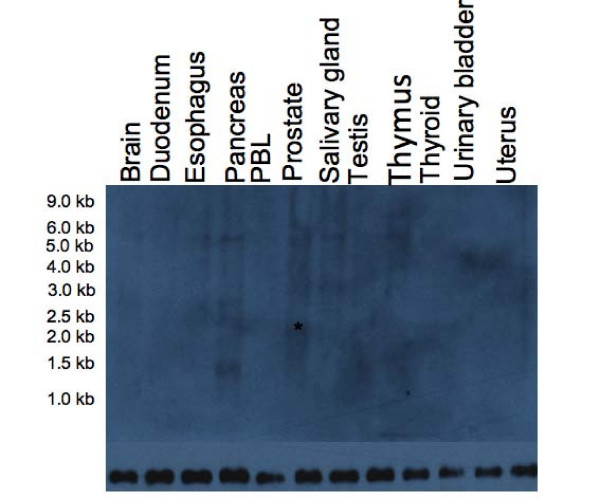
In order to examine the size and tissue distribution of GHRLOS transcripts, we performed Northern blot analysis of mRNA from human stomach and 12 normal, human tissues. With a riboprobe designed to span exons I, II, 1 and 2 of GHRLOS, a weak, smeared signal ranging from approximately 1.0 to 2.0 kb in size was observed in the human stomach upon a lengthened exposure time (data not shown). A riboprobe spanning exon 1 alone (a region common to all GHRLOS RNA isoforms) resulted in signals ranging from 1.0 to 5.5 kb in size in the pancreas, prostate, salivary gland and thymus (Fig. 2). Upon longer exposure times, a smear in the 1.0 to 5.5 kb range was seen in all tissues, except for peripheral blood leukocytes and urinary bladder (data not shown). This suggests that GHRLOS is a fully processed transcript consisting of many mRNA isoforms.
Figure 2.
Northern blot of poly(A)+ RNA from normal, human tissues with strand-specific, DIG-labelled riboprobe. Northern blot of 2 μg poly(A)+ RNA from 12 normal tissues (OriGene) probed for exon 1, which is an exon common to all known GHRLOS mRNA isoforms (top panel). The position of the RNA size markers is indicated (RNA Millennium Markers, OriGene). A pound mark (#) indicates a putative 1.7–1.8 kb (approximately 2.0 kb with polyadenylation tail) full-length transcript in the prostate, as deduced from RLM-RACE and RT-PCR experiments. Several other transcripts 1–5.5 kb in size could be seen. The same membrane was stripped and hybridized with a β-actin cRNA probe as a loading control (bottom panel).
GHRLOS exons are highly polymorphic
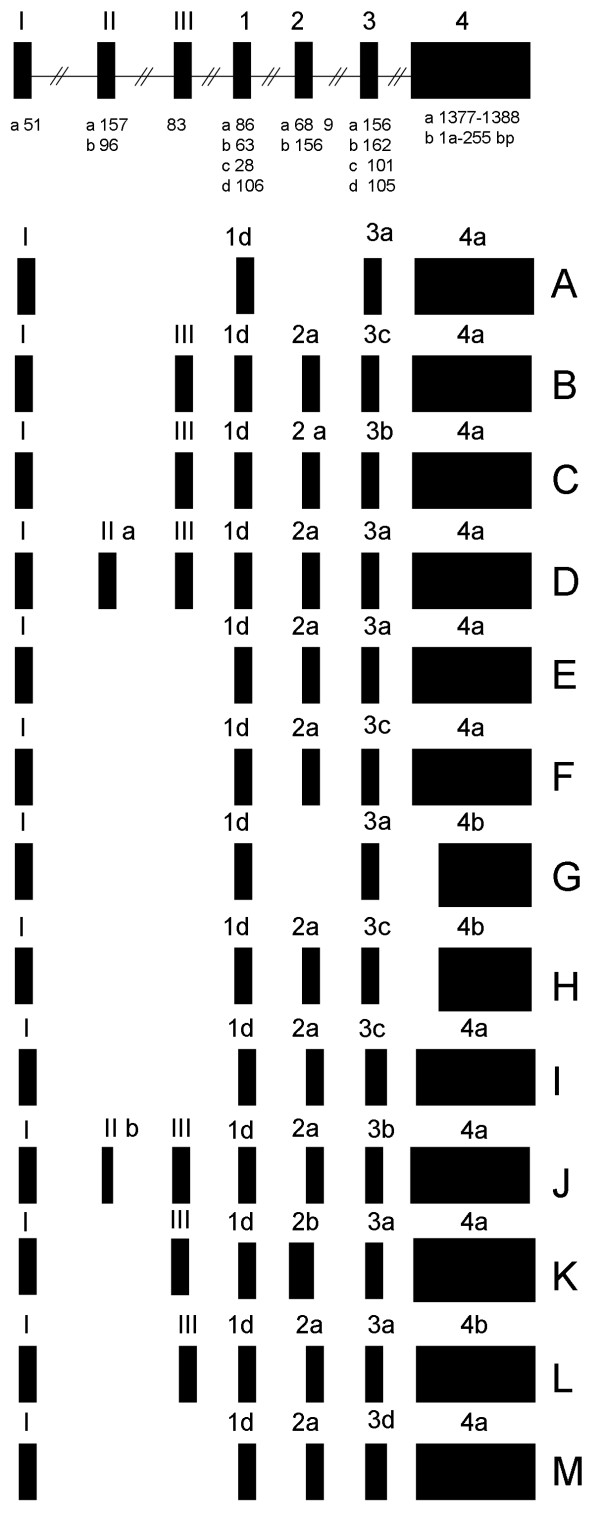
The RACE, inverse RT-PCR and Northern blotting experiments indicate that GHRLOS is extensively spliced and that isoforms range greatly in size (from approximately 1.0–5.5 kb). To examine the alternative splicing pattern of GHRLOS in greater detail, we performed RT-PCR using a range of human tissues and cell lines. We used a forward primer common to exon Ia (identified via 5' RLM-RACE) and a reverse primer in a region common to exons 4a and b. Sequence analysis indicated that, with the exception of exons 1 and 4, which are common to all known GHRLOS isoforms, GHRLOS exons are highly polymorphic in size and exon skipping occurs frequently. A representative amplicon banding pattern is shown in [Additional File 2]. The highly variable splicing pattern revealed by RT-PCR is consistent with the diffuse and broad signal which we detected by Northern blotting. In total we obtained 13 different GHRLOS splice variants (Fig. 3). Analysis using GMAP, a genomic mapping and alignment program for mRNA and EST sequences [14], indicated that all GHRLOS exons are flanked by canonical splice donor and acceptor sites (GT/AG), except for exons 3a and 3c where the splice junction is the common non-canonical splice pair GC/AG [15]. GHRLOS exons and introns identified are listed in [Additional File 3].
Figure 3.
Complex pattern of GHRLOS variant exon expression. GHRLOS transcripts A to M were demonstrated by RT-PCR, while the 5' end (exon I) and 3' end (exon 4) were determined by RACE, inverse PCR and by examining publically available Affymetrix exon array data and expressed sequence tag (EST) data. Exons are represented as boxes, introns as horizontal lines and sizes (bp) are indicated below each exon. The size of exon 4a is listed as 1377–1388 and corresponds to a transcript which is polyadenylated 20–30 bp from the same polyadenylation signal (data not shown). Exon 4b harbours the same polyadenlation site, but is 5' truncated (250 bp). GhrelinOS variants A to M have been deposited to GenBank: A-F, stomach [GenBank:EU789534, EU789535, EU789536, EU789537, EU789538, and EU789539]; G-H, prostate [GenBank:EU789540, and EU789541]; I, foetal brain [GenBank:EU789542]; J-M, heart [GenBank:EU789543, EU789544, EU789545, and EU789546].
Extending the known length of GHRLOS
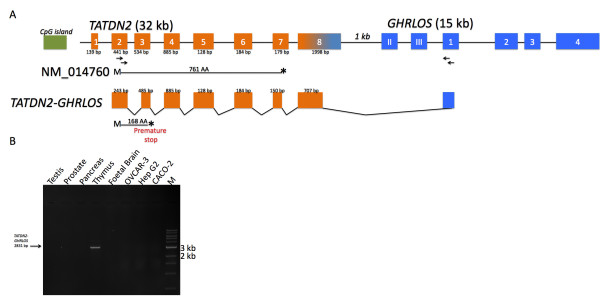
Northern blotting demonstrated GHRLOS transcripts 1–5.5 kb in size, however, further RT-PCR and 3' RACE analysis with a number of different primer combinations failed to reveal GHRLOS transcripts larger than 2 kb (data not shown). For this reason, we hypothesised that the ghrelin antisense gene may extend further upstream. Capped RLM-RACE of stomach tissue and uncapped inverse PCR of the PC3 prostate cancer cell line (Fig. 1) indicated that a GHRLOS promoter (exon I) was present at the very end of the 2 kb 3' untranslated exon 8 of TATDN2. Moreover, we discovered that multiple Cap Analysis of Gene Expression (CAGE) tags are clustered approximately 1.5 kb upstream of the GHRLOS transcription start sites found via 5' RLM-RACE. CAGE tags are an average of 20–21 nucleotides and are produced by large-scale sequencing of concatemers derived from the 5' ends of capped mRNA [16,17]. The CAGE method, therefore, detects the most 5' site of the mRNA transcripts (the transcription start site) and gives an unbiased and comprehensive picture of the positions and usage of transcription start sites [18]. To confirm if this region belongs to GHRLOS, we employed nested RT-PCR using thymus tissue, foetal brain tissue and Hep G2 hepatocarcinoma cell line cDNA (with RNA reverse transcribed using oligo(dT) primers). The forward primers were present immediately downstream of the CAGE tag cluster (which is 1.5 kb upstream of the 51 bp exon I of GHRLOS) and the reverse primers spanned exon 1 (which is common to all known GHRLOS variants). Sequence analysis revealed several novel exon I variants, which were approximately 1601 bp, 994 bp and 526 bp in length. All of these variants spliced into the expected acceptor site of the 106 bp exon 1 (Fig. 1 and [Additional File 4]). The end of the 1601 bp exon I (termed exon Ib) spanned the 51 bp exon I (exon Ia) identified via capped RLM-RACE of the stomach, while the other exons (Ic-d) employed different upstream donor sites, suggesting that they are alternative first exons. The data demonstrate that there are at least two first GHRLOS exon regions in the untranslated region of TATDN2. This suggests that the GHRLOS promoter is a broad-type, TATA-less promoter that initiates transcription at many sites [19].
The length of GHRLOS transcripts initiating in exon I (present in the 3' UTR of TATDN2) is 1.3–3.6 kb, corresponding in size to the Northern data. However, the potential identity of the approximately 5.5 kb transcript seen in the Northern blot with a full-length, 106 nt exon 1 probe (Fig. 2) was not determined. After prolonged exposure of the Northern blot, the 5.5 kb transcript was observed in all tissues except the urinary bladder and uterus (data not shown). TATDN2 is only approximately 1 kb upstream of exon II and 4.5 kb from exon 1 of GHRLOS, suggesting that transcription-induced chimaeras (TICs) may be generated [20] and give rise to large GHRLOS transcripts. Because TICs must contain a first exon of the upstream gene [20], we employed primers in exon 2 (immediately after the start codon) of TATDN2 and exon 1 of GHRLOS (which was also the sequence of our Northern riboprobe). Using nested RT-PCR (on cDNA reverse transcribed using oligo(dT) primers), we isolated a 2831 bp TATDN2-GHRLOS amplicon from the thymus [GenBank:EU789553] (Fig. 4). This transcript has canonical splice donor and acceptor sites (GT/AG) and splices into the expected acceptor site of the 106 bp exon 1. This variant harbour significant open reading frames corresponding to the TATDN2 protein, but contains alternative exons and a premature termination codon more than 50 bp upstream of the final coding exon 7 of TATDN2 (Fig. 4). This is likely to result in degradation of the mRNA by nonsense-mediated RNA decay (NMD), a surveillance mechanism that detects and degrades mRNA that may encode truncated proteins with dominant-negative or deleterious gain-of-function activities [21].
Figure 4.
Identification of a TATDN2-GHRLOS chimaeric transcript by nested RT-PCR. (A) Overview of the gene structures of TATDN2 (orange) and GHRLOS exons (blue). TATDN2 exon 8 contains first exons of GHRLOS and is therefore shown in orange and blue. Exons are represented as boxes, introns as horizontal lines and sizes (bp) are indicated below each exon. A TATDN2 CpG island is shown as a green box. The location of RT-PCR primers employed to detect chimaeras are shown below each exon as arrows. The reported 761 amino acid TATDN2 open reading frame (encoded by [GenBank:NM_014760]) is shown as a straight line (M = initiating methionine; * = stop codon). A chimaeric TATDN2-GHRLOS transcript, its open reading frame and locations of a premature stop codon (*) is shown. Not drawn to scale. (B) Ethidium bromide stained agarose gel of nested RT-PCR product for the TATDN2-GHRLOS chimaera. Arrows indicates the amplicon schematically displayed in (A). M = 1 kb DNA Ladder (NEB).
We have identified several novel GHRLOS transcripts. This includes overlapping GHRLOS transcripts initiating in the TATDN2 3' UTR and putative transcription-induced chimaeras of TATDN2 and GHRLOS. These findings extend the previously reported length of GHRLOS by ~37 kb. We propose that GHRLOS harbour several promoters with start sites in exon 1 (of GHRLOS), and in the 3' UTR and the first exon of TATDN2.
Results of in silico analysis indicate that GHRLOS is a non-coding RNA gene
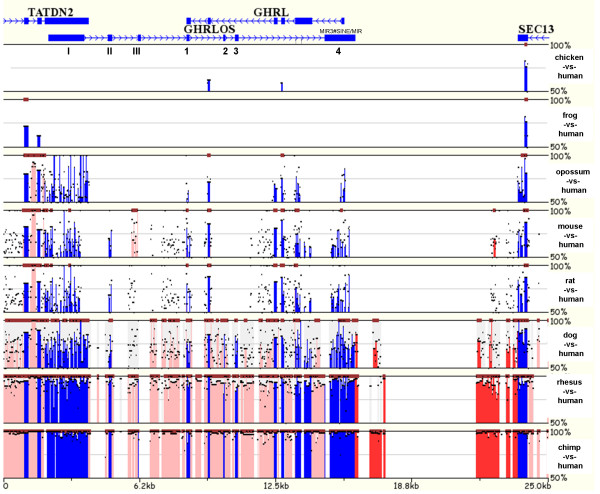
Sequence analysis of GHRLOS (excluding the putative TATDN2-GHRLOS transcription-induced chimaeras that are likely to result in nonsense mediated decay) reveals that GHRLOS transcripts do not harbour protein coding potential, but rather have several features of non-coding RNA genes. GMAP analysis showed that GHRLOS spans approximately 44 kb on 3p25.3 (data not shown). The Mulan sequence conservation profile for the human and putative vertebrate GHRLOS orthologues indicated that most GHRLOS exons (I, II, III, 2, 3) are not highly conserved compared to the putative mouse and rat orthologues (Fig. 5). The 106 bp exon 1 (which spans exon 4 of the ghrelin gene) and approximately 90 bp of the 1.4 kb exon 4 (which spans exon -1 of the ghrelin gene) are conserved, however [8]. RT-PCR experiments revealed no evidence for antisense transcription from exon I to exon 1 (of GHRLOS) in the mouse (data not shown), suggesting that ghrelin antisense transcription (at these specific locations) may not be conserved. Furthermore, there is very low sequence similarity between the chicken, frog, opossum and human GHRLOS sequences (Fig. 5). Interestingly, exons II, 2 and 3 appear to be highly conserved between human and dog [Additional File 5], with 70–75% homology for exons 2 and 3. However, no significant open reading frames (ORFs) are conserved between dog and human GHRLOS sequence (data not shown). Taken together, this suggests that GHRLOS has evolved rapidly and may be unique to primates.
Figure 5.
Mulan sequence conservation profile for vertebrate GHRLOS loci. The horizontal axis displays the input sequence from human (UCSC hg18), chicken [UCSC galGal3], frog [UCSC xenTro2], opossum [UCSC monDom4], mouse [UCSC mm9], rat [UCSC rn4], dog [UCSC canFam2], rhesus macaque [UCSC rheMac2], and chimpanzee [UCSC PanTro2] GHRLOS loci. Evolutionary conserved regions (ECRs, > 70% identity; ≥ 99 bp) are depicted as dark red bars above each pairwise alignment. Exons (blue), intergenic elements (red) and intron sequence (pink) are marked and the vertical axis shows the percent similarity of vertebrate GHRLOS orthologues to the human sequence. GHRLOS exon numbers are shown. A transposable element in exon 4 (MIR3#SINE/MIR) is shown above exon 4.
Our study suggests that GHRLOS is a non-coding RNA. In silico translation revealed that the heterogeneous GHRLOS RNAs contain multiple stop codons, resulting in lack of extensive reading frames, and the putative ORFs do not span conserved regions (data not shown). Moreover, no significant sequence similarity to any known proteins was observed (data not shown). Finally, screening of GHRLOS sequence against a reference collection of repeats (RepbasE) using CENSOR [22] identified a 203 bp overlap of exon 4 with the extinct 224 bp MIR3 SINE element, which is present in all vertebrates [23] (Fig. 5). Interestingly, the presence of repeat elements in exons of non-coding RNAs has been reported previously [24-27]
The GHRLOS terminal exon 4 and a putative SEC13 exon overlap in an antisense manner
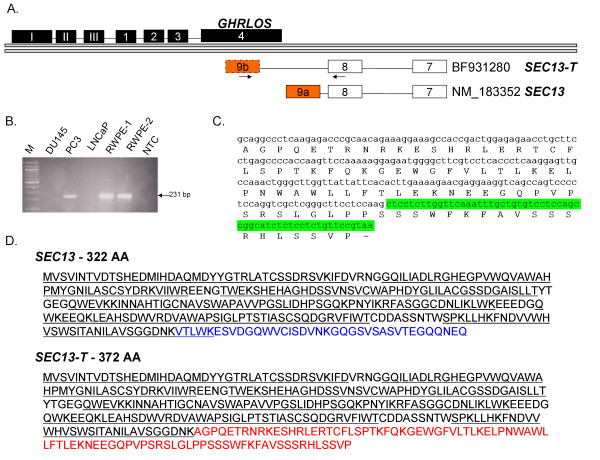
We have discovered that exon 4 of GHRLOS is also on the opposite strand of a novel terminal exon of the neighbouring SEC13 gene in a tail-to-tail, 3' to 3', fashion (Fig. 6A) [Additional File 6]. SEC13 plays an important role in protein transport and is a component of the COPII complex [28,29]. BLAST analysis identified a brain tumour EST [GenBank:BF931280] [30], which contains exon 7 and 8 of SEC13, as well as 206 bp sequence corresponding to a novel exon 9b of SEC13 10.7 kb upstream of exon 8. We verified the expression of this EST by RT-PCR (using cDNAs reverse transcribed with oligo(dT) primers) (Fig. 6B) and by sequencing [GenBank:EU789555]. We have named this SEC13 variant, SEC13-tentative (SEC13-T). SEC13-T may encode a 372 amino acid protein. The first 285 amino acids are identical to SEC13, while the normal C-terminal 37 amino acids have been replaced by 87 amino acids encoded by the alternative terminal exon 9b (Fig. 6C and 6D). The putative C-terminal-coding exon of the SEC13-T isoform appears to be conserved only in primates (data not shown). Therefore SEC13-T alternative splicing is likely to be human-specific or primate-specific.
Figure 6.
GHRLOS overlaps a putative SEC13 isoform, SEC13-T, in an antisense manner. (A) Diagrammatic representation of GHRLOS, SEC13, and a novel SEC13 isoform, named SEC13-tentative (SEC13T). SEC13-T contains a novel terminal exon 9, termed exon 9b, which overlaps exon 4 of GHRLOS in an antisense manner. In the figure, exon 9b of SEC13-T is dotted to indicate that its complete 3' end has not been verified. Exons are represented as boxes and introns as horizontal lines. The locations of the primers employed to verify the SEC13T variant are displayed as arrows. SEC13 and SEC13-T GenBank accession numbers are shown. (B) Ethidium bromide stained agarose gel showing the verification of a SEC13-T EST [GenBank:BF931280] transcription by RT-PCR in prostate (RWPE-1 and RWPE-2) and prostate cancer derived cell lines (DU-145, PC3, LNCaP). NTC = no template control (water). M= DNA Molecular Weight Marker VI (Roche). Primer locations are depicted in (A). A 231 PCR fragment was amplified and sequenced, confirming the EST. (C) RNA and amino acid sequence of exon 9b of SEC13-T. Additional downstream sequence corresponding to 18 in-frame codons and a stop codon is highlighted in green. (D) SEC13 and SEC13-T amino acid sequences. SEC13 encodes a 322 AA protein with six WD-40 repeats (also known as WD or beta-transducin repeats) are underlined. Amino acid sequence of SEC13 which is different to SEC13T is shown in blue. SEC13-T may encode a 372 AA protein: Its first 285 amino acids are identical to SEC13 and the normal C-terminal 37 amino acids have been replaced by 87 amino acids (shown in red) encoded by exon 9b.
GHRLOS is expressed in many tissues and cell lines and the level of expression shows great variability
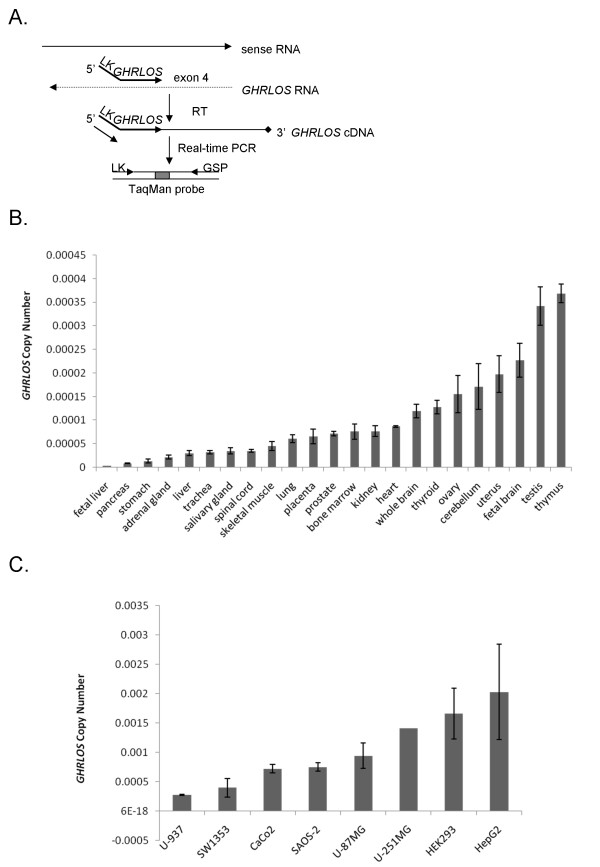
RT-PCR analysis (with primers spanning GHRLOS terminal exons) and Northern blotting (which is only suitable for high copy number transcripts) demonstrated that the size of GHRLOS transcripts is highly variable, resulting in significant transcript heterogeneity. We, therefore, employed a quantitative, real-time RT-PCR approach in order to more precisely gauge the expression of GHRLOS in a range of tissues and cell lines. As the number of alternatively spliced GHRLOS transcripts makes it impossible to generate real-time RT-PCR primers that are unique to each splice variant (data not shown), a strand-specific quantitative RT-PCR assay with primers in exon 4 (which is common to all GHRLOS variants) was designed to detect total GHRLOS RNA expression (Fig. 7A).
Figure 7.
Strand-specific GHRLOS real-time RT-PCR. (A) Strand-specific real-time RT-PCR approach to quantify GHRLOS expression. To select for RNA generated from the antisense strand, a linker sequence (LK) (not present in the genomic sequence of GHRLOS) is attached to a GHRLOS gene specific primer and employed in reverse transcription. The resulting cDNA is then combined in a real-time PCR, combining a gene-specific primer (GSP) and a primer containing the linker region from reverse transcription primer only. An internal TaqMan probe (depicted as a blue box) was employed to increase the specificity and sensitivity of the assay. (B) Relative total GHRLOS expression in a range of human human tissues. (C) Relative total GHRLOS expression in human cell lines (U-937 non-Hodgkin's lymphoma, SW1353 chondrosarcoma, CaCo-2 colorectal adenocarcinoma, SAOS-2 osteosarcoma, U-87 MG and U-251 MG glioblastoma, HEK293 human embryonic kidney and OVCAR-3 ovarian cancer). Calculations of GHRLOS expression levels were performed using the standard curve method (correlating the threshold cycle number (CT values) and copy numbers of GHRLOS), and normalised to the expression of 18S ribosomal RNA. Each bar presents the mean ± standard deviation of duplicate reactions.
The level of total GHRLOS transcript expression varied greatly in different human tissues, with high levels in the thymus, testis, foetal brain, uterus, cerebellum, ovary, thyroid, and whole brain. Very low levels of total GHRLOS RNA expression were detected in the stomach, foetal liver and pancreas (Fig. 7B). In the thymus the level of expression was approximately 133 fold higher than in the stomach (P < 0.01) and 110-fold higher than in the foetal liver (P < 0.01). Furthermore, the level of expression in the adult liver was 9-fold higher than in the foetal liver (P < 0.05), indicating differential expression according to developmental stage in this tissue. The level of GHRLOS in the foetal brain was two-fold higher than the adult brain, but this was not statistically significant (P > 0.05).
Expression of GHRLOS in a number of continuous cell lines was also examined using real-time RT-PCR (Fig. 7C). Levels of GHRLOS expression in the U-937 (Non-Hodgkin's lymphoma) and SW1353 (chondrosarcoma) cell lines were similar to the cerebellum, uterus, foetal brain, testis and thymus. High levels of expression were observed in the HEK293 human embryonic kidney cell line and in the CaCo-2 (colon adenocarcinoma), SAOS-2 (osteosarcoma), U-87 MG and U251-MG (glioblastoma), OVCAR-3 (ovarian adenocarcinoma) and Hep G2 (hepatocarcinoma) cancer cell lines. It is interesting to note that multiple Hep G2 CAGE tags have been identified in the exon I region of GHRLOS (see [Additional File 4]). GHRLOS RNA appears to be significantly upregulated in the Hep G2 cell line compared to adult and foetal liver (P < 0.05) (Fig. 7B and 7C). This observation could indicate that GHRLOS may be a specific target in the development of liver and other cancers.
Comparison of total GHRLOS and total GHRL expression
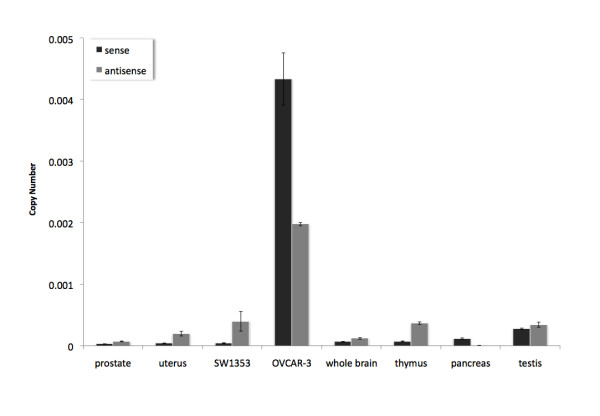
We examined the expression levels of the GHRL-GHRLOS cis-natural antisense transcript (cis-NAT) pair via quantitative real-time RT-PCR assays detecting total transcription from the ghrelin gene (GHRL). As expected [31], the highest level of GHRL expression was found in the stomach (data not shown), followed by the testis and pancreas (Fig. 8). When comparing total ghrelin and GHRLOS RNA expression in the stomach, GHRL was expressed at 2300 fold higher levels than GHRLOS, with GHRLOS expression almost undetectable (P < 0.001). The levels of GHRLOS expression were higher than GHRL in the thymus, whole brain, the SW1353 chondrosarcoma cell line, uterus and prostate. However, total GHRL RNA levels were higher than GHRLOS in the pancreas and the OVCAR-3 ovarian cancer cell line.
Figure 8.
Comparison of strand-specific GHRL and GHRLOS real-time RT-PCR. Relative total GHRL (sense, black) and GHRLOS (antisense, grey) RNA transcript expression levels in human tissues and cell lines. Calculations of RNA expression levels were performed using the standard curve method (correlating the threshold cycle number (CT values) and copy numbers of GHRL and GHRLOS) normalised to the expression of 18S ribosomal RNA. Each bar presents the mean ± standard deviation of duplicate reactions.
Discussion
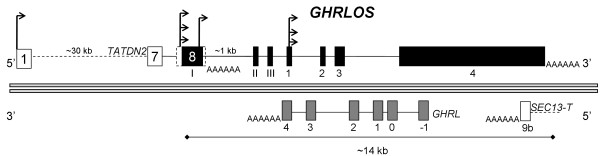
Our study demonstrates that GHRLOS gives rise to long, extensively spliced, mRNA-like, 5' capped and 3' polyadenylated transcripts suggesting that they are genuine products of RNA polymerase II mediated transcription [32]. We have shown that the GHRLOS gene gives rise to transcripts 1.0 to 5.5 kb in size and has many broadly distributed transcription starts sites (TSSs) (Fig. 9). This includes several TSSs in exon 1, TSSs overlapping the 3' UTR of TATDN2 and evidence of transcription induced chimaeras employing TSSs in the first exon of TATDN2. The ghrelin locus, therefore, gives rise to many antisense transcripts that are currently annotated as a single gene, GHRLOS. A well-described example of such complex architecture is the imprinted murine Gnas locus, which gives rise to multiple coding and non-coding sense and antisense transcription units [33].
Figure 9.
Structure of genes at the GHRLOS locus. Structure of the human GHRLOS (top) and ghrelin (GHRL) (bottom) genes. Exons are shown as boxes and introns as lines. Genes on the positive DNA strand are displayed above the double-lines, while genes on the negative DNA strand are shown below. Arrows indicate transcription start sites of GHRLOS transcripts. Ghrelin (GHRL) exons are shown in grey, and GHRLOS in black. A novel exon of a putative SEC13 variant, SEC13-T is shown as white boxes. The terminal exon 8 of TATDN2 is indicated by a dotted line.
First reported in 1987 [34] and originally thought to be a rarity, it has recently been established that promoters in 3' untranslated regions (3' UTRs) are not uncommon and may be independently transcribed and regulated from their upstream "host gene" [35]. The ENCODE (ENCyclopedia Of DNA Elements) consortium has recently demonstrated that two-thirds of the loci in their dataset contain new putative first exons, which frequently overlap upstream genes [36]. However, it is currently not known how promoters that overlap 3' UTRs are regulated and coordinated [37]. We have demonstrated GHRLOS transcription start sites in the 3' UTR of TATDN2 via RLM-RACE. The sequence upstream of exon I of GHRLOS contains no apparent TATA boxes (data not shown), indicating that GHRLOS has a broad type promoter, with many potential transcription start sites in the 2.1 kb 3' UTR of TATDN2. This may allow the transcription of numerous tissue-specific and developmental stage-specific transcripts [19]. In addition, multiple CAGE tags are present in the 3' UTR of TATDN2, indicating that GHRLOS transcripts initiate in this region.
Interestingly, we also report the joining of exons of the neighbouring genes TATDN2 and GHRLOS. Similar chimaeric transcripts (not caused by chromosomal translocation) have been reported in lower eukaryotes [38,39], but were until recently assumed to be relatively rare in mammals [20,40-43]. It is not known how chimaeric transcripts arise, but transcriptional read-through, followed by canonical cis-splicing is the most likely mechanism [40]. Alternatively, chimaeric transcripts could arise through trans-splicing, but the existence of this mechanism has not been well-established in mammals [44]. We, therefore, suggest that the chimaeric TATDN2-GHRLOS transcripts probably result from a single pre-mRNA, creating transcription-induced chimaeras (TICs) which are spliced in cis [40]. The TATDN2-GHRLOS TIC which was identified in the normal thymus harbours premature stop codons (PTCs) and this is likely to result in nonsense-mediated decay (NMD). NMD is an RNA surveillance mechanism (present in eukaryotes) which targets mRNA with premature stop codons that, if translated, encode truncated proteins with dominant-negative or deleterious gain-of function activities [21]. Whilst we are not aware of any studies of the translated product of TATDN2 (TatD DNase domain containing 2), TatD is a cytoplasmic protein that harbours magnesium-dependent DNase activity in E. coli [45]. Proteins with a TatD domain, therefore, belong to the large superfamily of metalloenzymes. This suggests that the chimaeric transcripts may play a role in auto-regulation of the TATDN2 protein product, which we hypothesise is likely to be a DNase associated with apoptosis. We speculate that TATDN2-GHRLOS chimaeras may be used to tightly regulate the programmed cell death of T-cells by downregulating the translation of the putative DNase TATDN2. Hillman and colleagues have termed this system of auto-regulation, where alternative splicing results in mRNA isoforms that are not translated, regulated unproductive splicing and translation (RUST) [46]. Interestingly, it has very recently been reported that transcripts of the tumour suppressor protein E-cadherin that result in non-sense mediated decay are upregulated in gastric cancer, suggesting that NMD may also promote disease [47].
We previously reported that GHRLOS completely overlaps the ghrelin (GHRL) gene [8]. Here we also show that the 3' terminal exon 4 of GHRLOS is present on the opposite (antisense) strand to a novel, 3' terminal SEC13 exon (Fig. 8). SEC13 is a protein that forms a part of the coat protein complex II (COPII) [28]. COPII proteins are required for the trafficking of nascent proteins from the endoplasmic reticulum (ER) to the Golgi apparatus. It also plays a role in the selection and concentration of cargo proteins for transport [29]. SEC13 [28], therefore, has a core endocrine function. GHRLOS overlaps a novel SEC13 variant, SEC13-tentative (SEC13-T). SEC13-T may encode a 372 amino acid protein with a unique C-terminus. Future studies are required to confirm the presence of full-length transcripts and to elucidate the functional role of SEC13-T protein. We suggest that transcription of SEC13-T may have implications for COPII function in health and disease states. As GHRLOS overlaps the terminal exons of both GHRL and SEC13-T, the GHRLOS natural antisense transcripts may regulate one or both of these genes.
GHRLOS, a candidate non-coding RNA
Non-coding RNAs are frequently not conserved between species, suggesting that they are either biological noise (non-functional transcription), or that they have species specific-functions. Species-specific non-coding transcripts have been observed and there is strong evidence that non-coding transcripts are functionally significant [48-52]. Interestingly it has recently been observed, using in silico analysis, that even between closely related Drosophila species non-coding RNAs are not conserved [53]. While the number of protein coding genes in distant eukaryotes (such as worms, mice, and humans) is approximately equal, the relative amount of non-coding DNA increases in proportion to eukaryotic complexity [54,55]. Mattick and colleagues explain this paradox by hypothesising that non-coding RNAs have evolved to enable the emergence of organisms with increasingly complex higher levels functions [55,56]. Our in silico analysis suggests that GHRLOS exons show very low sequence conservation in vertebrate species. Furthermore, exon 4 of GHRLOS contains a transposable element, a feature observed in many non-coding RNAs [25-27].
Our bioinformatic studies indicate that GHRLOS does not encode a protein and, therefore, is a non-coding RNA. Taking into account the full-length sequence of GHRLOS, the length of the open reading frames is very short. Moreover, GHRLOS RNAs are numerous (due to extensive splicing), contain a large number of stop codons and there is little nucleotide and putative amino acid conservation between vertebrates, making it less likely that this gene encodes proteins. Although it has been suggested that many small peptides may be translated [57], the majority of small peptides are processed from larger precursor proteins, as is ghrelin itself which is processed from preproghrelin. Therefore, while it cannot be excluded that GHRLOS encodes short ORFs (that are not conserved in the mouse) it appears unlikely that GHRLOS encodes biologically active peptides.
We examined the GHRLOS expression profile to strengthen the hypothesis that it is a candidate non-coding RNA. It has been demonstrated that mammalian long non-coding RNAs are expressed in a tissue-specific manner, indicating that they are biologically significant [50,58-61]. In both humans and mice, the major tissues of non-coding expression are the complex organs; the brain, testis, and thymus [50,61]. This also holds true in Drosophila, where the majority of the candidate non-coding RNAs are expressed in the central nervous system [53]. Indeed, non-coding RNAs are emerging as important regulators of complex systems, such as the central nervous system (brain) and intricate processes, including spermatogenesis in the testis [62-65]. We demonstrated high levels of GHRLOS in the thymus, brain, testis, uterus, ovary and thymus, while the expression levels in the stomach, where GHRL is highly expressed [1], were almost undetectable. Our data demonstrate that GHRLOS is predominantly expressed in a limited number of tissues and cell types, suggesting that these transcripts have physiological functions in distinct cell types and tissues. Moreover, real-time RT-PCR showed extremely low levels of GHRLOS in the foetal and adult liver and high levels in the Hep G2 hepatocarcinoma cell line, suggesting that GHRLOS expression may be altered in liver cancer. Indeed, it has recently been reported that non-coding RNA expression is frequently altered in cancer [58,66]. This indicates that non-coding RNAs may have specific functions in normal cells. Much like protein-coding transcripts, ncRNAs may act as tumour suppressors, or be upregulated in cancer and act as oncogenes. An examination of GHRLOS expression in cancer would, therefore, be of great interest.
Here we have characterised the structure and organisation of GHRLOS, a ghrelin antisense gene, suggesting that GHRLOS has multiple first exons. Therefore, it is possible that GHRLOS could be a part of very large, continuous ncRNA species in the 3p25 chromosomal region and beyond. In the absence of hallmarks, such as large open reading frames, mapping complex non-coding RNA genes remains a complex task. For example, two RNAs in the FMR1 locus, the cis-NAT ASFMR1 [67] and the non-coding RNA gene FMR4 (found just upstream of FMR1 [51]) may be one continuous RNA isoform [51].
What is the function of GHRLOS?
It is currently difficult and costly to determine the mechanism of function of long non-coding RNAs. The roles of non-coding RNAs are likely to be diverse, and there is strong evidence that they play a role in regulating important pathways. Many ncRNAs are expressed during development, neural differentiation, during macrophage activation and in cancer, indicating that they have key functions in these processes [12,63,68]. Non-coding RNAs have been found to play a role in the silencing of overlapping genes in cis [69], in the silencing of distant chromosome regions in trans [70], in nuclear trafficking [71], apoptosis [51,72], promoter repression [73], and can act as tumour suppressors [74]. Moreover, ncRNAs are emerging as markers for complex human disease, including lung cancer [75], heart disease [76], and a range of other pathologies [68,77]. GHRLOS may also serve as a host gene for snoRNA (small nucleolar RNAs) genes [78] or GHRLOS RNA transcripts may be precursors for short RNAs, such as micoRNAs [77], endogenous siRNAs [79,80], piRNAs [81] and other novel, short non-coding RNA species [82,83].
Although the understanding of natural antisense transcripts (NATs) remains in its infancy, they have been associated with a range of regulatory mechanisms that are not necessarily mutually exclusive. This includes transcriptional interference, RNA masking and dsRNA mediated gene-silencing via direct interaction between the sense and antisense transcripts [9,84,85].
While it is difficult to predict GHRLOS function, the fact that all spliced GHRLOS variants share exon 1, which overlaps the 3' untranslated exon 4 of GHRL is striking. Our findings suggest that GHRLOS functions as a non-coding RNA. There are a few examples that suggest that antisense transcripts are important in the regulation of endocrine hormone receptors, including a thyroid hormone receptor [86,87], and the luteinising hormone/choriogonadotropin receptor [88] gene and in the regulation of growth factors [89]. Interestingly, it has been recently suggested that the invertebrate (insect) polypeptide hormone allatostatin may be regulated by cis-NATs [90]. To date, however the physiological and pathophysiological roles of natural sense/antisense pairs have not been elucidated for any vertebrate endocrine hormone. Further studies are necessary to reveal the function and molecular mechanisms regulating the candidate non-coding RNA gene GHRLOS.
Conclusion
In the present study, we have characterised GHRLOS, which gives rise to endogenous ghrelin natural antisense transcripts. GHRLOS exhibits features which are common to many non-coding RNA genes, including extensive splicing, lack of significant and conserved open reading frames, differential expression and lack of conservation in vertebrates. Our data also reveal that GHRLOS contains multiple first exons and that it overlaps both GHRL and a novel SEC13 exon in the antisense direction, suggesting that GHRLOS may have a role in regulating these genes. Moreover, we report TATDN2-GHRLOS chimaeras that may function to regulate the translation of the putative DNase TATDN2. Additional studies are underway to elucidate the functions of GHRLOS and to investigate, in particular, its overlapping genomic arrangement with the ghrelin gene. These studies may provide a new, physiologically relevant model system for investigating the roles of antisense gene and non-coding RNA regulation and the mechanisms involved, as well as establishing whether GHRLOS RNAs may be useful markers for diagnosis and prognosis of complex disease.
Methods
Bioinformatics
Multiple sequence alignments were generated using the MUltiple sequence Local AligNment and conservation visualization tool (Mulan) [91]. Human [UCSC hg18], chicken [UCSC galGal3], frog [UCSC xenTro2], opossum [UCSC monDom4], mouse [UCSC mm9], rat [UCSC rn4], dog [UCSC canFam2], rhesus macaque [UCSC rheMac2], and chimpanzee [UCSC PanTro2] genomic sequences were obtained via the Evolutionary Conserved Regions (ECRs) Browser [92] and forwarded to Mulan to generate a full local alignment of the GHRLOS locus. GHRLOS was annotated based on the exons sequenced in this study.
To identify putative antisense exons and transcripts, we examined the publicly available Affymetrix Human Exon 1.0 ST Array tissue panel dataset consisting of 11 tissues (breast, cerebellum, heart, kidney, liver, muscle, pancreas, prostate, spleen, testes, and thyroid) using the Affymetrix All Exon track in the UCSC Genome Browser [93] and the Integrated Genome Browser (IGB) from Affymetrix [94].
To locate transcription start sites in the putative first exons of GHRLOS, CAGE (Cap Analysis of Gene Expression) tags (deposited by the RIKEN consortium and its collaborators) were obtained via the Genome Network Platform Viewer [95]. We then recovered the RNA library information for each CAGE tag starting site. Briefly, each CAGE tag was individually queried against the 1.4 GB CAGE tag sequencing file, (release date 13.11.2006) available on the Genome Network Platform website using the UNIX grep command [96].
The exon-intron-structure of ESTs and mRNA entries identified from BLAST searches, as well as sequenced PCR amplicons obtained in this study, were analysed against the human genome (NCBI release 35) using GMAP [14]. Presence of open reading frames was analysed by NCBI ORF Finder [97], Fickett's TestCode [98,99] and ESTScan2 [100,101]. The presence of transposable elements in GHRLOS sequence was examined using CENSOR v4.2.8 [22]. Protein domain analysis was performed using the SMART database [102].
Cell culture and RNA extraction
The following cell lines (originally obtained from the American Type Culture Collection/ATCC, Rockville, MD unless specified) were cultured in their recommended media: Prostate and/or prostate cancer derived cell lines DU145 (ATCC HTB-81), RWPE-1 (ATCC CRL-11609), RWPE-2 (ATCC CRL-11610), LNCaP (ATCC CRL-1740), 22Rv1 (ATCC CRL-2505) and PC3 (ATCC CRL-1435), HEK293 human embryonic kidney (ATCC CRL-1573), SAOS-2 osteosarcoma (ATCC HTB-85), Hep G2 hepatocarcinoma (ATCC HB-8065), U-87 MG and U-251 MG glioblastoma (ATCC HTB-14 and JCRB Cell Bank # IFO50288, respectively), CaCo-2 colorectal adenocarcinoma (ATCC HTB-37), SW1353 chondrosarcoma (ATCC HTB-94), and OVCAR-3 ovarian cancer (ATCC HTB-161). All cells were grown in T80 or T175 flasks (Nagle Nunc International, Roskilde, Denmark) in 95% CO2 in a Sanyo incubator at 37°C. Total RNA was harvested from cultured cells at 70% confluence using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Human RNA samples
Tissue Total RNA was obtained from the stomach, prostate (FirstChoice, Ambion, Austin, TX), foetal liver, adrenal gland, liver, trachea, salivary gland, spinal cord, skeletal muscle, lung, placenta, bone marrow, kidney, heart, whole brain, thyroid, cerebellum, uterus, foetal brain, testis, thymus (Human total RNA Master Panel II, Clontech, Mountain View, CA) and pancreas (Clontech).
5' and 3' RACE mapping of GHRLOS transcripts
To further characterise the 5' end of the putative ghrelin antisense RNAs, 5' RACE was undertaken using FirstChoice RLM-RACE-Ready human stomach cDNA (Ambion) according to the manufacturer's instructions. The first round PCR was performed with an adapter-specific sense primer (5'adapter-out-F, Table 1) and an exon 2-specific antisense primer (5'OS-out-R in Table 1). PCR product (1 μl) was used in a secondary, nested PCR with a gene specific primer in exon 2 (5'adapter-in-F and 5'OS-in-R, Table 1). PCRs were performed in a total reaction volume of 50 μl using 1 U of Platinum Taq Polymerase High Fidelity (Invitrogen) according to the manufacturer's instructions.
Table 1.
Designations and sequences of oligonucleotides
| Name | Sequence (5'-3') | GHRLOS Exon | Ta (°C) | PCR Cycles |
| 5'2-out-F | GATGGCGATGAATGAACACTG | N/A | ||
| 5'2-out-R | AATCATCTCAGGAATACCTGGA | 2 | 60 | 35 |
| 5'2-in-F | ATGAATGAACACTGCGTTTGC | N/A | ||
| 5'2-in-R | AAATGGAAGAGATGAGGCGC | 2 | 61 | 35 |
| Ex1-cRNA-F | CATACAGTTTGAACATTTATTCGCCTCC | 1 | ||
| Ex1-cRNA-R | CTAATACGACTCACTATAGGGAGACTCTCTCTAAGTTTAGAAGCGCTCATCTG | 1 | 62 | 25 |
| 3'2OF | GAGAGCGCCTCATCTCTTCC | 2 | ||
| 3'2OR | GCGAGCACAGAATTAATACGACTC | N/A | 63 | 35 |
| 3'2IF | ATGATTTATTGGAGCTCAAAGC | 2 | ||
| 3'2IR | GAATTAATACGACTCACTATAGGT | N/A | 57 | 15 |
| 3'4 | TACGGAACAGAGGAGAGATGC | 4 | 60 | 35 |
| 3' -RACE-adapter | GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN | N/A | N/A | |
| IPCR-out-F | AAATCCCACCTTTAGTCCCA | 4 | 60 | 35 |
| IPCR-in-F | CTGCCACCTGAGTGTAGAC | 4 | 60 | 20 |
| IPCR-ALL-R | CACAGGCTTGGAGACTTCC | I | ||
| Pthioate-hex | NNNNsNsN | N/A | 30 | N/A |
| Phospho-dT | P- GGCCACGCGTCGACTAGTAC(T)18 | N/A | 55 | N/A |
| GHRL-Real-RT-LK | CGACTGGAGCACGAGGACACTGAGCCAGAGAGCGCTTCTAAACTTA | N/A | N/A | |
| GHRL-Real-F | GCCCCAGCCGACAAGTG | N/A | 60 | 40 |
| Ito4-F | CATGGAAGTCTCCAAGCCTG | I | ||
| Ito4-R | CTGCTCTACTGCCTCAATGTC | 4 | 63 | 34 |
| GHRLOS-Real-RT-LK | CGACTGGAGCACGAGGACACTGACAATCCTCCCTGAGGTTGATCT | 4 | N/A | |
| GHRLOS-Real-F | CATTGAGGCAGTAGAGCAGTTGA | 4 | ||
| LK | CGACTGGAGCACGAGGACACTGA | N/A | 60 | 40 |
| Ex4-TaqMan | FAM-TGCCGAATGACCACCTACCCTGACTT- BHQ1 | 4 | ||
| 18S-Real-F | TTCGGAACTGAGGCCATGAT | N/A | ||
| 18S-Real-R | CGAACCTCCGACTTTCGTTCT | N/A | 60 | 40 |
| F_CAGE | GGGACTGCCTGTAATAGCAC | I | ||
| R_CAGEout | CACGACTGTTGTACAAGCTC | 1 | 60 | 35 |
| R_CAGEin | GGAGGCGAATAAATGTTCAAACTG | 1 | 61 | 30 |
| ChiOut-F | TGAAAGCCCAGAAGGAGGA | N/A | ||
| ChiOut-R | TCTAAGTTTAGAAGCGCTCATCTG | 1 | 63 | 35 |
| ChiIn-F | CAGAAGGAGGACGATGTGG | N/A | ||
| ChiIn-R | CACGACTGTTGTACAAGCTC | 1 | 62 | 30 |
| UIS231-F | ACAAGTTCAACGATGTGGTG | N/A | ||
| UIS231-R | CAAGTGTGAATAATAACCAAGCCC | N/A | 55 | 40 |
T7 promoter sequence in PCR primers is underlined. Linker sequence (LK) in the primer GHRLOS-Real-RT-LK and GHRL-Real-RT-LK is shown in bold. In primer 3' -RACE-adapter V denotes an A/G/C residue and N denotes A/G/C. Lowercase letters in primer Pthioate-hex denotes phosphothioate linkages. An uppercase P in primer Phospho-dT indicates that the synthetic oligonucleotide was 5' phosphorylated. Annealing temperatures (Ta) of oligonucleotides employed in PCR is shown. The GHRL or GHRLOS exon location of nucleotides are listed, while oligonucleotides spanning synthetic sequences (adapters and linkers) or genes other than GHRL and GHRLOS are denoted as N/A.
For GHRLOS 3' RACE, human stomach and PC3 prostate cancer cell line total RNA was reverse transcribed using Transcriptor reverse transcriptase (Roche Applied Science Applied Science, Penzberg, Germany) and 10 μM adapter primer (3'-RACE-adapter, Table 1) from the FirstChoice RLM-RACE Kit (Ambion). 3' RACE was performed with 2 μl of this cDNA. Two 3' RACE reactions were performed – one combined an exon 4 GHRLOS-specific forward primer and an adapter-specific reverse primer (3'4F and 3'2OR, Table 1), and the other used an adapter-specific reverse primer and an exon 2-specific forward primer 3'2OR/F, Table 1). PCR products were then diluted and used in a secondary, nested PCR with a gene-specific forward and a reverse adapter primer (3'2IF/R, Table 1). PCR products were purified using a High Pure PCR purification kit (Roche Applied Science), cloned into pCR-XL-TOPO (Invitrogen) or pGEM-T Easy (Promega, Madison, WI), transformed into DH5α chemical competent cells (Invitrogen) and sequenced at the Australian Genome Research Facility (AGRF, Brisbane, Australia) using BigDye III (Applied Biosystems, AB, Foster City, CA).
Determination of GHRLOS transcription start and polyadenylation sites by Rolling Circle Amplification Rapid Amplification of cDNA Ends (RCA-RACE)
To simultaneously obtain the 5' and 3' ends of GHRLOS transcripts, we employed Rolling Circle Amplification-RACE (Rapid Amplification of cDNA Ends [103], an improved inverse PCR approach. Briefly, 3 μg stomach, prostate, RWPE-1 cell line and PC3 prostate cancer cell line total RNA were reverse transcribed using 10 U of Transcriptor reverse transcriptase (Roche Applied Science) and 100 μM HPLC-purified 5'-end phosphorylated oligo d(T)-adapter primer (Phospo-dT, Table 1) (Proligo, Boulder, CO) according to the manufacturer's instructions. The single-stranded cDNA was purified using a High Pure PCR purification kit (Roche Applied Science) and eluted in 50 μl elution buffer (10 mM Tris-HCl, pH 8.5). Next, 25 μl purified linear cDNA was circularised using 100 U of CircLigase (EPICENTRE Biotechnologies, Madison, WI) and purified as before. After self-ligation, 15 μl circular cDNA was added to a rolling circle amplification reaction with 10 U of φ 29 DNA polymerase (NEB) and 10 μM HPLC-purified random hexamer primers with two phosphothioate linkages on their 3'ends (Pthioate-hex, Table 1) (Proligo). Following a 21 h incubation at 30°C in a waterbath, RCA-products were subjected to two rounds of inverse PCR with 1 U of Platinum Taq HIFI polymerase (Invitrogen), as per manufacturer's instructions. PCRs were performed in a reaction volume of 50 μl using a PTC-200 thermocycler (MJ Research, Waltham, MA). For the first round amplification, the RCA reaction was diluted 1/100 in water, and 1 μl used in an outer PCR with a forward primer at the start of exon 4 and a reverse primer in exon I (IPCR-out-F and IPCR-ALL-R, Table 1). After 35 cycles at a 60°C annealing temperature, the outer PCR product was diluted 100 times in water and 1 μl was used in a hemi-nested PCR of 20 cycles, with annealing at 60°C (IPCR-in-F and IPCR-ALL-R, Table 1). Amplification products were eluted from agarose gels in 50 μl water overnight, reamplified, cloned into pCR-XL-TOPO (Invitrogen), transformed into One Shot MAX Efficiency DH5α-T1R chemically competent cells (Invitrogen) and sequenced at the Australian Genome Research Facility (AGRF, Brisbane, Australia).
Northern blot hybridisation
Initially, a cRNA probe spanning exon I, II, 1 and 2 of GHRLOS was employed. Briefly, a 5' RACE clone in pGEM-T Easy was linearised with SalI restriction enzyme and a cRNA probe was synthesised using T7 RNA polymerase and a digoxigenin (DIG) RNA labelling kit (Roche Applied Science). Probe concentration was estimated by dot blot comparison with digoxigenin-labelled standards. 500 ng stomach poly(A)+ RNA (FirstChoice, Ambion) was separated on a 1.2% formaldehyde gel and blotted, as described previously [103]. Samples were electrophoresed with 50 ng RNA Molecular Weight Marker II (Roche Applied Science). The blot was hybridised to 50 ng/mL DIG-labelled cRNA probe overnight. Prehybridisation and hybridisation was performed with DIG-Easy Hyb (Roche Applied Science) at 65°C. The membranes were washed twice for 5 min at room temperature with 1 × Saline-Sodium Citrate (SSC), 0.1% sodium dodecyl sulfate (SDS) and then washed three times for 10 min at 65°C with 0.1 × SSC, 0.1% SDS. The membrane was then reacted with an alkaline phosphatase (AP)-conjugated anti-DIG antibody (Roche Applied Science). AP activity was detected using a chemiluminescence method using CDP-Star (Roche Applied Science).
A second cRNA probe, which spanned exon 1 (which is common to all known GHRLOS mRNA isoforms) was synthesised from 100 ng human stomach genomic DNA (BioChain, Hayward, CA) using the PCR method [104] (Ex1-cRNA-F/R, Table 1). The PCR product was purified using a High Pure PCR Product Purification Kit (Roche Applied Science) and the DIG-labelled cRNA probe synthesised and quantified as detailed above. A multi-tissue membrane containing poly(A)+ RNA from 12 human tissues (brain, duodenum, oesophagus, pancreas, PBL/leukocytes, prostate, salivary gland, testis, thymus, thyroid, urinary bladder and uterus) was purchased from OriGene (Rockville, MD). Prehybridisation and hybridisation were performed as described above, except that ULTRAhyb Ultrasensitive Hybridization Buffer (Ambion) was used instead of DIG-Easy Hyb (Roche Applied Science). Equivalent loading between tissues on the blot was determined by rehybridising with 20 ng/mL DIG-labelled β-actin cRNA probe (Roche Applied Science).
Isolation of alternatively spliced GHRLOS mRNAs via non-quantitative RT-PCR
For non-quantitative RT-PCR analysis of GHRLOS splicing, RT-PCRs were performed with a forward primer in a region common to the 5' terminal exon Ia/b and a reverse primer in the 3' terminal exon 4 of GHRLOS (Ito4-F/R, Table 1). cDNA was synthesised in a final volume of 20 μl from 3 μg total RNA from tissues and cell lines using 10 U of Transcriptor reverse transcriptase (Roche Applied Science), 20 U of RNasin Plus RNase Inhibitor (Promega) and a 3' RACE adapter primer (3'-RACE-adapter, Table 1) at 55°C according to the manufacturer's instructions. PCR amplicons from the stomach, prostate, foetal brain, heart, thymus, testis, and pancreas were purified, sub-cloned and sequenced as described above.
Long-range RT-PCR to detect putative chimaeric TATDN2-GHRLOS transcripts
To detect long, chimaeric transcripts, we employed RT-PCR with a forward primer in exon 2 of TATDN2 (ChiOut-F, Table 1) and a reverse primer in exon 1 of GHRLOS (ChiOut-R, Table 1). PCR was carried out with 1 U of Platinum Taq HIFI polymerase (Invitrogen) as per manufacturer's instructions, extending at 68°C for 2.5 minutes per cycle. cDNA was synthesised as above in a final volume of 20 μl from 2 μg total RNA, from the Hep G2 hepatocarcinoma cell line, CaCo-2 colorectal adenocarcinoma cell line, OVCAR-3 ovarian cancer cell line, and from a range of normal tissues (testis, prostate, pancreas, thymus, and foetal brain). RT-PCR products were sub-cloned and sequenced as described above.
CAGE-aided cDNA primer walking
To determine if the identified upstream CAGE tag starting sites transcribe exons that belong to GHRLOS, we employed RT-PCR using a forward primer designed to the region immediately after a CAGE cluster in the ~2 kb 3' untranslated region of the adjacent gene TATDN2 (TSS ID T03F009D1927) and a reverse primer in exon 1, an exon which is common to all known GHRLOS variants (F_CAGE and R_CAGEout in Table 1, respectively). cDNA was synthesised in a final volume of 20 μl from 2 μg total RNA, from the Hep G2 hepatocarcinoma cell line and the thymus and foetal brain, using 10 units Transcriptor reverse transcriptase (Roche Applied Science), 20 U of RNasin Plus RNase Inhibitor (Promega) and oligo(dT)18 primers (Proligo) according to the manufacturer's instructions. The cDNAs were subjected to 35 cycles of PCR with a two-minute extension time per cycle, and then diluted 1/100 in water and subjected to a hemi-nested 30-cycle PCR with a nested primer in exon 1 (R_CAGEin, Table 1). PCRs were performed in a total reaction volume of 50 μl using 10 U of Platinum Taq Polymerase High Fidelity (Invitrogen) according to the manufacturer's instructions. Entire PCR products were purified using the High Pure PCR Product Purification Kit (Roche Applied Science), subcloned into pCR-XL-TOPO (Invitrogen) and transformed into One Shot MAX Efficiency DH5α-T1R chemical competent cells (Invitrogen). Insert-positive, purified clones were sequenced by the Australian Genome Research Facility (AGRF, Brisbane, Australia) using the AB PRISM BigDye Terminator Cycle Sequencing Kit v3.1 protocol (AB).
Identification of novel SEC13 exon
To verify the presence of a novel SEC13 exon identified in a brain tumour EST [GenBank:BF931280], cDNAs reverse transcribed with an oligo(dT) primer (as described above) were challenged by RT-PCR with primers in exon 8 of SEC13 and a reverse primer in the novel exon (231-F/R, Table 1, respectively).
Strand-specific, quantitative real-time RT-PCR
To allow strand-specific and RNA-specific amplification [105-107] of GHRLOS transcripts, reverse transcription was performed using a gene-specific primer in exon 4 with a linker (LK) [107] sequence attached to the 5' end of the primer (GHRLOS-Real-RT-LK, Table 1). cDNA was generated from 1 μg total RNA using 40 U of AMV reverse transcriptase (Roche Applied Science) at 42°C, according to the manufacturer's instructions. The strand-specific, real-time RT-PCR was performed with an exon 4 specific forward primer, a reverse primer with the LK sequence only (GHRLOS-Real-F and LK, Table 1) and a TaqMan probe (Ex4-TaqMan, Table 1). To detect sense GHRL transcripts, we employed a strand-specific RT-PCR approach, with a reverse transcription primer spanning the 3' terminal exon 4 of the ghrelin gene (GHRLex4_RT_LK, Table 1) followed by PCR with an exon 4 specific forward primer (GHRLex4_F, Table 1) and a linker-specific reverse primer. (LK, Table 1). The relative quantification of GHRLOS and GHRL transcripts was estimated by direct normalisation to the threshold cycle (CT) of the housekeeping gene, 18S ribosomal RNA (18S-Real-F/R, Table 1). 18S PCRs were used to normalise real-time data. As reported for GAPDH [107], 18S RNAs self-primes efficiently in reverse transcription reactions without the addition of random or gene-specific primers. All primers were designed using the Primer Express version 2.0 software (AB).
PCRs were performed in a total reaction volume of 20 μl using Platinum Quantitative PCR SuperMix-UDG w/ROX (Invitrogen) for GHRLOS, while GHRL and the housekeeping gene 18S ribosomal RNA were amplified using 2 × SYBR green master mix (AB). Controls included the use of cDNA, which was reverse transcribed using random hexamers as primers, as well as the reverse transcription of RNA in the absence of primer. Real-time RT-PCR was performed using the AB 7000 sequence detection system (AB) and data analysed using the absolute standard curve method (User Bulletin #2, AB) to determine expression levels in a range of tissues and cell lines. Briefly, we calculated values from duplicate reactions for each sample from standards, which were constructed from PCR products. Statistical significance was determined using the Student's t-test and, where applicable, one-way analysis of variance (ANOVA) with Tukey post-hoc analysis. P-values of < 0.05 were considered to be statistically significant. Data are represented as mean ± standard deviation (S.D.).
Authors' contributions
IS conceived and designed the study and carried out all experiments except quantitative real-time RT-PCR. SLC carried out real-time RT-PCR and the resulting statistical analyses. ACH and LKC participated in its design and coordination. All authors participated in interpreting the data, wrote, read and approved the final manuscript).
Supplementary Material
Overview of GHRLOS exon 1. This is a TIFF file showing GHRLOS transcription start sites and exon 1 sequence. Exon 1 and exons I, II and III, all of which splice into the 106 bp exon 1, are depicted as black boxes. Exons with transcription start sites are indicated by arrows in the direction of transcription. The sequence of the 106 bp exon 1, which splices into upstream exons, is shaded in grey. Transcription start sites in exon 1, previously determined via 5' RACE (Δ) and CAGE (Cap Analysis of Gene Expression) (*), are indicated, and the exact transcription start site nucleotides are underlined and in bold. For comparison, the sequence of the reference (sense) exon 4 of the ghrelin gene [GenBank:NM_016362] is shown in red. GHRLOS exon 1 sequence is shown in upper case, while intron sequence is shown in lower case.
Ethidium bromide stained agarose gel electrophoresis of GHRLOS exon I a/b to 4 non-quantitative RT-PCR amplicons from cultured cells and normal prostate and stomach tissue. This is a TIFF file showing the expression profile of exon I a/b to 4 amplicons in various human tissues and cell lines, indicating a complex splice pattern. M = MassRuler Express DNA ladder (Fermentas, Burlington, Canada).
Compilation of exons of GHRLOS transcripts. This is a PDF file listing exons of GHRLOS. Exon and intron sizes (bp) are indicated. Experimental evidence from this study and/or external references for each exon are shown.
Overview of CAGE-aided primer walking amplicons and CAGE tags in the 3' untranslated exon 8 of TATDN2. This is a PDF file showing (A) The expression pattern of transcripts spanning exon 8 of TATDN2 (orange) and exon 1 of GHRLOS (blue) revealed via cDNA primer walking (exon shown as boxes, introns as lines, and sizes in bp indicated above each exon). (B) CAGE tags present in the 3' untranslated region of TATDN2 (H51BA34H0302-testis; H22BA47L0809-Hep G2 hepatocarcinoma; H21BB07A0112-Hep G2; H52BA49L1810-adrenal gland; H04BB25B1106-cerebrum; H63BA79F2401-heart; H59BA94L1109-SK-N-AS neuroblastoma cell line; H08BA37K2001-kidney malignancy; H21BA90I0206-HepG2 hepatocellular liver carcinoma; H62BB07A0901-SK-N-AS). TATDN2 exon sequence is shown in beige, intron sequence in lowercase and grey, GHRLOS sequence deduced from cDNA primer walking in orange, and the TATDN2 polyadenylation signal (AATAAA) is shown in red.
Comparison of human (h) and putative dog GHRLOS exon sequences. This is a JPEG file showing the comparison of human and putative dog GHRLOS exon sequences. The alignments were generated by the ClustalW program and drawn by BOXSHADE http://www.ch.embnet.org/software/BOX_form.html. Black shading indicates conserved nucleotides. (A) Exon II (B) Exon 2 (C) Exon 3.
Overview of GHRLOS exon 4 and overlapping exons. This is a TIFF file with an overview of GHRLOS exon 4 (highlighted in grey) showing the overlap with sense GHRL and SEC13-T exons (boxed). GT/AG intron splice sites are underlined. The exon 4 polyA signal is depicted in orange font. A stretch of poly(A) in the genomic sequence (exon 4) resulting of frequent oligo(dT) mispriming during cDNA synthesis is highlighted in pink.
Acknowledgments
Acknowledgements
This work was supported by grants from the Cancer Council Queensland (to LKC and ACH), the Faculty of Science, Queensland University of Technology and a QUT International Doctoral Scholarship (to IS). We thank Dr Yaowu He (IHBI, Kelvin Grove, Australia) for the U-937, U-87 MG and U-251 MG cell lines; Carina Walpole (IHBI) for the OVCAR-3 cell line; and Dr Penny Jeffery (Mater Medical Research Institute, South Brisbane, Australia) for the CaCo-2 colorectal adenocarcinoma cell line.
Contributor Information
Inge Seim, Email: i.seim@qut.edu.au.
Shea L Carter, Email: sl.carter@qut.edu.au.
Adrian C Herington, Email: a.herington@qut.edu.au.
Lisa K Chopin, Email: l.chopin@qut.edu.au.
References
- Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402:656–660. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- Heijboer AC, Hoek AM van den, Parlevliet ET, Havekes LM, Romijn JA, Pijl H, Corssmit EP. Ghrelin differentially affects hepatic and peripheral insulin sensitivity in mice. Diabetologia. 2006;49:732–738. doi: 10.1007/s00125-006-0138-2. [DOI] [PubMed] [Google Scholar]
- Bednarek MA, Feighner SD, Pong SS, McKee KK, Hreniuk DL, Silva MV, Warren VA, Howard AD, Ploeg LH Van Der, Heck JV. Structure-function studies on the new growth hormone-releasing peptide, ghrelin: minimal sequence of ghrelin necessary for activation of growth hormone secretagogue receptor 1a. J Med Chem. 2000;43:4370–4376. doi: 10.1021/jm0001727. [DOI] [PubMed] [Google Scholar]
- Neary NM, Druce MR, Small CJ, Bloom SR. Acylated ghrelin stimulates food intake in the fed and fasted states but desacylated ghrelin has no effect. Gut. 2006;55:135. [PMC free article] [PubMed] [Google Scholar]
- Tack J, Depoortere I, Bisschops R, Delporte C, Coulie B, Meulemans A, Janssens J, Peeters T. Influence of ghrelin on interdigestive gastrointestinal motility in humans. Gut. 2006;55:327–333. doi: 10.1136/gut.2004.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigman JM, Nakano Y, Coppari R, Balthasar N, Marcus JN, Lee CE, Jones JE, Deysher AE, Waxman AR, White RD, Williams TD, Lachey JL, Seeley RJ, Lowell BB, Elmquist JK. Mice lacking ghrelin receptors resist the development of diet-induced obesity. J Clin Invest. 2005;115:3564–3572. doi: 10.1172/JCI26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritos NA, Kokkotou EG. The physiology and potential clinical applications of ghrelin, a novel peptide hormone. Mayo Clin Proc. 2006;81:653–660. doi: 10.4065/81.5.653. [DOI] [PubMed] [Google Scholar]
- Seim I, Collet C, Herington AC, Chopin LK. Revised genomic structure of the human ghrelin gene and identification of novel exons, alternative splice variants and natural antisense transcripts. BMC Genomics. 2007;8:298. doi: 10.1186/1471-2164-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) and the FANTOM Consortium The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210:1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- Nagase T, Seki N, Ishikawa K, Ohira M, Kawarabayasi Y, Ohara O, Tanaka A, Kotani H, Miyajima N, Nomura N. Prediction of the coding sequences of unidentified human genes. VI. The coding sequences of 80 new genes (KIAA0201-KIAA0280) deduced by analysis of cDNA clones from cell line KG-1 and brain. DNA Res. 1996;3:321–329. doi: 10.1093/dnares/3.5.321. 341–354. [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Burset M, Seledtsov IA, Solovyev VV. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, Fukuda S, Sasaki D, Podhajska A, Harbers M, Kawai J, Carninci P, Hayashizaki Y. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. PNAS. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbers M, Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nat Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- Frith MC, Ponjavic J, Fredman D, Kai C, Kawai J, Carninci P, Hayashizaki Y, Sandelin A. Evolutionary turnover of mammalian transcription start sites. Genome Res. 2006;16:713–722. doi: 10.1101/gr.5031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R, Novik A, Sorek R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Chen W, Bocker W, Brosius J, Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183:345–351. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Yoon H, He H, Nagy R, Davuluri R, Suster S, Schoenberg D, Pellegata N, Chapelle Ade L. Identification of a novel noncoding RNA gene, NAMA, that is downregulated in papillary thyroid carcinoma with BRAF mutation and associated with growth arrest. Int J Cancer. 2007;121:767–775. doi: 10.1002/ijc.22701. [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Bata-Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy-Szabo A, Molnar G, Szentpali K, Bari L, Megyeri K, Mandi Y, Dobozy A, Kemeny L, Szell M. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280:24159–24167. doi: 10.1074/jbc.M501704200. [DOI] [PubMed] [Google Scholar]
- Moh MC, Lee LH, Yang X, Shen S. Identification of a novel gene HEPT3 that is overexpressed in human hepatocellular carcinoma and may function through its noncoding RNA. Int J Oncol. 2007;31:293–301. [PubMed] [Google Scholar]
- Hughes H, Stephens DJ. Assembly, organization, and function of the COPII coat. Histochem Cell Biol. 2008;129:129–151. doi: 10.1007/s00418-007-0363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan C, Stagg SM, Lapointe P, Balch WE. The COPII cage: unifying principles of vesicle coat assembly. Nat Rev Mol Cell Biol. 2006;7:727–738. doi: 10.1038/nrm2025. [DOI] [PubMed] [Google Scholar]
- Dias Neto E, Correa RG, Verjovski-Almeida S, Briones MR, Nagai MA, da Silva W, Jr, Zago MA, Bordin S, Costa FF, Goldman GH, Carvalho AF, Matsukuma A, Baia GS, Simpson DH, Brunstein A, de Oliveira PS, Bucher P, Jongeneel CV, O'Hare MJ, Soares F, Brentani RR, Reis LF, de Souza SJ, Simpson AJ. Shotgun sequencing of the human transcriptome with ORF expressed sequence tags. Proc Natl Acad Sci USA. 2000;97:3491–3496. doi: 10.1073/pnas.97.7.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanapavan S, Kola B, Bustin SA, Morris DG, McGee P, Fairclough P, Bhattacharya S, Carpenter R, Grossman AB, Korbonits M. The tissue distribution of the mRNA of ghrelin and subtypes of its receptor, GHS-R, in humans. J Clin Endocrinol Metab. 2002;87:2988. doi: 10.1210/jc.87.6.2988. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5'-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes R, Williamson C, Peters J, Denny P, Wells C. A comprehensive transcript map of the mouse Gnas imprinted complex. Genome Res. 2003;13:1410–1415. doi: 10.1101/gr.955503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshimaru M, Nakanishi S. Identification of a new type of mammalian myosin heavy chain by molecular cloning. Overlap of its mRNA with preprotachykinin B mRNA. J Biol Chem. 1987;262:14625–14632. [PubMed] [Google Scholar]
- Carninci P. Constructing the landscape of the mammalian transcriptome. J Exp Biol. 2007;210:1497–1506. doi: 10.1242/jeb.000406. [DOI] [PubMed] [Google Scholar]
- Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J, Dike S, Wyss C, Henrichsen CN, Holroyd N, Dickson MC, Taylor R, Hance Z, Foissac S, Myers RM, Rogers J, Hubbard T, Harrow J, Guigo R, Gingeras TR, Antonarakis SE, Reymond A. Prominent use of distal 5' transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras TR. The multitasking genome. Nat Genet. 2006;38:608–609. doi: 10.1038/ng0606-608. [DOI] [PubMed] [Google Scholar]
- Manak JR, Dike S, Sementchenko V, Kapranov P, Biemar F, Long J, Cheng J, Bell I, Ghosh S, Piccolboni A, Gingeras TR. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Williams BA, Slamovits CH, Patron NJ, Fast NM, Keeling PJ. A high frequency of overlapping gene expression in compacted eukaryotic genomes. Proc Natl Acad Sci USA. 2005;102:10936–10941. doi: 10.1073/pnas.0501321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigo R. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006;16:37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- Wexler M, Sargent F, Jack RL, Stanley NR, Bogsch EG, Robinson C, Berks BC, Palmer T. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J Biol Chem. 2000;275:16717–16722. doi: 10.1074/jbc.M000800200. [DOI] [PubMed] [Google Scholar]
- Hillman RT, Green RE, Brenner SE. An unappreciated role for RNA surveillance. Genome Biol. 2004;5:R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam R, Carvalho J, Bruno I, Graziadio C, Senz J, Huntsman D, Carneiro F, Seruca R, Wilkinson MF, Oliveira C. The NMD mRNA surveillance pathway downregulates aberrant E-cadherin transcripts in gastric cancer cells and in CDH1 mutation carriers. Oncogene. 2008 doi: 10.1038/onc.2008.62. [DOI] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M, Jr, Vanderhaeghen P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Sasaki YT, Sano M, Ideue T, Kin T, Asai K, Hirose T. Identification and characterization of human non-coding RNAs with tissue-specific expression. Biochem Biophys Res Commun. 2007;357:991–996. doi: 10.1016/j.bbrc.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile × syndrome. PLoS ONE. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan MD, Hong CC, Lai GM, Cheng AL, Lin YW, Chuang SE. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet. 2005;14:1465–1474. doi: 10.1093/hmg/ddi156. [DOI] [PubMed] [Google Scholar]
- Inagaki S, Numata K, Kondo T, Tomita M, Yasuda K, Kanai A, Kageyama Y. Identification and expression analysis of putative mRNA-like non-coding RNA in Drosophila. Genes Cells. 2005;10:1163–1173. doi: 10.1111/j.1365-2443.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- Taft R, Mattick J. Increasing biological complexity is positively correlated with the relative genome-wide expansion of non-protein-coding DNA sequences. Genome Biology. 2003;5:P1. doi: 10.1186/gb-2003-5-1-p1. [DOI] [Google Scholar]
- Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Forrest AR, Nourbakhsh E, Pang KC, Kai C, Kawai J, Carninci P, Hayashizaki Y, Bailey TL, Grimmond SM. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, Streng PS, Smith DI. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, Grimmond SM, Hume DA, Hayashizaki Y, Mattick JS. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, Mattick JS, Suzuki H. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, 3rd, Wahlestedt C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 2007;30:612–621. doi: 10.1016/j.tins.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Non-coding RNAs in the nervous system. J Physiol. 2006;575:333–341. doi: 10.1113/jphysiol.2006.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- Okada Y, Tashiro C, Numata K, Watanabe K, Nakaoka H, Yamamoto N, Okubo K, Ikeda R, Saito R, Kanai A, Abe K, Tomita M, Kiyosawa H. Comparative expression analysis uncovers novel features of endogenous antisense transcription. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn051. [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the 'genome complexity' conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Tsang WP, Wong TW, Cheung AH, Co CN, Kwok TT. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. Rna. 2007;13:890–898. doi: 10.1261/rna.359007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Nakamura Y, Tanaka T. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. Biochim Biophys Acta. 2005;1756:65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Kim M, Patel B, Schroeder KE, Raza A, Dejong J. Organization and transcriptional output of a novel mRNA-like piRNA gene (mpiR) located on mouse chromosome 10. Rna. 2008;14:1005–1011. doi: 10.1261/rna.974608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G. In search of antisense. Trends Biochem Sci. 2004;29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Osato N, Suzuki Y, Ikeo K, Gojobori T. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–1306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar MA, Hodin RA, Cardona G, Chin WW. Gene expression from the c-erbA alpha/Rev-ErbA alpha genomic locus. Potential regulation of alternative splicing by opposite strand transcription. J Biol Chem. 1990;265:12859–12863. [PubMed] [Google Scholar]
- Hastings ML, Ingle HA, Lazar MA, Munroe SH. Post-transcriptional regulation of thyroid hormone receptor expression by cis-acting sequences and a naturally occurring antisense RNA. J Biol Chem. 2000;275:11507–11513. doi: 10.1074/jbc.275.15.11507. [DOI] [PubMed] [Google Scholar]
- Chambers AE, Banerjee S. Natural antisense LHCGR could make sense of hypogonadism, male-limited precocious puberty and pre-eclampsia. Mol Cell Endocrinol. 2005;241:1–9. doi: 10.1016/j.mce.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Barclay C, Li AW, Geldenhuys L, Baguma-Nibasheka M, Porter GA, Veugelers PJ, Murphy PR, Casson AG. Basic fibroblast growth factor (FGF-2) overexpression is a risk factor for esophageal cancer recurrence and reduced survival, which is ameliorated by coexpression of the FGF-2 antisense gene. Clin Cancer Res. 2005;11:7683–7691. doi: 10.1158/1078-0432.CCR-05-0771. [DOI] [PubMed] [Google Scholar]
- Yin GL, Chen Q, Yang WJ. Naturally occurring antisense RNA of allatostatin gene in the prawn, Macrobrachium rosenbergii. Comp Biochem Physiol B Biochem Mol Biol. 2007;146:20–25. doi: 10.1016/j.cbpb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, Hardison RC, Stubbs L, Miller W. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004:W280–286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pedersen JS, Hsu F, Hinrichs AS, Harte RA, Diekhans M, Clawson H, Bejerano G, Barber GP, Baertsch R, Haussler D, Kent WJ. The UCSC genome browser database: update 2007 10.1093/nar/gkl928. Nucl Acids Res. 2007;35:D668–673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix Integrated Genome Browser https://www.affymetrix.com/support/developer/tools/IGB_User_Guide.pdf
- Genome Network Platform Viewer http://genomenetwork.nig.ac.jp/public/sys/gnppub/
- Haertel M. GNU grep-2.0. Internet Archive: Usenet archive comp. sources.reviewed. 1996.
- NCBI ORF Finder http://www.ncbi.nlm.nih.gov/projects/gorf/
- Fickett JW. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TestCode http://www.bioinformatics.org/SMS/testcode.html
- Lottaz C, Iseli C, Jongeneel CV, Bucher P. Modeling sequencing errors by combining Hidden Markov models. Bioinformatics. 2003;19:ii103–112. doi: 10.1093/bioinformatics/btg1067. [DOI] [PubMed] [Google Scholar]
- Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol. 1999:138–148. [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polidoros AN, Pasentsis K, Tsaftaris AS. Rolling circle amplification-RACE: a method for simultaneous isolation of 5' and 3' cDNA ends from amplified cDNA templates. Biotechniques. 2006;41:35–36, 38, 40 passim.. doi: 10.2144/000112205. [DOI] [PubMed] [Google Scholar]
- Chopin LK, Veveris-Lowe TL, Philipps AF, Herington AC. Co-expression of GH and GHR isoforms in prostate cancer cell lines. Growth Horm IGF Res. 2002;12:126–136. doi: 10.1054/ghir.2002.0271. [DOI] [PubMed] [Google Scholar]
- Lin L, Fevery J, Hiem Yap S. A novel strand-specific RT-PCR for detection of hepatitis C virus negative-strand RNA (replicative intermediate): evidence of absence or very low level of HCV replication in peripheral blood mononuclear cells. J Virol Methods. 2002;100:97–105. doi: 10.1016/S0166-0934(01)00399-8. [DOI] [PubMed] [Google Scholar]
- Pinto FL, Svensson H, Lindblad P. Generation of non-genomic oligonucleotide tag sequences for RNA template-specific PCR. BMC Biotechnol. 2006;6:31. doi: 10.1186/1472-6750-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of GHRLOS exon 1. This is a TIFF file showing GHRLOS transcription start sites and exon 1 sequence. Exon 1 and exons I, II and III, all of which splice into the 106 bp exon 1, are depicted as black boxes. Exons with transcription start sites are indicated by arrows in the direction of transcription. The sequence of the 106 bp exon 1, which splices into upstream exons, is shaded in grey. Transcription start sites in exon 1, previously determined via 5' RACE (Δ) and CAGE (Cap Analysis of Gene Expression) (*), are indicated, and the exact transcription start site nucleotides are underlined and in bold. For comparison, the sequence of the reference (sense) exon 4 of the ghrelin gene [GenBank:NM_016362] is shown in red. GHRLOS exon 1 sequence is shown in upper case, while intron sequence is shown in lower case.
Ethidium bromide stained agarose gel electrophoresis of GHRLOS exon I a/b to 4 non-quantitative RT-PCR amplicons from cultured cells and normal prostate and stomach tissue. This is a TIFF file showing the expression profile of exon I a/b to 4 amplicons in various human tissues and cell lines, indicating a complex splice pattern. M = MassRuler Express DNA ladder (Fermentas, Burlington, Canada).
Compilation of exons of GHRLOS transcripts. This is a PDF file listing exons of GHRLOS. Exon and intron sizes (bp) are indicated. Experimental evidence from this study and/or external references for each exon are shown.
Overview of CAGE-aided primer walking amplicons and CAGE tags in the 3' untranslated exon 8 of TATDN2. This is a PDF file showing (A) The expression pattern of transcripts spanning exon 8 of TATDN2 (orange) and exon 1 of GHRLOS (blue) revealed via cDNA primer walking (exon shown as boxes, introns as lines, and sizes in bp indicated above each exon). (B) CAGE tags present in the 3' untranslated region of TATDN2 (H51BA34H0302-testis; H22BA47L0809-Hep G2 hepatocarcinoma; H21BB07A0112-Hep G2; H52BA49L1810-adrenal gland; H04BB25B1106-cerebrum; H63BA79F2401-heart; H59BA94L1109-SK-N-AS neuroblastoma cell line; H08BA37K2001-kidney malignancy; H21BA90I0206-HepG2 hepatocellular liver carcinoma; H62BB07A0901-SK-N-AS). TATDN2 exon sequence is shown in beige, intron sequence in lowercase and grey, GHRLOS sequence deduced from cDNA primer walking in orange, and the TATDN2 polyadenylation signal (AATAAA) is shown in red.
Comparison of human (h) and putative dog GHRLOS exon sequences. This is a JPEG file showing the comparison of human and putative dog GHRLOS exon sequences. The alignments were generated by the ClustalW program and drawn by BOXSHADE http://www.ch.embnet.org/software/BOX_form.html. Black shading indicates conserved nucleotides. (A) Exon II (B) Exon 2 (C) Exon 3.
Overview of GHRLOS exon 4 and overlapping exons. This is a TIFF file with an overview of GHRLOS exon 4 (highlighted in grey) showing the overlap with sense GHRL and SEC13-T exons (boxed). GT/AG intron splice sites are underlined. The exon 4 polyA signal is depicted in orange font. A stretch of poly(A) in the genomic sequence (exon 4) resulting of frequent oligo(dT) mispriming during cDNA synthesis is highlighted in pink.