Abstract
Evaluation of the transfer efficiency of a rat heme oxygenase-1 (HO-1) transgene into mice requires differentiation of rat and mouse HO-1. However, rat and mouse HO-1 have 94% homology; antibodies and enzyme activity cannot adequately distinguish HO-1. We designed a qRT-PCR method to monitor HO-1 transcription relative to a housekeeping gene, GAPDH. The ratio of rat and mouse HO-1 mRNA could be estimated through restriction fragment length polymorphism (RFLP) analysis of the PCR products. In vitro, murine AML12 hepatocytes were transfected with rat HO-1. After 40 h, total HO-1 mRNA was enriched 2-fold relative to control cells and rat HO-1 comprised 84% of HO-1 cDNA. In vivo, the rat HO-1 transgene was cloned into a Sleeping Beauty transposase (SB-Tn) construct and injected hydrodynamically into a mouse model of sickle cell disease (SCD). After 21 days, there was a 32% enrichment of HO-1 mRNA relative to control mice and the rat transgene comprised 88% of HO-1 cDNA. After 21 days, HO-1 protein expression in liver was increased 2.5-fold. In summary, qRT-PCR RFLP is a useful and reliable method to differentiate transgene from host gene transcription, especially when the host and transgene protein are identical or highly homologous. This method has translational applications to the design, delivery and monitoring of gene therapy vectors.
Introduction
The study of a rat HO-1 transgene in a mouse system is complicated by the inducibility of host HO-1 and the near identity of the two species at the nucleic acid and amino acid levels. Rat and mouse HO-1 are highly conserved with only 16 out of 289 amino acids being different (94% homology) and 93% identity at the nucleic acid level. The high degree of amino acid similarity makes it difficult to differentiate rat and mouse HO-1 using primary antibodies. As an alternative to measuring protein expression and enzyme activity, mRNA can be measured by qRT-PCR and coupled with RFLP analysis to differentiate host and transgene transcription. This method is more sensitive and accurate than antibody or activity assay methods for distinguishing endogenous and transgene expression in host tissues. RFLP can accurately distinguish single nucleotide differences at restriction endonuclease sites between target DNA sequences. The method developed in this study is applicable to all stages of gene transfer studies and provides a general tool for the detection of transgenes.
We developed a qRT-PCR RFLP method to evaluate rat HO-1 transgene expression in murine tissues. HO-1 is the rate-limiting enzyme in the catabolism of heme. It breaks down the porphyrin ring to yield equal molar amounts of biliverdin, free iron and carbon monoxide (1). HO-1 is highly inducible in response to multiple stimuli including heme, oxidative stress, heavy metals, hypoxia, and cytokines (1). HO-1 and its products protect tissues from oxidative stress and inflammation, reduce vasoconstriction, and inhibit vascular injury (2, 3). Therefore, HO-1 is a potential therapeutic target for vascular disorders. Sickle cell disease (SCD) is a candidate for HO-1 gene therapy because of marked oxidative stress, inflammation and vasculopathy, caused by the release of hemoglobin and heme into the vasculature (4-6).
The cDNA clone for rat HO-1 was isolated from a rat spleen cDNA library and has been used to study HO-1 cytoprotection in transfected cells and tissues (7-10). In SCD, ideally HO-1 would have persistent overexpression and the HO-1 gene delivery system would be non-inflammatory. To transfer the rat HO-1 gene into mice, we have cloned the rat HO-1 gene into a Sleeping Beauty transposase (SB-Tn) system. The SB-Tn construct contains the cargo HO-1 transgene flanked by two direct repeat (DR) binding sites within two inverted repeat (IR) (11) sequences, that act as essential binding sites for the transposase. The SB-Tn system was developed by resurrecting nonfunctional remnants of an ancient vertebrate transposable element from salmonid fish (12). SB-Tn has features that make it particularly attractive as a vector for gene therapy. It can accommodate a much larger transgene than viral vectors; it is nonviral; it does not induce an immune inflammatory response in rodent models; and it mediates efficient stable transgene integration that exhibits persistent expression.
Detection of the rat HO-1 transgene in the mouse is desirable to evaluate the success of HO-1 gene transfer. We have designed a reproducible and efficient qRT-PCR RFLP method in which the relative amounts of transgene and host HO-1 gene transcription can be estimated. This method can be applied to other transgenes and is especially useful when subtle differences exist between endogenous and exogenous gene products. We have used this method to detect the relative amounts rat and mouse HO-1 mRNA in murine hepatocytes in vitro and the livers of sickle mice in vivo. The transfer and detection of a highly homologous rat gene in a mouse model provides proof of concept of a qRT-PCR RFLP transgene detection method that can be applied to other gene transfer studies relevant to translational research.
Materials and Methods
Materials
Materials were obtained from Sigma (St Louis, MO) unless indicated otherwise.
Animals
Animal experiments were approved by University of Minnesota's Institutional Animal Care and Use Committee. We utilized male and female C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME), Sprague Dawley rats (Harlan, Indianapolis, IN) and in house S+S-Antilles sickle mice as a model for SCD. The S+S-Antilles mice are homozygous for deletion of the mouse βmajor globin locus and express human α, βS and βS-Antilles globin transgenes.(13) βS-Antilles globin contains, in addition to the βS mutation at β6, a second mutation at β23 (Val→Ile). The animals were 6 - 12 weeks of age and maintained on standard chow diet.
Animal Tissue Collection
The mice and rats were asphyxiated in CO2. The livers and spleens removed, flash frozen in liquid nitrogen and stored at -80°C for RNA and protein analysis.
Cell Culture
AML12 hepatocytes, a well-differentiated murine cell line transgenic for human transforming growth factor-α, were used for transfection (14). Cells were maintained in DMEM/F12 media with 10% FBS, 14.3 mM sodium bicarbonate, 15 mM HEPES, 10 mM pyruvate, 48 mM L-glutamine, 0.5 mg/ml insulin, 0.5 mg/ml transferrin, 0.5 ng/ml selenium, 1 U/ml penicillin G, 1 μg/ml streptomycin, and 0.25 μg/ml amphotericin B. Cultures were grown at 37°C in humidified 5% CO2 atmosphere and passaged at 90-100% confluence.
Rat HO-1 Plasmids
A rat HO-1 plasmid was a generous gift from Dr. Leo Otterbein and Dr. Jawed Alam. It had a pBluescript backbone containing the Friend's Spleen Focus-Forming Virus (SFFV) LTR promoter, SV40 enhancer, and SV40 polyadenylation. The backbone contained the entire coding region of rat HO-1 (10). A second plasmid containing the rat HO-1 coding sequence was cloned into pORF5 (Invivogen, San Diego, CA) by directional sticky end cloning using PCR linker compatible ends. The pORF5 clone was comprised of the rat HO-1 coding region driven by an EF 1 α/eIF 4g hybrid promoter and SV40 polyadenylation.
For in vivo experiments in mice, rat HO-1 was blunt cloned between IR/DR elements into a 2 kb, albumin (Alb)-driven, SB10 transposase plasmid. The rat HO-1 pBluescript plasmid was digested with HindIII and SalI and the resulting promoter, coding region, and polyadenylation segment was separated from vector backbone by agarose gel electrophoresis and purified using a Qiagen™ (Valencia, CA) gel purification kit. The recovered rat HO-1 DNA was treated with Klenow to generate blunt ends. Purified Alb/SB10/IRDR plasmid containing transposase prepared from a pCMV SB10 using EcoRI and XhoI (12) and inserted outside IR/DRs was purified using a Qiagen™ plasmid isolation kit, opened at a unique EcoRI site between the IR/DRs and phosphatase treated. The prepared SB10 vector and rat HO-1 DNA were ligated overnight at 16°C. Clones were screened by restriction digest mapping and sequencing followed by protein expression in tissue culture and Western blot analysis.
Transfection of AML12 Hepatocytes
AML12 murine hepatocytes were transfected with plasmid pORF5-rat HO-1, pBluescript-rat HO-1 or an empty pORF5 (irrelevant transgene) using Lipofectamine and Plus Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Media was changed at 24 h post-transfection and cells were harvested for RNA and protein isolation after 40 h.
Transfection of Mice with Rat HO-1 using a Sleeping Beauty Transposase Delivery System
Sleeping Beauty transposase (SB-Tn) (12), was utilized to deliver the rat HO-1 gene to S+S-Antilles sickle mice. The SB-Tn plasmid containing the rat HO-1 gene was delivered by hydrodynamic injection of either 12.5 or 25 μg of SB-Tn-HO-1 plasmid DNA in 2.5 ml of sterile lactated Ringers solution (LRS) into the tail veins of the mice (15). Control mice were injected with LRS without any DNA or an irrelevant transgene. Mouse livers were removed either 96 h or 21 days after injection, frozen in liquid nitrogen and stored at -80° C for RNA and protein analysis.
RNA Isolation and Reverse Transcription (RT) to cDNA
RNA was extracted from tissues and cells using Trizol LS Reagent (16). RNA was quantified by spectrophotometry at 280 and 260 nm. RNA was digested with DNase I (New England Biolabs, Ipswich, MA) and type II restriction endonuclease ApaI to remove any contaminating DNA. ApaI enzyme inactivation and RNA linearization was accomplished by heating this product to 65°C for 5 min followed by holding at 4°C. cDNA was generated immediately with gene-specific reverse transcription primers to HO-1 and GAPDH using Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science, Indianapolis, IN) in the presence of Thermostable Protector RNase Inhibitor and RNase H RNA:DNA degradation enzyme.
qRT-PCR
qRT-PCR for HO-1 and GAPDH was performed in triplicate using cDNA created from 25 ng total RNA and LightCycler® FastStart DNA MasterPLUS SYBR Green I on a LightCycler® (Roche Applied Bioscience, Foster City, CA). Four primers (Integrated DNA Technologies, Coralville, IA) were selected for the qRT-PCR reaction: 5′-TGA ACG GGA AGC TCA CTG G-3′ and 5′-TCC ACC ACC CTG TTG CTG TA-3′ for GAPDH; and 5′-AAC AAG CAG AAC CCA GTC TAT GC-3′ and 5′-GGC CCA CGC ATA TAC CCG CTA CCT-3′ for HO-1. The primers amplify 287 base pairs (bp) and 212 bp, respectively. Candidate HO-1 primer pairs and housekeeping gene GAPDH primers were evaluated for dilution linearity and peak melt temperature by qRT-PCR. Primer pairs chosen for HO-1 and GAPDH generated similar curve shapes, heights, fluorescence intensities, and crossing points in rat and mouse samples. The primers selected demonstrated single cycle linearity of dilution over the range of 50 ng to 6.25 ng template by serial halving of template. Intra-assay variance over this template range was 1%. Reaction times and temperatures were optimized to produce single melt peaks. Both the housekeeping primer pair and the HO-1 primer pair demonstrated approximately a 10 cycle crossing point difference and a higher melt temperature than the no template control reactions.
Quantitation and RFLP Analysis
HO-1/GAPDH ratios were determined by equal fluorescence cycle number following the relationship of 2 (cycle number HO-1) - (cycle number GAPDH). Fluorescence of intercalated cyber green, proportional to the increase in double stranded DNA, was measured at 530 nm through 40 cycles of amplification using ABI LightCycler software (Roche Applied Bioscience). The contents of the qRT-PCR reaction were recovered and digested with type II restriction endonuclease ApaI (1 U/reaction) as described by the vendor (New England Biolabs). Digested products were visualized on 2% agarose gel and intercalated ethidium bromide band image captured by gel-doc and band density quantified using NIH Image J software. The percentage of native and transgene HO-1 was quantified according to the equation:
Western Blots
Western blots for HO-1 protein were performed on AML12 hepatocyte extracts and liver microsomes as previously described (7-9) using antibodies that recognize HO-1 (Assay Designs, Ann Arbor, MI). Gels were scanned on a Storm® gel and blot imaging system (Molecular Dynamics, Sunnyvale, CA) and band density relationships were quantified using NIH Image J software. After HO-1 staining, gels were stripped (Thermo Electron, Rockford, IL) and restained for GAPDH protein as a loading control.
Results
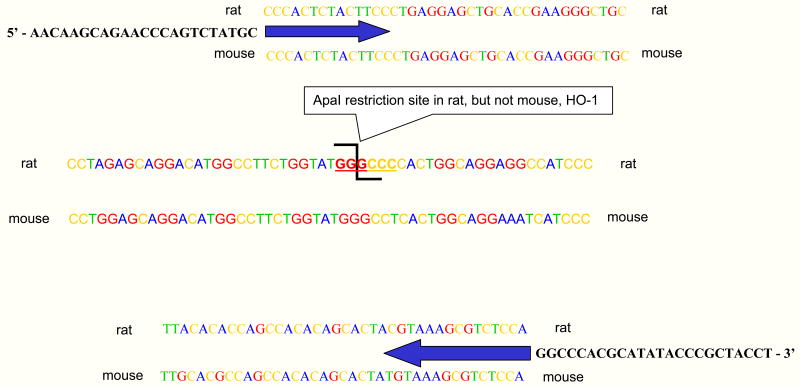
We designed primers from areas of concordance such that the same primer pair amplifies both mouse and rat HO-1 generating a single mass product of 212 bp (Fig 1). The PCR products can be distinguished by RFLP. A discordant type II restriction endonuclease site (ApaI) was identified in the cDNA of the transgene (rat), but not the host (mouse). The ApaI restriction enzyme recognizes the palindrome GGGCCC. The mouse sequence at the same location is GGGCCT which is not recognized by ApaI. Digestion of rat HO-1 cDNA with ApaI generates fragments of 92 and 120 bp. The mouse HO-1 cDNA remains undigested by ApaI with a length of 212 bp.
Fig 1. Mouse and rat DNA sequence differences provide an opportunity to distinguish rat from mouse HO-1 by RFLP.
Primers were made from areas of concordance such that the same primer pair amplifies both native and transgene, generating a single mass product of 212 bp. The PCR products can be distinguished by RFLP. A discordant type II restriction endonuclease site (ApaI) was identified in the cDNA of rat, but not mouse, HO-1 that generates cDNA fragments of 92 and 120 bp in the rat. Species mismatches between rat and mouse are underlined in the mouse DNA sequence. The PCR primers are shown next to the arrows at the 5′ and 3′ ends.
In data not shown, RNA isolated from rat and mouse livers had 11.8- and 17.6-fold more GAPDH than HO-1, respectively, by qRT-PCR. In contrast, rat and mouse spleens had approximately equal quantities of HO-1 and GAPDH by qRT-PCR. The HO-1 qRT-PCR products from rat and mouse liver could be distinguished after digestion with an ApaI (Fig 2). Both mouse and rat cDNA generate 212 bp PCR products without ApaI digestion. The mouse HO-1 cDNA remains 212 bp after ApaI digestion, but the rat HO-1 cDNA generates equimolar fragments of 92 and 120 bp.
Fig 2. RFLP analysis distinguishes RT-PCR products of HO-1 from rat and mouse.
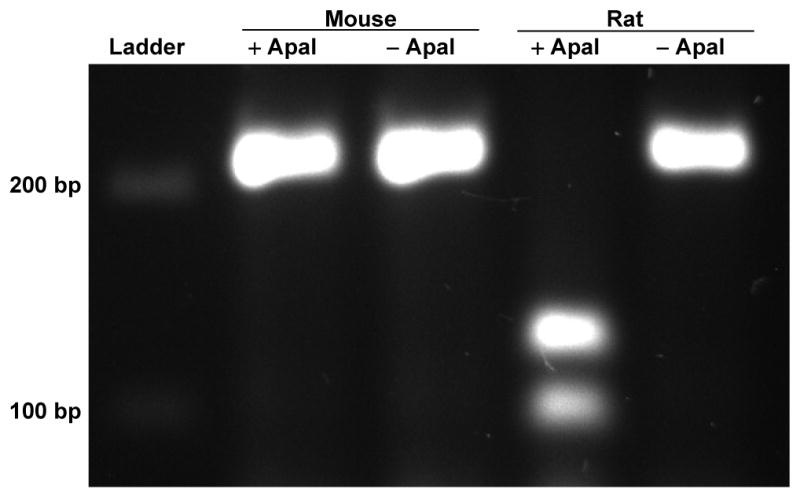
The HO-1 RT-PCR cDNA products from rat and mouse liver can be distinguished after digestion with ApaI restriction enzyme. Without ApaI digestion (-ApaI), both the rat and the mouse generate a cDNA with 212 bp. After digestion with ApaI (+ApaI), the mouse HO-1 cDNA remains 212 bp, but the rat HO-1 cDNA generates fragments of 92 and 120 bp. RFLP reactions contained 400 ng of cDNA PCR products that were incubated at 37°C with (+ApaI) and without (-ApaI) ApaI enzyme. Reaction products were separated by 2% agarose gel electrophoresis with ethidium bromide UV illumination. The data presented are representative of more than 3 experiments.
To further test the method in vitro, murine AML12 hepatocytes were transfected with a pORF rat HO-1 plasmid. RNA was isolated from AML12 cells after 40 h. HO-1 and GAPDH cDNAs were made by qRT-PCR. In data not shown, non-transfected AML12 cells demonstrated 3-fold more GAPDH mRNA than HO-1 mRNA by qRT-PCR. In contrast, AML12 cells transfected with the rat HO-1 transgene demonstrated 1.5-fold higher GAPDH mRNA than HO-1, consistent with a doubling in HO-1 transcription in HO-1 transfected cells. RFLP analysis was used to determine the relative contributions of rat HO-1 transgene and host mouse/AML12 HO-1. Both rat and mouse HO-1 bands can be seen at 212, 120 and 92 bp in ApaI digested cDNA from cells transfected with rat HO-1 (Fig 3A, lane 1). The mouse product is 212 bp and the rat products are 92 and 120 bp. AML12 cells transfected with an irrelevant transgene (lane 2) and non-transfected AML12 cells (lane 3) display only the mouse HO-1 band at 212 bp. Control HO-1 cDNA made from rat liver RNA and digested with ApaI displays only the rat bands at 92 and 120 bp (lane 4). Using the bands in lane 1, we calculated the rat transgene comprised 84.3% of HO-1 cDNA and the host HO-1 comprised 15.7%. The irrelevant transgene (lane 2) did not induce the endogenous mouse HO-1. A Western blot of HO-1 demonstrates the presence of an HO-1 band at 32 kDa (Fig 3B). AML12 cells transfected with a rat HO-1 plasmid had 2.3-fold more HO-1 protein expression than cells transfected with an irrelevant gene.
Fig 3. The rat HO-1 transgene is transcribed in transfected murine AML12 hepatocytes and can be differentiated from mouse HO-1 by RFLP analysis of RT-PCR products of HO-1.
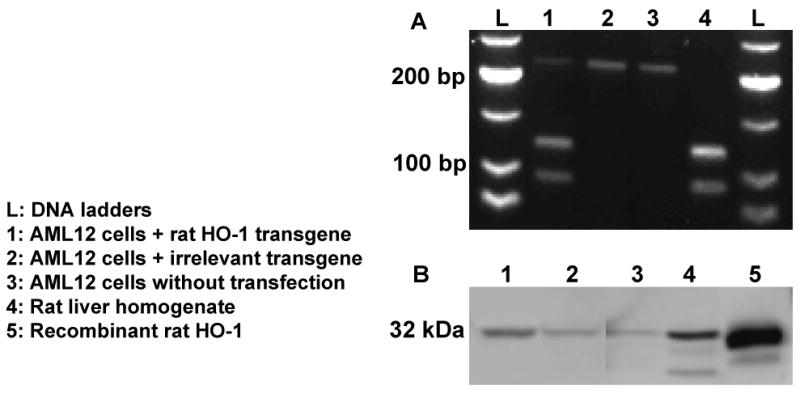
(A) Type II restriction endonuclease digestion of HO-1 RT-PCR products with ApaI allows determination of the relative contributions of the rat HO-1 transgene cDNA and the endogenous mouse HO-1 gene. AML12 cells were transfected using lipofectamine with either a rat HO-1 transgene or an irrelevant transgene. RNA was isolated from AML12 cells 40 h after transfection. RT-PCR was used to make HO-1 cDNA. The cDNA was digested with ApaI restriction enzyme. The resulting products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Lane 1 is loaded with cDNA from murine AML12 hepatocytes transfected with a rat HO-1 transgene. Lane 2 contains cDNA from AML12 hepatocytes transfected with an irrelevant transgene. Lane 3 contains cDNA from non-transfected AML12 cells. Lane 4 contains control HO-1 cDNA from rat liver. (B) AML12 cells transfected with a rat HO-1 transgene had more HO-1 protein expression on a Western blot than cells transfected with an irrelevant gene or non-transfected cells. Lanes 1-4 of the Western blot, transfected as described in (A), were loaded with 22 μg of AML12 protein. Lane 5 contains recombinant rat HO-1 protein (400 ng). The data presented are representative of 2 experiments.
To demonstrate applicability of the qRT-PCR RFLP method to monitoring gene transfer in vivo, a Sleeping Beauty transposon (SB-Tn), was utilized to deliver the rat HO-1 gene to S+S-Antilles sickle mice by hydrodynamic injection (15). Control mice were injected with LRS containing no DNA. Mouse livers were removed and frozen for analysis either 96 hours or 21 days after injection. In data not shown, qRT-PCR was used to measure GAPDH/HO-1 mRNA ratios in mouse liver RNA. After 96 hours and 21 days, respectively, there was a 1.2- and 1.3-fold enrichment of HO-1 mRNA in mice injected with SB-Tn-HO-1 DNA relative to LRS control mice. RFLP analysis of mouse liver HO-1 cDNA showed the rat transgene comprised 82 - 88% of HO-1 cDNA (92 and 120 bp) and the host HO-1 (212 bp) comprised 12 - 18%. after 96 hours and 21 days, respectively, in the mice injected with SB-Tn-HO-1 (Fig 4A). There was no rat HO-1 cDNA seen on the agarose gel in the mice injected with LRS only or hemin. The mouse injected with 12.5 μg of SB-Tn-HO-1 plasmid DNA showed 2.6-fold greater HO-1 protein expression in liver after 96 hours on Western blot relative to a control mouse injected with LRS only (Fig 4B). Similarly, a mouse injected with 25 μg of SB-Tn-HO-1 plasmid DNA had 2.5-fold greater HO-1 protein expression in liver after 21 days than a control mouse injected with LRS (Fig 4B). A mouse injected with hemin, a potent inducer of HO-1, showed 2.1-fold greater HO-1 protein expression in liver after 16 hours relative to a control mouse injected with LRS at 96 hours (Fig 4B).
Fig 4. The rat HO-1 transgene is transcribed in mouse liver and can be differentiated from mouse HO-1 by RFLP analysis of HO-1 cDNA.
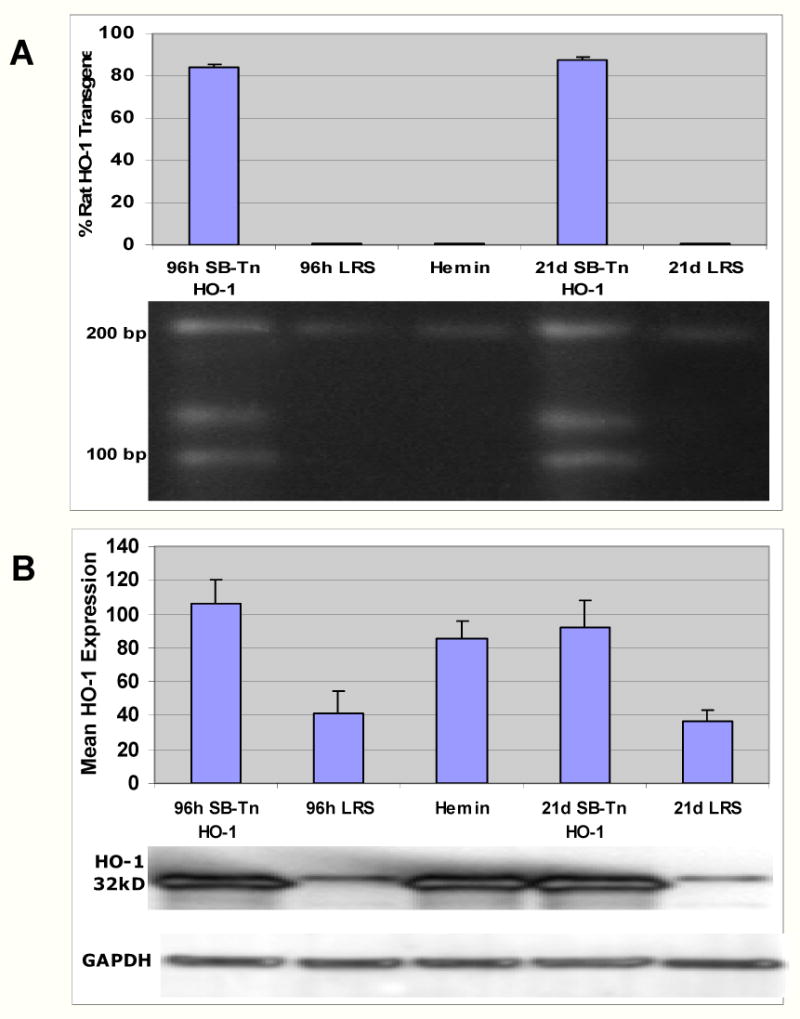
A Sleeping Beauty transposase (SB-Tn) was utilized to deliver the rat HO-1 gene to S+S-Antilles sickle mice. The SB-Tn plasmid containing the rat HO-1 gene was delivered by hydrodynamic injection of 12.5 μg or 25 μg of SB-Tn-HO-1 plasmid DNA in 2.5 ml of sterile lactated Ringers solution (LRS) into the tail veins of the mice. Control sickle mice were injected with LRS containing no DNA. After either 96 hours (12.5 μg DNA injected) or 21 days (25 μg DNA injected), livers were harvested and RNA was isolated for RT-PCR RFLP analysis of HO-1 transcription and microsomes were isolated for Western blot analysis of HO-1 protein expression. (A) RFLP digestion with ApaI of the HO-1 cDNA from these livers shows an enrichment of rat HO-1 cDNA (92 and 120 bp) relative to mouse HO-1 cDNA (212 bp) in the mice injected with SB-Tn-HO-1 at both 96 hours and 21 days after injection. There is only mouse HO-1 cDNA (212 bp) in the mice injected with LRS only and the mouse injected with hemin. Each lane in (A) was loaded with 100 μg of ApaI-digested HO-1 RT-PCR cDNA. The bar graph in (A) summarizes the RFLP data (n=3 gels/sample) and shows the mean (± SD) percentage of rat HO-1 transgene in mouse liver HO-1 RNA. (B) Western blots for HO-1 expression in these livers shows mice injected with SB-Tn-HO-1 express more HO-1 in the liver at both 96 hours and 21 days after injection than mice injected with LRS only. Each lane of the Western blot was loaded with 6 μg of liver microsomal protein. Bands for HO-1 (32 kDa) and GAPDH are shown in (B) from a representative Western blot. As a positive control for HO-1 protein expression in the Western, a C57BL/6 mouse was injected with hemin (40 μmol/kg i.p.) for 3 consecutive days and the liver was removed 16 h after the third injection (Hemin). The bar graph in (B) summarizes the Western blot quantitation (n=3 blots/sample) and shows the mean (± SD) HO-1 protein expression in the mouse livers.
Discussion
The areas of discordance between the rat HO-1 transgene and native mouse gene were not sufficient to monitor transgene expression through primary antibody differentiation or enzymatic activity. However, transcription analysis provided a method of establishing the level of gene transcription. We identified qRT-PCR primer sequences from areas of identity in the native and transgene sequence that generate cDNA of appropriate mass, have high temperature single melt peak profiles, linearly amplify the templates over the same template concentration range, and span a discordant type II restriction endonuclease site. Our GAPDH housekeeping gene primer pair was equally evaluated and selected. We were able to estimate the change in HO-1 transcription compared to GAPDH and the transgenic and native contribution to the change by linking qRT-PCR and RFLP.
This type of analysis has translational applicability to other gene transfer studies. The ultimate goal of gene transfer is the introduction of a gene that produces a product as identical in structure and function as possible to the native gene while guaranteeing the safety of the protocol (17). The currently utilized practice of selecting a highly conserved second species could be replaced with the creation of an RFLP detectable transgene from the native gene. Research involving vector expression of factor IX cDNA demonstrated that even highly homologous proteins can elicit negative immune reactions in non-human primates (18). Rhesus factor IX is >95% homologous to human factor IX cDNA and >97% identical by amino acid sequence. When given intravenously it was not found to be immunogenic. However, when administered as a transgene, both the factor IX and the adenoviral vector generated an immune response in the rhesus (19, 20). Furthermore, xenogenic transduced cells have shown shortened survival time based on circulating half lives (21) and xenogenic transgene expression of factor VIII and XI mRNA gradually decreased without loss of vector DNA (22). Although general agreement exists regarding the value of information gained from non-human primate studies, the expense and effort involved are huge impediments. To date, most non-human primate studies have been related to gene transfer optimization or hematopoiesis, while humanized, transgenic, and knock out/knock in mice have become the animal of choice for transgene work. Even in these new generation rodents in combination with non-viral vectors, immunotolerization issues remain (23). Researchers using non-human primates have monitored transfer efficiency through integration site detection, which unlike our model, does not indicate successful transcription of the transgene. Use of the qRT-PCR RFLP method to track transgene RNA addresses the issues of highly stringent, very low level detection of transcription product.
Toxicity and tolerance (24) issues remain incompletely studied nearly 10 years after early patient deaths. Naturally occurring polymorphisms suggest that polymorphic transgenes would produce a functional tolerated protein. Exploitation of the redundancy in the translation code allows the creation of a silent nucleotide change and the creation of a novel restriction site, a polymorphic transgene. For example, the human HO-1 gene has a leucine and lysine at amino acids 147 and 148, respectively. These are coded by the non-palindromic sequence CTC AAA in the native gene. Leucine and lysine can also be coded by the palindrome CTT AAG creating a restriction site for AflII. PCR primers can then be selected for locations 5′ and 3′ to this site and an RFLP protocol designed. Similarly, the same approach could be considered for the factor IX studies. Both the rhesus gene and the human gene have a CTC AAA sequence which could be changed through site directed mutagenesis to CTT AAG, an AflII restriction site, without changing the amino acid composition of the protein. This would allow gene therapy to the rhesus with a rhesus transgene and to the human with a human transgene without sacrificing tracking of the transgene.
In conclusion, qRT-PCR RFLP is a useful and reliable method to differentiate transgene from host gene expression, especially when the host and transgene protein are identical or highly homologous. This type of analysis has translational applications to the design, delivery and monitoring of gene therapy vectors.
Acknowledgments
The authors wish to acknowledge the contributions and comments of Betsy T. Kren and Alycia A. Trossen. This work was supported by NIH/NHLBI grant PO1 HL55552.
Abbreviations
- HO-1
heme oxygenase-1
- RFLP
restriction fragment length polymorphism
- SB-Tn
Sleeping Beauty transposase
- SCD
sickle cell disease
- DR
direct repeat
- IR
inverted repeat
- SFFV
spleen focus-forming virus
- Alb
albumin
- LRS
lactated Ringers solution
- RT
reverse transcription
- bp
base pairs
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maines MD. Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications. Faseb J. 1988 Jul;2(10):2557–68. [PubMed] [Google Scholar]
- 2.Otterbein LE, Soares MP, Yamashita K, Bach FH. Heme oxygenase-1: unleashing the protective properties of heme. Trends Immunol. 2003 Aug;24(8):449–55. doi: 10.1016/s1471-4906(03)00181-9. [DOI] [PubMed] [Google Scholar]
- 3.Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev. 2006 Apr;86(2):583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 4.Aslan M, Freeman BA. Oxidant-mediated impairment of nitric oxide signaling in sickle cell disease--mechanisms and consequences. Cell Mol Biol (Noisy-le-grand) 2004 Feb;50(1):95–105. [PubMed] [Google Scholar]
- 5.Hebbel RP, Osarogiagbon R, Kaul D. The endothelial biology of sickle cell disease: inflammation and a chronic vasculopathy. Microcirculation. 2004 Mar;11(2):129–51. [PubMed] [Google Scholar]
- 6.Wagener FA, Abraham NG, van Kooyk Y, de Witte T, Figdor CG. Heme-induced cell adhesion in the pathogenesis of sickle-cell disease and inflammation. Trends Pharmacol Sci. 2001 Feb;22(2):52–4. doi: 10.1016/s0165-6147(00)01609-6. [DOI] [PubMed] [Google Scholar]
- 7.Belcher JD, Mahaseth H, Welch TE, Otterbein LE, Hebbel RP, Vercellotti GM. Heme oxygenase-1 is a modulator of inflammation and vaso-occlusion in transgenic sickle mice. J Clin Invest. 2006 Mar;116(3):808–16. doi: 10.1172/JCI26857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee PJ, Alam J, Wiegand GW, Choi AM. Overexpression of heme oxygenase-1 in human pulmonary epithelial cells results in cell growth arrest and increased resistance to hyperoxia. Proc Natl Acad Sci U S A. 1996 Sep 17;93(19):10393–8. doi: 10.1073/pnas.93.19.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otterbein LE, Kolls JK, Mantell LL, Cook JL, Alam J, Choi AM. Exogenous administration of heme oxygenase-1 by gene transfer provides protection against hyperoxia-induced lung injury. J Clin Invest. 1999 Apr;103(7):1047–54. doi: 10.1172/JCI5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibahara S, Muller R, Taguchi H, Yoshida T. Cloning and expression of cDNA for rat heme oxygenase. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7865–9. doi: 10.1073/pnas.82.23.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002 May 17;318(5):1221–35. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 12.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997 Nov 14;91(4):501–10. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 13.Fabry ME, Sengupta A, Suzuka SM, Costantini F, Rubin EM, Hofrichter J, et al. A second generation transgenic mouse model expressing both hemoglobin S (HbS) and HbS-Antilles results in increased phenotypic severity. Blood. 1995 Sep 15;86(6):2419–28. [PubMed] [Google Scholar]
- 14.Wu JC, Merlino G, Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci U S A. 1994 Jan 18;91(2):674–8. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell JB, Podetz-Pedersen KM, Aronovich EL, Belur LR, McIvor RS, Hackett PB. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protoc. 2007;2(12):3153–65. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chomczynski P, S N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry. 1987;162(1):156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg LE, Schechter AN. Gene therapist, heal thyself. Science (New York, NY. 2000 Mar 10;287(5459):1751. doi: 10.1126/science.287.5459.1751. [DOI] [PubMed] [Google Scholar]
- 18.Herzog RW, Yang EY, Couto LB, Hagstrom JN, Elwell D, Fields PA, et al. Long-term correction of canine hemophilia B by gene transfer of blood coagulation factor IX mediated by adeno-associated viral vector. Nature medicine. 1999 Jan;5(1):56–63. doi: 10.1038/4743. [DOI] [PubMed] [Google Scholar]
- 19.Lozier JN, Metzger ME, Donahue RE, Morgan RA. The rhesus macaque as an animal model for hemophilia B gene therapy. Blood. 1999 Mar 15;93(6):1875–81. [PubMed] [Google Scholar]
- 20.Lozier JN, Metzger ME, Donahue RE, Morgan RA. Adenovirus-mediated expression of human coagulation factor IX in the rhesus macaque is associated with dose-limiting toxicity. Blood. 1999 Dec 15;94(12):3968–75. [PubMed] [Google Scholar]
- 21.Hanazono Y, Terao K, Ozawa K. Gene transfer into nonhuman primate hematopoietic stem cells: implications for gene therapy. Stem cells (Dayton, Ohio) 2001;19(1):12–23. doi: 10.1634/stemcells.19-1-12. [DOI] [PubMed] [Google Scholar]
- 22.Haack A, Poller W, Schneider-Rasp S, Thalheimer P, Schmitt C, Hanfland P, et al. Highly sensitive and species-specific assay for quantification of human transgene expression levels. Haemophilia. 1999 Sep;5(5):334–9. doi: 10.1046/j.1365-2516.1999.00324.x. [DOI] [PubMed] [Google Scholar]
- 23.Ohlfest JR, Frandsen JL, Fritz S, Lobitz PD, Perkinson SG, Clark KJ, et al. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood. 2005 Apr 1;105(7):2691–8. doi: 10.1182/blood-2004-09-3496. [DOI] [PubMed] [Google Scholar]
- 24.Brenner M. Gene transfer by adenovectors. Blood. 1999 Dec 15;94(12):3965–7. [PubMed] [Google Scholar]