Abstract
Using a methylated-DNA enrichment technique (methylated CpG island recovery assay, MIRA) in combination with whole-genome tiling arrays, we have characterized by MIRA-chip the entire B cell “methylome” of an individual human at 100-bp resolution. We find that at the chromosome level high CpG methylation density is correlated with subtelomeric regions and Giemsa-light bands (R bands). The majority of the most highly methylated regions that could be identified on the tiling arrays were associated with genes. Approximately 10% of all promoters in B cells were found to be methylated, and this methylation correlates with low gene expression. Notably, apparent exceptions to this correlation were the result of transcription from previously unidentified, unmethylated transcription start sites, suggesting that methylation may control alternate promoter usage. Methylation of intragenic (gene body) sequences was found to correlate with increased, not decreased, transcription, and a methylated region near the 3′ end was found in approximately 12% of all genes. The majority of broad regions (10–44 kb) of high methylation were at segmental duplications. Our data provide a valuable resource for the analysis of CpG methylation patterns in a differentiated human cell type and provide new clues regarding the function of mammalian DNA methylation.
Keywords: chromosome structure, DNA methylation, epigenetics, CpG island
Methylated cytosine (5-methylcytosine, 5MC) is the only modified base yet detected in mammalian cells and is found almost exclusively at the dinucleotide sequence 5′CG (CpG dinucleotide). The distribution of CpG dinucleotides along mammalian genomes is not uniform; for example, sequences near many promoters have a much higher frequency of CpG dinucleotides than the rest of the genome. These CpG-rich sequences are called CpG islands (1). Approximately 50% of promoters are within CpG islands, and methylation of these promoters commonly leads to gene inactivation (2, 3). Methylated DNA is often associated with inactive chromatin marks, for example deacetylated histones H3 and H4, histone H3 lysine 9 methylation, and histone H3 lysine 27 methylation (4). DNA methylation now is commonly thought to be a silencing mechanism more difficult to reverse than covalent histone modifications, but the function of mammalian DNA methylation is still not completely understood. Most current data point to a role of DNA methylation in gene regulation and/or control of repetitive elements (2, 5).
When DNA methylation was first proposed as a heritable, epigenetic mechanism for mammalian gene regulation, X chromosome inactivation, and development (6, 7), only the total level of 5MC could be assayed, usually by HPLC or other chromatography. Methylation-sensitive restriction enzymes were soon used to give sequence-specific information (8–10), and then bisulfite treatment followed by DNA sequencing (11) began to give methylation information at single-nucleotide resolution. Only recently have new technical advances made it possible to undertake genome-wide analysis of DNA methylation. Most of these studies have focused on CpG islands and promoters, which have been found to often undergo methylation changes during tissue development or tumorigenesis (2, 12). Most techniques that have been used for genome-wide methylation analysis of mammals depend on either cleavage by methylation-sensitive restriction endonucleases (e.g., HpaII, NotI) (13), differential sensitivity of cytosine and 5MC toward chemical modification (e.g., bisulfite sequencing) (14), or precipitation of methylated DNA with an antibody (15, 16). Using the antibody enrichment technique, a genome-wide analysis of DNA methylation in the Arabidopsis genome has been reported (17, 18). In addition, bisulfite treatment and high-throughput, massively parallel sequencing has been successfully used to give a high-resolution DNA methylation map of the Arabidopsis genome (19). However, the mammalian genome is 25 times larger, so a complete analysis of cytosine methylation in mammalian genomes is still needed. For mammalian genomes, although technical advances are being made (20), currently available high-resolution data on DNA methylation patterns are mostly limited to CpG islands and promoters (14, 21–25).
We recently developed a methylation detection technique (methylated CpG island recovery assay, MIRA) that makes use of the high affinity of the MBD2/MBD3L1 protein complex to enrich for methylated, double-stranded DNA. Combined with microarrays, this method (MIRA-chip) has been used to determine the DNA methylation status of large numbers of genes and chromosomal regions in normal and cancerous tissue (26–28). Here, we have used the MIRA technique in combination with whole-genome tiling arrays to derive the first comprehensive high-resolution methylation map of the human genome in CD19+ B cells.
Results and Discussion
High-Resolution Mapping of Global DNA Methylation Patterns.
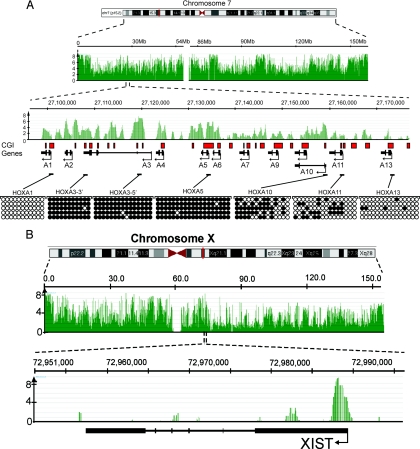
MIRA-chip has proven to be a sensitive, robust, and reproducible technique for mapping DNA methylation patterns in mammalian genomes (26–28). We previously performed MIRA-chip studies on a 140-Mb stretch of human chromosomes 7 and 8 in normal and lung tumor tissue (28). Methylation patterns are tissue/cell-type specific, and whole blood has several cell types, so for this study we prepared DNA from purified CD19+ B cells. For microarray analysis we used NimbleGen's HG18 whole-genome tiling arrays. The probe length on these arrays is 50–75 nucleotides, and the median probe spacing is 100 bp. After sonication, MIRA-enriched, methylated DNA samples and unfractionated input DNA samples from B cells were cohybridized onto the tiling microarrays. Log2 ratios and P value scores for methylation signals were provided by NimbleGen for each of the 21 million probes. The NimbleGen P value scores are derived from the Kolmogorov-Smirnov test comparing the log2 ratios (log2 of the ratio of MIRA vs. input signal) within a 750-bp window centered at each probe and the rest of the data on the array (NimbleScan software version 2.4) (29). For initial validation of the array data, we first analyzed the methylation signals at 2 well-characterized genomic regions harboring the CD19 and RNA polymerase II genes (Figs. S1 and S2). The CD19 gene is highly expressed in B cells, and its cell-surface product was used for the isolation of B cells. The RNA polymerase II gene is expressed ubiquitously in all cell types, and its promoter overlaps with a CpG island. Consistent with the general correlation between lack of methylation at promoters and active gene expression, their promoter regions showed as unmethylated, although neighboring regions were identified as densely methylated sequences. Additional confirmation was obtained by conducting bisulfite sequencing of promoter regions that scored as strongly, moderately, or weakly methylated in the microarray data sets. We analyzed the methylation status of several randomly chosen promoters that showed gradually decreasing average log2 ratios (the log2 of the ratio of MIRA signal and input signal). High log2 ratios corresponded to densely methylated regions, whereas lower ratios represented moderately methylated loci, as determined by sodium bisulfite sequencing (Fig. S3). We also focused on the promoters of the HOXA cluster genes on chromosome 7 because they are located close to each other but have different methylation status (Fig. 1A). Using bisulfite sequencing, we verified that a cluster of strong array signals (−log10 P value scores >3) corresponded to highly methylated regions. At the promoter of HOXA11, 30% of the CpGs are methylated and the −log10 P value score is ≈1.7–2.0. Sequences that have <10% of their CpGs methylated, such as the promoter of HOXA13, show no significant methylation signals in the array data. Furthermore, we analyzed the XIST gene located on the X chromosome. The XIST gene is unmethylated and expressed from the inactive X chromosome in females but is methylated and silenced on the active X chromosome (30–32). Because the B cells were derived from a male individual, the XIST promoter is silent, and we find a cluster of strong methylation signals in the promoter region (Fig. 1B).
Fig. 1.
Snapshots of MIRA data from whole-genome methylation analysis. (A) The HOXA gene cluster on chromosome 7 was analyzed in detail. The methylation signal is shown plotted along the chromosome as a −log10 P value score (green). Therefore, the minimum number on the y axis is 0 (when P = 1). The P value score was obtained by NimbleScan software and is derived from the Kolmogorov-Smirnov test comparing the log2 ratios (MIRA vs. input) within a 750-bp window centered at each probe and the rest of the data on the array. The HOXA genes and the direction of transcription are indicated. The red boxes show the CpG islands. Methylation patterns at several CpG islands were confirmed by bisulfite sequencing. Open circles represent unmethylated CpG dinucleotides, and black circles represent methylated CpGs. The MIRA signals correlate with the density of methylated CpGs. (B) Analysis of the XIST locus on the active X chromosome. The promoter of the XIST gene shows a strong MRI.
Identification and Mapping of Methylated Regions of Interest.
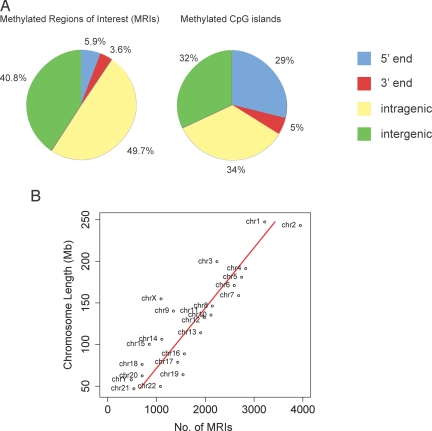
The log2 ratios of MIRA signal vs. input were provided by NimbleGen and used without further normalization. Each of the 38 arrays was analyzed separately. Probes were selected as positive if their log2 ratios were above the 95th percentile range on the array (P value cutoff of <0.05). For our analysis we defined a methylated region of interest (MRI) as a region with at least 4 positive probes (each with a P value cutoff of <0.05) covering a minimum length of 350 bp, allowing only a 1-probe gap. This stringent definition will give few false-positive results. MRIs were categorized on the basis of their location relative to known genes according to the University of California at Santa Cruz (UCSC) Genome Browser, HG18 RefSeq database. Using these definitions, a total of 42,844 MRIs were identified (Table S1). Table 1 and Fig. 2 show that 5.9% of the MRIs were located near the 5′ end of genes, defined as ±500 bp relative to the transcription start site. Additionally, a large fraction (3.6%) of all MRIs was found near the 3′ end of genes, defined as ±500 bp relative to the end of the RefSeq mRNA sequences (Table 1). Of all MRIs, 49.7% were intragenic, defined as between +500 bp downstream of the transcription start site and −500 base pairs upstream of the end of the corresponding RefSeq mRNA sequence, and 40.8% of all MRIs were intergenic. A total of 14.5% of the MRIs was located in proximity to short interspersed elements (SINE) or long interspersed elements (LINE). Although repetitive elements were not present on the tiling arrays, we could gain limited information regarding their methylation status by the hybridization of flanking unique DNA sequences to adjacent probes. We should note, however, that SINEs often occur in clusters, which makes quantitative methylation profiling of repetitive DNA more difficult. The analysis of LINE methylation status by MIRA-chip is not optimal for several reasons: probes are usually not present on the arrays, they are relatively long sequences (5 kb), tend to form clusters, and just a short promoter region of a LINE element is subject to DNA methylation. Thus, with the caveat that repetitive DNA methylation is not fully captured in our analysis, we observed that the majority of the identified MRIs on the tiling arrays (a total of 59.2%) was associated with genes (Table 1, Fig. 2A). These data suggest that DNA methylation, in addition to silencing repetitive DNA, has additional functions.
Table 1.
Summary of MRIs relative to genome annotation
| MRI location | No. of MRIs* (%) |
|---|---|
| 5′ end of a gene | 2,552 (5.9) |
| 3′ end of a gene | 1,578 (3.6) |
| Intragenic | 21,474 (49.7) |
| Intergenic | 18,054 (40.8) |
| Within 500 bp of LINE | 4,538 (10.5) |
| Within 500 bp of SINE | 1,751 (4.0) |
| Total MRIs | 42,844 (100) |
Probes were selected as positive if they fell into the more than 95th percentile range on the array.
A 350-bp minimum length was used to define an MRI. A 1-probe gap was allowed.
*A methylated region of interest is equivalent to 4 or more positive probes.
Fig. 2.
Distribution of DNA methylation along the B cell genome. (A) Distribution of MRIs and methylated CpG islands relative to annotated genes. A total of 42,944 MRIs were identified, and their location with respect to genes and the 5′ and 3′ ends of genes, defined as ±500 bp relative to the transcript start or end, was determined. Intragenic MRIs are located from +500 of transcription start to −500 of transcript end. Methylated CpG islands were defined as those CpG islands that overlap with an MRI. (B) Distribution of MRIs along individual chromosomes. The number of MRIs was plotted against the length of the chromosomes.
Methylated CpG islands are defined here as any CpG island overlapping with an identified MRI. Only 7,262 of the 42,844 MRIs (16.9%) mapped to CpG islands. The definition of a CpG island was according to Gardiner-Garden and Frommer (33) (i.e., length >200 bp, CpG observed to expected >0.6, G+C content >50%). These methylated CpG islands were categorized according to their location relative to known genes (HG18 RefSeq) (Table 2). Of a total of 28,226 CpG islands, 7,262 (25.7%) were methylated. This number is greater than one would have expected on the basis of the still-prevalent idea that almost all CpG islands in the human genome are unmethylated (1). However, recently it has become appreciated that there is a substantial fraction of methylated CpG islands in somatic differentiated tissues (21–23, 25, 34). We find that intragenic CpG islands show the highest percentage of methylation (35.7%), and CpG islands located near the 5′ end of genes are methylated at a rate of 17.5% (Table 2). Accordingly, 82.5% of the CpG islands at 5′ gene ends are unmethylated. The relative distribution of all methylated CpG islands relative to annotated genes is shown in Fig. 2A. It is clear that the majority of all methylated CpG islands is associated with genes (68%), and only 32% of all methylated CpG islands are intergenic.
Table 2.
Summary of methylated CpG islands relative to RefSeq genes (HG18)
| Location | All CpG islands | Methylated CpG islands | % Methylated |
|---|---|---|---|
| 5′ end of a gene | 12,091 | 2,121 | 17.5 |
| 3′ end of a gene | 1,188 | 335 | 28.2 |
| Intragenic | 7,349 | 2,626 | 35.7 |
| Intergenic | 8,558 | 2,347 | 27.4 |
| Total | 28,226 | 7,262 | 25.7 |
We did a functional analysis of the genes that had MRIs at their 5′ ends using the DAVID interface (http://david.abcc.ncifcrf.gov/). The functional classes of genes at the top of the list included those involved in cellular development, cell death, neuron development, cell migration, cadherins, transcriptional regulation, homeobox genes, and genes functioning in the WNT- and Frizzled-related signaling pathways and the cell cycle. Many gene products functioning in these pathways are probably not required in B cells, and therefore silencing of these genes by DNA methylation seems plausible.
Our analysis identified 10 particularly broad MRIs, ranging in size from 10 to 44 kb (Table S2). All of these MRIs are either segmental duplication regions or simple tandem repeats. Although we do not know whether all copies are methylated, silencing of these regions by DNA methylation is likely a common feature of duplicated (i.e., repetitive) regions. This situation is reminiscent of that in the filamentous fungus Neurospora crassa, in which duplicated genes are silenced by DNA methylation (35), and of plants and mammals in which the insertion of multiple copies of a transgene leads to methylation and loss of expression of some or all copies of the transgene (36–38).
DNA Methylation Patterns Along the Chromosomes.
The distribution of MRIs along chromosomes roughly followed chromosome size (R2 = 0.77, P < 0.001) (Fig. 2B). Exceptions were chromosomes 19 and 22, which had a higher MRI density than predicted from their length, and chromosomes 9, 15, 18, and X had among the lowest relative methylation levels (Table S1 and Fig. 2B). Chromosomes 19 (25.5 genes per Mb) and 22 (11.7 genes per Mb) are relatively gene rich. Chromosomes 9 (6.7 genes per Mb) and X (8.1 genes per Mb) are relatively gene poor. However, they are not the highest or lowest, respectively. The percentage of probe coverage does not fully explain the MRI density differences between chromosomes, and most likely, differences in chromosome structure dictate MRI density profiles.
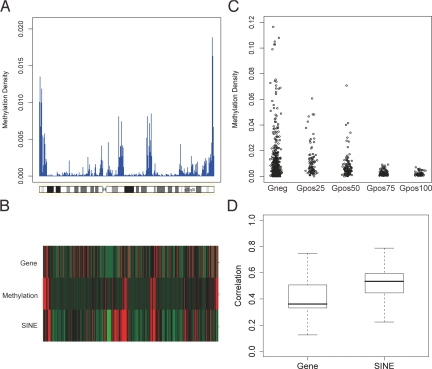
Recently published genome-wide, high-resolution DNA methylation studies have focused mainly on promoter regions and CpG islands (14, 16, 21–23, 25, 27, 39). The distribution of DNA methylation along whole chromosomes, including inter- and intragenic regions, has not been analyzed at high resolution in mammalian cells. Our methylation density maps were created for each chromosome by applying a 5-Mb sliding window and scanning the chromosomes in 500-kb steps (Fig. 3 and Figs. S4–S7). Methylation density was calculated by counting the number of nucleotides within MRIs in the sliding window and dividing it by the window size; this ratio was plotted along the chromosomes. Chromosome band information was obtained from the UCSC genome assembly HG18. Subtelomeric regions were often more densely methylated than other parts of the chromosomes (Fig. 3 and Figs. S4–S7). Upon closer examination of the data, we observed that the methylation density profiles show good correlation with the patterns of Giemsa staining of chromosomes. Giemsa staining has been used for many decades to visualize mitotic chromosomes and to detect different chromosomal aberrations, such as translocations and inversions. Darkly stained, late-replicating G-bands and lightly stained, early-replicating R-bands reflect different chromatin compaction, replication timing, gene density, and GC content (40). Efforts to trace back Giemsa staining to nucleotide sequence differences have been only moderately successful, and GC-content-based in silico band prediction algorithms produced just weak similarities (41). The relationship between nucleotide sequence, epigenetic status, and cytogenetic bands has not been fully characterized. To extend the original observation regarding the DNA methylation profile and Giemsa staining patterns, we analyzed the statistical correlation between them (Fig. 3C). A negative correlation (−0.25) was found between the staining intensity and DNA methylation status; the stronger the Giemsa staining, the less DNA methylation signal was detected. The more highly methylated, lightly staining R-bands represent constitutive euchromatin and are SINE-rich and gene-rich genomic regions. To investigate how SINE sequences and gene density contribute to the formation of the DNA methylation landscape, we analyzed the correlation between DNA methylation and SINE repeat or gene density. The density of SINEs or genes was determined by using a 5-Mb sliding window with 500-kb step size along each chromosome, and Spearman correlation between DNA MRI density and SINE or gene density was calculated for each chromosome (Fig. 3 B and D). The distribution of the correlations is plotted in Fig. 3D. This data set shows that there is a positive correlation between SINE-enriched regions and DNA methylation. Gene density also positively correlates with DNA methylation density, a finding that may be related to gene body methylation (Fig. 2A and see below).
Fig. 3.
Methylation patterns along human chromosomes. (A) Chromosomal methylation density map (see text for details). Chromosome 7 is shown as an example. All other chromosomes are displayed in Figs. S4-S7. (B) Chromosomal methylation density profile of chromosome 7 (Middle) aligned with gene density profile (Top) and density of SINE elements (Bottom). (C) Correlation of methylation density and Giemsa staining. Giemsa staining intensity (from UCSC database) was partitioned into 5 groups (Giemsa negative and 4 groups, increasing from the 25th to the 100th percentile). Giemsa light bands have higher levels of DNA methylation. (D) Correlation of DNA methylation density with SINE and gene density. Density maps were determined using a 5-Mb sliding window with 500-kb step size along each chromosome. The Spearman correlation between DNA methylation density and gene density or SINE density was calculated, and the distribution of the correlations is plotted.
DNA Methylation Status of Promoters.
DNA sequences spanning ±500 bp of transcription start sites in RefSeq genes were obtained from the UCSC HG18 assembly. As done by Weber et al. (22), the CpG observed/expected ratio was calculated for each promoter. High-CpG promoters (HCP) contain a 500-bp region with a CpG ratio >0.75 and GC content >55%. Low CpG promoters (LCP) do not contain a 500-bp region with a CpG ratio >0.48. Intermediate CpG promoters (ICPs) are neither HCP nor LCP (Fig. S8) (22). The methylation level is positively correlated with the number of CpGs in LCP promoters, consistent with a previous report (22). Promoters with an overlapping MRI were considered as methylated. HCPs and ICPs are overrepresented as methylated promoters (Fig. S8). LCPs are underrepresented as methylated promoters, most likely owing, at least in part, to their low CpG content.
HCP/ICP/LCP promoters were separated into 2 groups according to methylation status, either methylated or unmethylated (i.e., overlapping with an MRI or not). Expression values of these 2 groups of genes in CD19+ cells were retrieved from the Genomics Institute of the Novartis Research Foundation (GNF) SymAtlas database. The Kolmogorov-Smirnov test was applied to compare whether the 2 sets of expression values are from the same distribution. As shown in Table 3, expression of methylated genes is lower than expression of unmethylated genes, but this correlation is significant only for ICP and HCP promoters. Very similar data were obtained when the methylation status of genes was compared with the average expression level from 8 normal B cell datasets [Gene Expression Omnibus (GEO) dataset GDS2643] (Table 3).
Table 3.
Correlation between gene expression and promoter methylation
| Promoter class | Total | Methylated* | Unmethylated | Methylated with expression data | Unmethylated with expression data | P value† |
|---|---|---|---|---|---|---|
| GNF dataset | ||||||
| HCP | 12,850 | 1,521 | 11,329 | 1,329 | 9,123 | 8.11E-08 |
| ICP | 2,951 | 411 | 2,540 | 303 | 1,874 | 2.33E-04 |
| LCP | 5,344 | 293 | 5,051 | 220 | 3,561 | 0.3742 |
| GEO dataset | ||||||
| HCP | 12,850 | 1,521 | 11,329 | 1,105 | 7,900 | 8.86E-09 |
| ICP | 2,951 | 411 | 2,540 | 252 | 1,599 | 7.03E-03 |
| LCP | 5,344 | 293 | 5,051 | 192 | 2,916 | 0.68 |
*A methylated promoter overlaps with an MRI.
†P values were determined by the Kolmogorov–Smirnov test.
We find that 10.5% of the promoters in B cells are part of MRIs when a (±) 500-bp window size around the transcription start site is used for analysis (Table 3). According to the GNF expression atlas database, most of the methylated genes are not expressed or just weakly expressed in B cells. The expression level of 5 randomly chosen genes was checked using real-time RT-PCR (Fig. S9). We compared the relative expression levels in B cells, brain, heart, and testis and confirmed that promoter methylation in these genes in B cells occurs in silenced or weakly expressed genes. Together, the data strongly support the idea that DNA methylation at promoters participates in gene silencing.
Alternate Promoters.
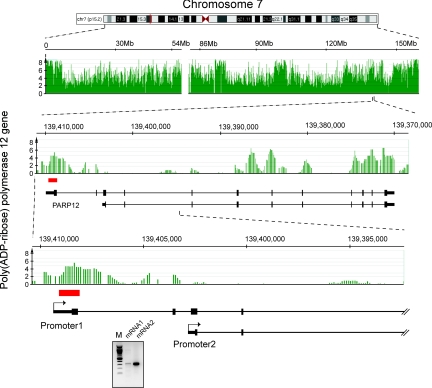
Of note, we found 4 genes among the top 100 most highly methylated gene targets that should be expressed in normal B cells according to the GNF database. For these genes, which apparently contradict the general rule, we conducted 5′ RACE experiments to map the transcription initiation sites. We detected alternative promoters/transcription start sites for 3 of the 4 genes (Fig. 4 and Figs. S10 and S11). In the case of the fourth gene (PPP1R2), we also detected mRNA transcript initiation from the authentic promoter region. Using the DNA methylation-positive sequence of the PPP1R2 gene as a probe, we conducted a BLAT (BLAST-like alignment tool) search on the human genome. We identified several different chromosomal loci showing 88.3%–98.0% nucleotide sequence identity with the query probe in an almost 200-bp-long region (data not shown). These highly homologous sequences are mapped onto segmental duplications, and in this way the methylation of the PPP1R2 gene can be considered as a potentially false-positive event that originated from mishybridization. The identification of new unmethylated promoters for the PARP12, MFHAS1, and MSL2L1 genes highlights the likely importance of DNA methylation for control of alternative promoter usage and can explain the origin of the large pool of tissue- and cell-specific alternative 5′ end transcripts.
Fig. 4.
Expression of methylated genes is often explained by alternative promoter usage. In this example, data for the PARP12 gene are shown. The methylation profile is in green, and the red box indicates a CpG island. Initial analysis showed that this gene contains a methylated region in the promoter, but the gene is expressed according to expression microarrays. Using 5′ RACE, we identified an alternative transcript that emanates from an unmethylated promoter. RT-PCR analysis of the 2 alternative transcripts is shown in the bottom gel panel. mRNA1 is initiated from promoter 1, whereas mRNA2 is initiated from promoter 2.
DNA Methylation at the 3′ End of Genes.
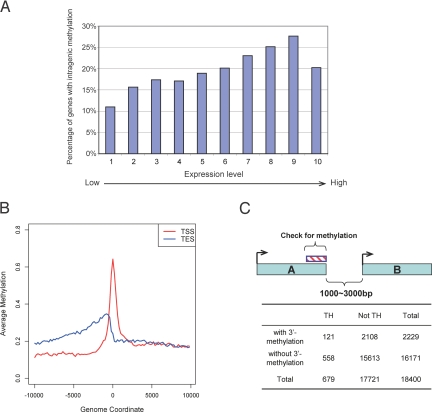
Using cluster analysis of all annotated genes, we determined that, as expected, MRIs are often found at the 5′ end of genes (Table 3, Fig. 5B). However, we also found frequent MRIs near the 3′ end of genes, before the end of corresponding RefSeq mRNA sequences (Fig. 5B). Using a window of 2 kb before these 3′ ends (Fig. 5C), we determined that 2,229 of 18,400 genes (12.1% of all annotated genes) had 3′-end MRIs (Fig. 5C). This 3′-end methylation occurred in 11.9% (2,108 of 17,721) of all singular genes, defined as genes not having 3′ neighboring genes within 3 kb of the end of the RefSeq mRNA sequence. However, 17.8% of genes (121 of 679) that have 3′ neighbors in tail-to-head orientation had a gene end MRI (P < 1 × 10−5; Fisher's exact test, 2-tailed) (Fig. 5C). No significant association was found between 3′-end methylation and gene pairs present in tail-to-tail orientation (P = 0.52). The data of Illingsworth et al. (21) for CpG islands also suggest methylation of 3′ ends. The biologic significance of 3′ gene end methylation remains to be determined. Although evidence is currently lacking, 3′-end methylation may be related to suppression of antisense transcripts. Another possibility is that this methylation is part of a mechanism that regulates polyadenylation and/or transcription termination. Precise termination of transcription is particularly important in those cases in which interference with expression of a neighboring gene needs to be prevented.
Fig. 5.
Intragenic methylation and methylation at the 3′ end of genes. (A) Correlation between intragenic CpG methylation and gene expression levels. Data for gene expression levels in B cells were obtained from the GNF and GEO databases and were partitioned into 10 groups according to expression level (see Materials and Methods). The y axis is the percentage of methylated genes within each group defined as having at least 1 MRI within the gene body. With increased gene expression levels, there is a tendency toward increased intragenic methylation. Only the most highly expressed genes differ from the trend. (B) Identification of an MRI at the 3′ end of genes. The composite profile of methylation densities along all human RefSeq genes shows MRIs at the 5′ end of genes and near the 3′ end. The y axis is the average log2 ratio of all of the genes at each corresponding genome coordinate window. TSS, transcription start site; TES, transcript end site. (C) Methylation at 3′ gene ends and flanking genes. Genes present in tail-to-head orientation in the configuration indicated were analyzed in more detail. Genes in tail-to-head orientation were 1.5-fold more likely to have 3′-end MRIs than singular genes (P < 0.00001).
DNA Methylation of the Gene Body and Gene Expression.
We compared gene expression levels with MRIs within the gene body (i.e., >500 bp downstream of the transcription start sites). Data for gene expression levels in B cells were obtained from the GNF and GEO databases and were partitioned into 10 groups according to expression levels (see Materials and Methods). Fig. 5A shows that the percentage of genes with at least 1 internal MRI increases with gene expression level, then becomes lower again at the most highly expressed genes. A similar correlation between methylation of gene bodies and transcript levels has been reported for the plant Arabidopsis thaliana (17, 18). However, loss of intragenic methylation leads to gene upregulation, and it was proposed that methylation interferes with transcript elongation in Arabidopsis (18). Similarly, incorporation of a transgene that became methylated into a region downstream of a promoter yielded a decrease in transgene expression in mammalian cells (42). It is not known whether methylation of gene bodies in mammalian cells has a role in upregulation or downregulation of expression of endogenous genes. Our data indicate that a positive correlation between intragenic methylation and transcription levels exists for human B cells (Fig. 5A). As reviewed by Jones (3), it has been known for some time that housekeeping genes rarely have internal CpG islands, whereas 49% of tissue-specific genes have such islands, which are often methylated. On the basis of these observations it was suggested that transcription may facilitate de novo methylation (3). Our results are consistent with this possibility. Another suggested possibility (2) is that intragenic (gene body) methylation represses the expression of antisense transcripts (perhaps representing transcriptional noise) that would downregulate expression of the sense transcript. Increased gene body methylation has previously been reported for the active X chromosome relative to the inactive X chromosome in humans (43). Although the biologic significance of this phenomenon is not known, it is potentially linked to upregulation of X-linked genes and dosage compensation in mammals (44). Our data suggest that intragenic methylation is indeed correlated with increased transcription and that this is not limited to CpG islands or genes on the X chromosome.
Materials and Methods
Genomic DNA Purification.
CD19+ B cells from one of the authors (A.D.R.) were isolated from whole blood, using an institutional review board-approved protocol. The Dynabeads CD19 pan B cell kit (Invitrogen) was used for B cell isolation according to the company's protocol. Genomic DNA was purified from B cells by standard procedures by using proteinase K digestion, phenol chloroform extraction, and ethanol precipitation.
Enrichment of the Methylated CpG Sequences.
Genomic DNA was fragmented by sonication to ≈750-bp average size. Sonicated genomic DNA was treated with T4 DNA polymerase (New England Biolabs), and a double-stranded linker (5′-GCGGTGACCCGGGAGATCTGAATTC-3′ and 5′-GAATTCAGATC-3′) was ligated onto the ends. Enrichment of methylated DNA by the MIRA reaction was performed as described previously (26, 27). Four MIRA binding reactions were set up with 500 ng of end-treated genomic DNA each and incubated overnight at 4 °C on a rotating platform. The fraction representing the methylated DNA was collected from the binding reaction by magnetic beads and washed 3 times with a 700-mM NaCl-containing buffer. The methylated fraction was eluted from the beads by using a Qiagen PCR purification kit and amplified by ligation-mediated PCR. The labeling of amplicons, microarray hybridization, and scanning were performed by the NimbleGen Service Group (Reykjavik, Iceland). NimbleGen genomic tiling arrays covering the entire human genome (HG18 Tiling–Whole Human Genome, 38 Array Set) were used in the DNA methylation profile analysis. MIRA-enriched DNA fractions were compared with input DNA.
MIRA enrichment requires only 2 mCpGs within ≈50 to 100 bp for efficient pulldown (26), but, as for other methods for enrichment of methylated DNA (20), there is a relationship between CpG density and enrichment as measured by log2 ratios. For confirmation in the present data set, we identified several regions of high CpG density (>0.05) that were highly methylated by bisulfite sequencing (Fig. 1 and Fig. S3). Their log2 ratios (MIRA/input) plotted against CpG density show a good correlation (R2 = 0.761; data not shown). Additionally, Fig. S8B shows a log2 ratio vs. CpG density plot, grouped by HCP/ICP/LCP promoters. According to Weber et al. (22), most CpGs in LCP promoters (CpG density <0.05) are methylated, and we do find a linear relationship (R2 = 0.62) between log2 ratio and CpG density (Fig. S8B). These results indicate that MIRA gives useful data for regions of both high and low CpG density, especially when several adjacent probes are considered together.
Identification and Annotation of Methylated Regions.
Log2 values of MIRA-enriched DNA vs. input DNA were determined with NimbleScan software and provided by NimbleGen. Data from each of the 38 arrays were analyzed separately. MRIs were defined as described in Results and Discussion. Identified MRIs were mapped relative to known transcripts defined in the UCSC genome browser HG18 RefSeq database (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/). MRIs falling into ±500 bp of transcription start sites were defined as 5′-end MRIs; MRIs falling within ±500 bp of RefSeq transcript end sites were defined as 3′-end MRIs, and those falling within gene bodies (from +500 of transcription start to −500 from transcript end) were defined as “intragenic” MRIs. MRIs that are not close to any known transcripts were defined as “intergenic” MRIs. The MRIs were also mapped to the 28,226 CpG islands defined as such (at http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/) if the MRIs overlapped with known CpG islands. These CpG islands were also mapped relative to known transcripts in the HG18 RefSeq database using the same approach described above.
Methylation Density Analysis.
To examine the methylation profile at the entire chromosome level, a condensed density profile was generated for each chromosome. Each chromosome was divided into a series of 5-Mb windows with a step size of 500 Kb. The methylation density of each window was determined by counting the number of nucleotides within MRIs in that window and dividing that number by the window size. Density of genes and SINE elements along each chromosome were calculated by the same approach. This transformation gives us lower resolution data for easy visualization and comparison between genomic features. Spearman correlation was used to represent the relationship between methylation density, gene density, and SINE density. Heatmaps of these density data were generated for each chromosome using Partek Genomic Suite version 6.3. To examine whether there is a correlation between methylation and chromosome Giemsa staining patterns, the methylation density within each chromosome band was calculated by averaging the log2 ratios of probes within that band. A jittered scatterplot was generated by adding a small random number to the methylation density data within each chromosome band.
Promoter Classification, Gene Body Methylation, and Expression Analysis.
Promoters were classified into 3 categories (HCP, ICP, and LCP) according to Weber et al.'s approach (22). We retrieved the genomic sequences spanning ±500 bp of 26,855 genes defined in the HG18 RefSeq database. Only 1 promoter was retained for genes sharing the same transcription start sites, which resulted in 21,891 promoters. We determined the GC content and the ratio of observed vs. expected CpG dinucleotides in sliding 500-bp windows with a 5-bp offset, as described in Results and Discussion and by Weber et al. (22). The 3 categories of promoters were determined as follows: HCPs contain a 500-bp region with a CpG ratio >0.75 and GC content >55%; LCPs do not contain a 500-bp region with a CpG ratio >0.48; and ICPs are neither HCPs nor LCPs. To determine a methylation parameter for each promoter, we used an approach slightly different from MRI identification, because not all of the promoter regions have MRIs. For each of the 1,000-bp promoter regions, a methylation level was determined by the maximum average ratios of adjacent 4 probes with 1-probe step size within the region. This value should reflect the relative methylation level of each promoter. To determine the relationship between methylation of promoters and expression level of the genes, we separated each HCP, ICP, and LCP promoter category into 2 subcategories, methylated and unmethylated. Gene expression data of CD19+ cells within each subcategory for each promoter class were obtained from the GNF SymAtlas database and GEO dataset GDS2643. The average log2 intensity value of the 2 replicates in the GNF dataset and 8 replicates in the GEO dataset were used to represent the gene expression level of the genes. For each promoter class, the Kolmogorov-Smirnov test was applied to compare whether the expression values of the genes having methylated promoters are significantly smaller than those of genes having unmethylated promoters. To examine how gene body methylation might affect gene expression, gene expression data from GNF or GEO were stratified into 10 groups. The 10 groups were stratified according to equally spaced log2 intensity (low to high). For each expression group, the presence of intragenic MRIs was examined for each gene within the group.
DNA Methylation at Closely Adjacent Genes.
Genes that are closely adjacent to each other in tail-to-head orientation were identified using the following 2 criteria: (i) both genes are on the same strand, and (ii) the transcription end of the upstream gene and the transcription start site of the immediate downstream gene must be >1 kb and <3 kb apart. The presence of MRIs within the region up to −2 kb upstream of the transcription end was determined (Fig. 5B). Gene pairs with tail-to-tail orientation were identified using the following criteria: (i) the 2 genes are on the opposite strand, and (ii) the transcription end site of the upstream gene and the transcription end site of the downstream gene must be <3 kb apart and without overlap. Fisher's exact test was used to estimate whether there are more 3′-end MRIs of the upstream gene in tail-to-head or tail-to-tail orientated gene pairs vs. other genes that have no neighbors as defined above.
DNA Methylation Analysis Using Bisulfite Sequencing.
DNA was treated and purified with the EpiTect Bisulfite kit (Qiagen). PCR primer sequences for amplification of specific targets in bisulfite-treated DNA are available upon request. The PCR products were cloned into the pDrive PCR cloning vector (Qiagen), and 5–10 individual clones were sequenced.
RNA Isolation and Quantitative RT-PCR.
Total RNA was isolated from CD19+ B cell with the RNEasy Mini Kit (Qiagen). Reference total RNAs from brain, heart, and testis samples were purchased from Ambion. cDNA was created by using SuperScript III First-Strand Synthesis System (Invitrogen). cDNA was amplified with transcript-specific primers (PCR primer sequences are available upon request). A relative standard curve method was used to perform real-time quantitative PCR. Gene transcription was normalized to GAPDH expression in all samples.
Rapid Amplification of cDNA Ends.
Mapping of transcription start sites was conducted with the FirstChoice RLM-RACE kit (Ambion). PCR-amplified 5′ ends were cloned into the pDrive PCR cloning vector (Qiagen) and sequenced.
Supplementary Material
Acknowledgments.
This research was supported by a grant of the National Cancer Institute Grant CA084469 (to G.P.P.).
Footnotes
The authors declare no conflict of interest.
Data deposition: the data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE13700).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812399106/DCSupplemental.
References
- 1.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 4.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 5.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 6.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 7.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 8.Bird AP, Southern EM. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978;118:27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- 9.Singer J, Roberts-Ems J, Riggs AD. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science. 1979;203:1019–1021. doi: 10.1126/science.424726. [DOI] [PubMed] [Google Scholar]
- 10.Waalwijk C, Flavell RA. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: Tissue-specific variations. Nucleic Acids Res. 1978;5:4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello JF, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 14.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 16.Keshet I, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 19.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Down TA, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 23.Shen L, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farthing CR, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakyan VK, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 27.Rauch T, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci USA. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rauch TA, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci USA. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 31.Gartler SM, Goldman MA. Biology of the X chromosome. Curr Opin Pediatr. 2001;13:340–345. doi: 10.1097/00008480-200108000-00010. [DOI] [PubMed] [Google Scholar]
- 32.Norris DP, et al. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 33.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 34.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selker EU. Gene silencing: Repeats that count. Cell. 1999;97:157–160. doi: 10.1016/s0092-8674(00)80725-4. [DOI] [PubMed] [Google Scholar]
- 36.Flavell RB. Inactivation of gene expression in plants as a consequence of specific sequence duplication. Proc Natl Acad Sci USA. 1994;91:3490–3496. doi: 10.1073/pnas.91.9.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 38.Matzke MA, Mette MF, Matzke AJ. Transgene silencing by the host genome defense: Implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol Biol. 2000;43:401–415. doi: 10.1023/a:1006484806925. [DOI] [PubMed] [Google Scholar]
- 39.Ladd-Acosta C, et al. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmquist GP, Ashley T. Chromosome organization and chromatin modification: Influence on genome function and evolution. Cytogenet Genome Res. 2006;114:96–125. doi: 10.1159/000093326. [DOI] [PubMed] [Google Scholar]
- 41.Niimura Y, Gojobori T. In silico chromosome staining: Reconstruction of Giemsa bands from the whole human genome sequence. Proc Natl Acad Sci USA. 2002;99:797–802. doi: 10.1073/pnas.022437999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 43.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.