Abstract
Protein degradation is an essential cellular function that, when dysregulated or impaired, can lead to a wide variety of disease states. The two major intracellular protein degradation systems are the ubiquitin-proteasome system (UPS) and autophagy, a catabolic process that involves delivery of cellular components to the lysosome for degradation. While the UPS has garnered much attention as it relates to neurodegenerative disease, important links between autophagy and neurodegeneration have also become evident. Furthermore, recent studies have revealed interaction between the UPS and autophagy, suggesting a coordinated and complementary relationship between these degradation systems that becomes critical in times of cellular stress. Here we describe autophagy and review evidence implicating this system as an important player in the pathogenesis of neurodegenerative disease. We discuss the role of autophagy in neurodegeneration and review its neuroprotective functions as revealed by experimental manipulation in disease models. Finally, we explore potential parallels and connections between autophagy and the UPS, highlighting their collaborative roles in protecting against neurodegenerative disease.
Keywords: autophagy, ubiquitin-proteasome system, neurodegeneration, HDAC6, p62
A precarious balance
The energy expenditure needed to produce protein is substantial; thus, the degradation of these macromolecules comes at a high cost. Nevertheless, protein turnover is essential for removing defective proteins and for contributing to the pool of amino acids required for continued protein synthesis in times of limited nutrient availability. Furthermore, many essential cellular functions –including cell division, transcription, and signal transduction – are regulated by fluctuation in protein levels accomplished by altering the balance of protein synthesis and degradation. The role of protein catabolism in protecting cells from defective, misfolded proteins has been the subject of increased attention as its relevance to human disease has become apparent. A substantial fraction of newly synthesized proteins are translated incorrectly or fold incorrectly due to errors in synthesis or genetic mutations [1–4]. Oxidative or nitrosylative damage adds to the burden of defective proteins. Efficient degradation of these proteins is essential, as cells cannot risk the long-term accumulation of proteins that engage in aberrant protein-protein interactions, form insoluble aggregates, or acquire other toxic properties. Considering the importance of protein catabolism in maintaining cell homeostasis, it is not surprising that dysregulation of protein turnover is associated with myriad disease states such as cancer and neurodegeneration [5].
Two major pathways accomplish regulated protein catabolism: the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal system. The UPS serves as the primary route for degradation for thousands of short-lived proteins and provides the exquisite specificity and temporal control needed for fine-tuning the steady state levels of many regulatory proteins [6]. UPS-mediated catabolism is also essential to mainten amino acid pools in acute starvation and contributes significantly to the degradation of defective proteins [1, 2, 7]. Autophagy, by contrast, is primarily responsible for degrading long-lived proteins and maintaining amino acid pools in the setting of chronic starvation, although its contribution to the degradation of defective proteins may equal that of the UPS. Though it has received less attention than the UPS historically, breakthroughs in the molecular genetics of autophagy have led to a renaissance of interest in this catabolic pathway and has revealed many surprising insights about its regulation, function, and contribution to protein degradation, in both normal and disease states. This review will (1) highlight the parallels between the UPS and autophagy in their roles and regulation, (2) explore the role of autophagy in neurodegeneration, noting parallels with the UPS, and (3) discuss emerging evidence of a functional relationship between the UPS and autophagy and its relevance to neurodegeneration.
The basics: roles and regulation
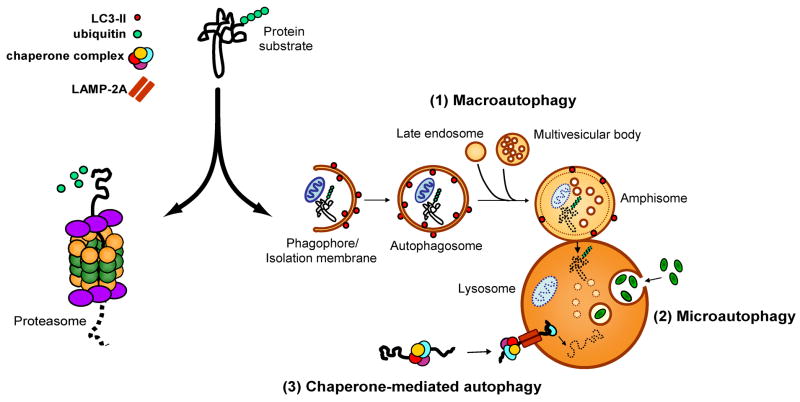
“Autophagy”, literally “self-eating”, describes a catabolic process in which cell constituents such as organelles and proteins are delivered to the lysosomal compartment for degradation. Autophagy is an evolutionarily conserved process whose primary task in lower organisms is the maintenance of metabolic homeostasis in the face of changing nutrient availability [8]. This role in recycling is complementary to that of the UPS, which degrades proteins to generate oligopeptides that are subsequently degraded into amino acids while replenishing the cell’s supply of free ubiquitin. Recent advances have demonstrated that autophagy also serves a surprisingly diverse array of additional functions, including organelle clearance, antigen presentation, elimination of microbes, as well as regulation of development and cell death [9]. Traditionally, autophagy has been considered a less selective degradative pathway than the UPS and is frequently illustrated as the engulfment of large portions of cytoplasm (including nearby cytosolic bystanders) and delivery of the contents to the lysosome in bulk. This view of autophagy as a crude, nonselective form of catabolism has been challenged by the appreciation of specialized forms of autophagy that are distinguished by the identity of the substrates and the route by which these substrates reach the lysosomal compartment (Figure 1). Microautophagy consists of direct engulfment of small volumes of cytosol by lysosomes [10], whereas chaperone-mediated autophagy (CMA) involves selective, receptor-mediated translocation of proteins into the lysosomal lumen [11]. These processes are distinguished from macroautophagy, in which an isolation membrane expands to engulf a portion of the cell, eventually fusing to form a new autophagic vacuole that subsequently fuses with a lysosome [12]. Even within the category of macroautophagy, there appears to be capability for selectivity, as autophagic processes have been observed that appear to be specific for mitochondria (mitophagy), portions of the nucleus (nucleophagy), peroxisomes (pexophagy), endoplasmic reticulum (reticulophagy), microorganisms (xenophagy), ribosomes (ribophagy) or protein aggregates (aggrephagy) (reviewed in [5]).
Figure 1.
The UPS and the autophagy-lysosomal systems are the two main protein degradation systems in the cell. Protein that are tagged with polyubiquitin chains are generally considered to be substrates for the UPS, which feeds unfolded proteins through the barrel of the 26S proteasome and generates small digested peptides. Recent evidence suggests that some ubiquitinated substrates can also be degraded via the autophagy-lysosomal system. This system is comprised of (1) chaperone-mediated autophagy (CMA), in which soluble substrates associated with a specific chaperone complex are translocated into the lysosome through the LAMP-2A lysosomal receptor, (2) microautophagy, in which small volumes of cytosol are directly engulfed by lysosomes, and (3) macroautophagy, in which cytosolic components are engulfed and delivered to the lysosome in bulk. Macroautophagy involves a series of maturation steps: first, a portion of cytoplasm is surrounded by an expanding elongation membrane or phagophore. The phagophore seals to form an autophagosome, which in mammals fuses with late endosomes and multivesicular bodies to form an amphisome. The amphisome then fuses with a lysosome to form an autolysosome, in which cytosolic cargo is degraded by lysosomal hydrolases. LC3-II is a protein that associates with the inner and outer surfaces of autophagic membranes and provides a histological marker of autophagic vacuoles.
While the molecular regulation of microautophagy remains obscure, there has been substantial insight into the regulation of CMA and macroautophagy. CMA is a process in which proteins harboring a pentapeptide motif related to the sequence KFERQ are specifically recognized by a cytosolic chaperone, the heat shock cognate protein of 70 kDa (hsc70). The substrate-chaperone complex is then targeted to the lysosome by binding to lysosome-associated membrane protein 2A (LAMP-2A) which carries out receptor-mediated translocation of the substrate into the lysosome for degradation [11, 13]. Up to 30% of all cytosolic proteins harbor the CMA recognition motif and are potentially subject to degradation by this catabolic pathway during long-term nutrient deprivation [11, 14].
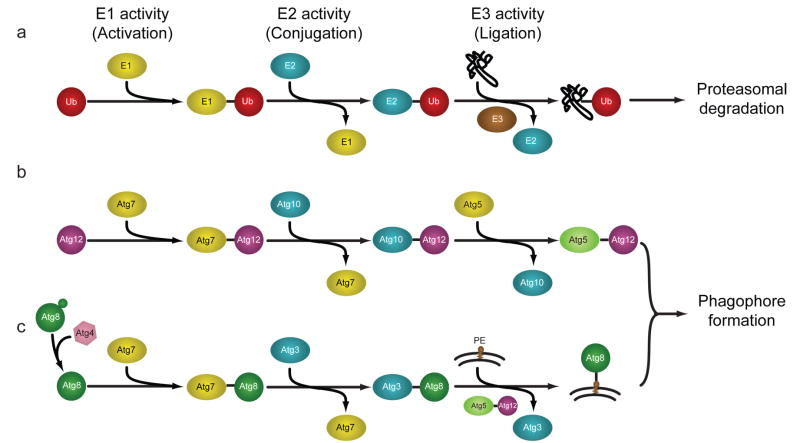
While it remains unclear how substrates are specifically marked for degradation by macroautophagy (hereafter referred to as autophagy), the identification of a family of autophagy-related (Atg) genes in yeast and their homologues in higher organisms has permitted minute dissection of the general process by which autophagy engulfs and degrades its targets. The initial step in autophagy involves expansion of a membranous structure called the “isolation membrane” or “phagophore” that engulfs a portion of the cell; the membrane eventually fuses to form a new double-membraned structure known as an autophagosome (Figure 1). The process of autophagy is controlled by parallel activation cascades that involve ubiquitin-like (UBL) protein modification, strikingly similar to the activation cascade that regulates the UPS (Figure 2a). In the first arm of the Atg conjugation system, phagophore membrane elongation is triggered through the sequential action of an E1-like protein (Atg7) and an E2-like protein (Atg10) leading to an isopeptide linkage between the C-terminal glycine the UBL protein Atg12 and a lysine residue of Atg5 (Figure 2b). These Atg12-Atg5 conjugates are further cross-linked to Atg16 to form a large (~350 kDa) multimeric complex, which has been thought to act as a structural support for membrane expansion [15]. More recent work has demonstrated that the Atg12-Atg5 conjugate can function as an E3-like enzyme in the second arm of the Atg conjugation cascade to promote lipidation of Atg8 [16]. In a second arm of the Atg conjugation system, Atg4 cleaves the UBL protein Atg8 to promote interaction with Atg7. Atg8 is then conjugated with the phospholipid phosphotidylethanolamine (PE) by the concerted action of the E2-like Atg3 and the E3-like Atg12-Atg5 conjugate. Of note, this E3-like activity results in a protein-lipid conjugation, in contrast to the classical E3 protein-protein conjugation of the UPS. As PE is a component of the autophagosomal membrane, the lipidation reaction results in studding of the inner and outer membranes of autophagosomes with Atg8 (Figure 2c).
Figure 2.
Assembly and elongation of autophagic membranes are accomplished via sequential action of UPS-like E1-E2-E3 cascades. In each case, an E1 enzyme activates a ubiquitin-like protein (UBL) such as ubiquitin, Atg12, or Atg8. The UBL is then transferred to an E2 conjugating enzyme, followed by an association with an E3 ligase that promotes association of the UBL and its target. (a) In the UPS, ubiquitination of substrates is accomplished by an E1-activating enzyme, E2-conjugating enzyme, and an E3-ligase. (b) In the first arm of the Atg conjugation pathway, Atg12 associates with the E1-like Atg7, is transferred to the E2-like Atg10, and is subsequently conjugated to Atg5. No E3-like protein has been identified in this pathway. (c) In the second arm of the Atg conjugation pathway, Atg8 associates with the E1-like Atg7, is transferred to the E2-like Atg3, and is conjugated to PE via the E3-like action of the Atg12-Atg5 complex. Adapted from [101] with permission.
Once formed, new autophagosomes move through a stepwise maturation process that culminates with fusion to a lysosome permitting degradation of the lumenal contents. In mammals, autophagosomes first fuse with endosomes and multivesicular bodies to form amphisomes, which subsequently fuse with lysosomes to create degradative vacuoles termed autolysosomes [17]. Autophagosomes and autolysosomes can be distinguished morphologically, as autophagosomes contain contents with densities similar to cytosol, while autolysosomes appear as electron-dense material with a hollow rim beneath the limiting membrane. However, because of occasional ambiguity in distinguishing autophagosomes, amphisomes, and autolysosomes morphologically, the term “autophagic vacuole” frequently appears in the literature to refer to all three structures.
Four metazoan homologs of Atg8 have been identified: MAP-LC3 (microtubule-associated protein light chain 3), GABARAP (γ-aminobutyric-acid-type-A-receptor-associated protein), GATE-16 (Golgi-associated ATPase enhancer of 16 kDa), and Atg8L, although GABARAP, GATE-16, and Atg8L have not been extensively characterized. While GABARAP and GATE-16 may also be conjugated to PE in experimental systems, at present MAP-LC3 (typically abbreviated LC3) is the only protein that is known to remain associated with the autophagosome in higher eukaryotes. Pro-LC3 is cleaved co-translationally to create a form of LC3 denoted “LC3-I”. LC3-I becomes conjugated to PE to form “LC3-II” and thereby covalently associates with the phagophore. Consequently, the generation and turnover of LC3-II is used as an index of autophagy induction and/or flux [18]. LC3-II staining is also used as a primary histological marker of autophagosomes. Because LC3-II remains on the inner membrane of autophagosomes until lysosomal enzymes degrade it, increased steady-state levels of LC3-II may be due to induction of autophagosome formation, a blockade in their maturation, or both. Distinguishing between these possibilities for experimental purposes is readily accomplished with the use of chemical inhibitors of maturation [18].
A role for autophagy in neurodegeneration
Many neurodegenerative diseases are characterized by accumulation of misfolded protein deposits in affected brain regions, suggesting a failure in the cell’s degradative capacity [19]. Neurons, as highly metabolically active, post-mitotic cells, are especially vulnerable to the accumulation of defective proteins, and this may account for the frequency with which conformational diseases affect the nervous system. In most cases, these proteinaceous deposits are composed of ubiquitin conjugates, suggesting a failure in the clearance of proteins targeted for proteasomal degradation. Indeed, experimental evidence indicates that neurodegeneration is frequently associated with impaired UPS function, although whether this is a cause or consequence of neurodegeneration is a contested issue, as is reviewed elsewhere in this special issue. It has also been suggested that autophagy plays a role in the initiation or progression of some neurodegenerative diseases [20]. This suggestion originates from the observed accumulation of autophagic vacuoles in neurons from affected brain regions in a number of neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, Creutzfeldt-Jakob disease, and many of the polyglutamine diseases [21–24]. This notion has since been validated by experimental evidence and insights provided by human genetics, as described below.
Neurodegeneration is frequently characterized by increased frequency of autophagic vacuoles
Huntington’s and Alzheimer’s diseases are among the best-studied examples where histopathology implicates autophagy as playing a role in disease pathogenesis. Alzheimer’s disease pathology features massive accumulation of autophagic vacuoles within large swellings along dystrophic and degenerating neurites in neocortical and hippocampal pyramidal neurons [21]. In Huntington’s disease, affected neurons show accumulation of huntingtin in cathepsin-D-positive vacuoles [24]. Cathepsin-D is a lysosomal protease enriched in neuronal tissues, suggesting that these are autolysosomes. However, the observed increased frequency of autophagic vacuoles in disease brain is ambiguous with respect to whether autophagy is induced or whether autophagy flux is impaired. Furthermore, autophagosomes are frequently observed in dying neurons, where it is unclear whether autophagy is operating as a futile cytoprotective response, whether autophagy mediates cell death, or whether it is induced secondarily in a cell already otherwise committed to dying. Insight into the role of autophagy in neurodegeneration has been provided by studies indicating that: 1) some neurodegenerative disease-related proteins are degraded by autophagy, 2) impairment of autophagy promotes neurodegeneration in animal models and several human neurodegenerative diseases, and 3) manipulation of autophagy modifies phenotypes in animal models of neurodegeneration.
Neurodegenerative disease-related proteins are degraded by autophagy
That neurodegenerative disease-causing proteins are frequently degraded by autophagy was demonstrated by a series of in vitro studies which showed that pharmacological induction or inhibition of macroautophagy alters the rate of turnover of a number of disease-related proteins including polyglutamine-expanded proteins, polyalanine-expanded proteins, as well as wild type and mutant forms of α-synuclein [25, 26]. Moreover, ultrastructural analysis by immuno-electron microscopy showed that in cell culture models, disease-related proteins are delivered to autophagic vacuoles [27, 28]. CMA has also been found to contribute to the degradation of α-synuclein [29]. Collectively, these studies suggested that autophagy contributes to the degradation of multiple disease proteins and the efficiency of this pathway could relate to the onset or progression of disease. Of note, there is evidence that many of these same disease-causing proteins are also degraded by the UPS [30–32], suggesting that more than one degradative route may be available to them. In the case of α-synuclein, for example, Webb et al. concluded that soluble forms of the disease protein are efficiently degraded by the UPS, while aggregated or oligomeric α-synuclein require autophagy for clearance [26]. These observations have led to the suggestion that autophagy provides an alternate, compensatory route of degradation when clearance by the UPS and CMA are compromised. The relative contribution of autophagy and the UPS to degrading disease-related substrates, and the relationship of this to the onset and progression of various diseases, remains to be elucidated -- and this may differ amongst different diseases.
Impairment of autophagy promotes neurodegeneration
It is becoming increasingly evident that the autophagy-lysosomal system is essential to neuronal homeostasis, and may in some settings be neuroprotective. The consequences of impaired lysosome function, for example, may be observed in cathepsin D knockout mice and Drosophila melanogaster cathepsin D mutants which show neurodegeneration and associated accumulation of autophagosomes and lysosomes [33–35]. The importance of autophagy to neuronal homeostasis is further illustrated by characterization of mice with conditional knockout of Atg genes. These mice die prematurely with extensive neurodegeneration and ubiquitin-positive pathology [36, 37]. On the basis of these observations one might predict that impairment of autophagy could contribute to neurodegenerative disease in humans. Indeed, primary lysosomal dysfunction in inherited congenital “lysosomal storage disorders” has long been recognized to cause severe neurodegenerative phenotypes characterized pathologically by accumulations of lysosomes and autophagic vacuoles [38]. For example, the neuronal ceroid lipofuscinoses (NCLs) are a heterogeneous group of inherited, neurodegenerative disorders with onset ranging from infancy to late adulthood that are caused by a variety of defects in lysosomal function. Furthermore, a growing list of adult-onset, familial neurological diseases have been linked to mutations expected to have an impact on autophagy-lysosomal function (reviewed in [38]), including Kufor-Rakeb syndrome (a form of early-onset parkinsonism with dementia) [39], Charcot-Marie-Tooth type 2B (CMT2B) [40], and distal-spinobulbar muscular atrophy (distal-SBMA) [41]. Mutations in CLN3, a transmembrane protein that localizes to the late endosomal/lysosomal membrane, cause a form of NCL. CLN3-related neurodegeneration appears to be a consequence of reduced autophagosome-lysosome fusion [42]. Mutations in ATP13A2, which encodes a primarily neuronal lysosomal ATPase, were recently found to cause Kufor-Rakeb syndrome (previously designated PARK9). Disease-causing mutations in ATP13A2 result in protein retention in the endoplasmic reticulum and enhanced proteasomal degradation, suggesting that neurodegeneration could be caused by overwhelming the UPS and/or loss of function in lysosomal protein degradation [39]. Mutations in Rab7 cause the dominantly inherited axonal neuropathy CMT2B. Rab7 participates in trafficking autophagosomes and fusion with lysosomes and disease-causing mutations are predicted to impair this process [43–45]. Mutations in p150/dynactin are responsible for the motor neuron disease distal-SBMA. Microtubule-based vesicular trafficking is essential for delivery of autophagosomes to lysosomes and subsequent fusion [46], and impaired dynein-mediated trafficking is associated with impaired autophagosome/lysosome fusion and reduced protein turnover [47, 48]. Thus, motor neuron loss in distal-SBMA may result from impaired autophagosome trafficking and/or fusion with lysosomes. Indeed, a mouse model of distal-SBMA that expresses mutant p150/dynactin is characterized by accumulation of ubiquitin-positive aggregates and autophagic vacuoles in affected neurons [49].
Manipulation of autophagy modifies neurodegenerative phenotypes in animal models
While deficiency in autophagy results in neurodegeneration, a separate question concerns the role of autophagy in the context of diseases initiated by mutations in genes unrelated to autophagic function. To investigate this perspective, researchers have turned to animal models of common neurodegenerative diseases that are amenable to genetic and pharmacological manipulation of autophagy. These studies have largely shown that reduced autophagy worsens disease phenotypes whereas augmented autophagy provides benefit, leading to the conclusion that autophagy is cytoprotective. For example, in a Drosophila model of X-linked spinobulbar muscular atrophy (SBMA), a polyglutamine disease, degeneration was strongly enhanced by genetic inhibition of autophagy [50]. Similarly, in transgenic mice expressing amyloid precursor protein, a mouse model of Alzheimer’s disease, genetic inhibition of autophagy by heterozygous depletion of beclin-1 results in enhancement of neurodegeneration [51]. In both of these studies it was determined that autophagy deficiency results in greater accumulation of the offending, disease-related protein [50, 51], suggesting that autophagy was needed to degrade cytotoxic proteins. This provided the rationale for investigating whether increasing autophagic activity might provide benefit. Pharmacological upregulation of autophagy can be accomplished using the drug rapamycin, which works by inhibiting TOR (target of rapamycin), a pleiotropic molecule that negatively regulates autophagy, among other functions. Indeed, treatment with rapamycin ameliorates the degenerative phenotype in a Drosophila model of SBMA, as well as in Drosophila and mouse models of Huntington’s disease [48, 50, 52]. In addition, inducing autophagy in an TOR-independent manner using lithium [53] or trehalose [54–56] has been shown to accelerate clearance of disease proteins in vitro [56] and protect against neurodegeneration in mouse and Drosophila models of Huntington’s disease [53, 54]. These exciting results have opened the door to the possibility that pharmacological upregulation of autophagy by rapamycin, lithium, trehalose, or a newer generation of small molecules might be of therapeutic benefit for patients with neurodegenerative disease. Recently, high throughput screening efforts have identified small molecule activators of autophagy. Some of these compounds inhibit TOR and activate autophagy in a manner analogous to rapamycin, but other compounds are TOR-independent and reflect multiple points of potential therapeutic intervention [57]. There have been fewer efforts to manipulate UPS function for therapeutic benefit in neurodegenerative disease, but it was recently shown that use of a proteasome activator enhanced survival in an in vitro model of Huntington’s disease [58], suggesting that augmenting other routes of protein degradation may also provide neuroprotection.
However, it should be pointed out that the relationship of autophagy to the accumulation of disease protein may not always be straightforward. There is evidence that in some cases cellular attempts to degrade cytotoxic protein aggregates interfere with normal autophagy function leading to lysosomal “indigestion” that ultimately compromises cell function or viability. For example, α-synuclein is degraded at least in part by CMA [29]. Mutations in α-synuclein that are causative of familial Parkinson’s disease are poorly transferred to the lysosomal lumen and accumulate on the lysosomal surface, resulting in blockade of receptor-mediated translocation. This results in disrupting degradation of other CMA substrates [29, 59]. With respect to Alzheimer’s disease, an even more complex story is emerging. Several lines of evidence suggest that there is an impairment of autophagy resulting from impaired autophagosome-lysosome fusion combined with decreasing efficiency of the lysosomal system [60]. These vacuoles may further contribute to pathogenesis by interfering with normal intracellular trafficking and/or by leaking undigested toxic contents into the cytosol, or more generally by disrupting normal metabolic turnover required for neuronal homeostasis. Recent evidence suggests that the autophagic turnover of amyloid beta precursor protein (APP) may underlie the generation of toxic amyloid-β species [61]. Thus, the relationship of autophagy to Alzheimer’s disease progression is complex, with autophagy-related production of toxic amyloid-β that culminates in impaired autophagy and exacerbation of disease.
Links between UPS and autophagy
The UPS and autophagy were long viewed as independent, parallel degradation systems with no point of intersection. This view was initially challenged by the observation that monoubiquitination operates as a key signal in endocytosis, a process important for numerous cell functions including lysosomal biogenesis [62]. Subsequently, several lines of evidence have developed suggesting that the UPS and autophagy are functionally interrelated catabolic processes [50, 63, 64]. Specifically, these degradation systems share certain substrates and regulatory molecules, and show coordinated and, in some contexts, compensatory function. Thus, in contrast to the traditional notion of the UPS and autophagy providing discrete routes of degradation for short-lived and long-lived proteins, respectively, it is increasingly clear that a substantial subset of proteins may be degraded by either pathway. Short-lived proteins normally degraded by the UPS can be selectively degraded by autophagy under certain conditions [65, 66], while longer-lived proteins can also be degraded by the UPS [67]. The neuronal protein α-synuclein, for example, can be degraded by the UPS, macroautophagy and chaperone-mediated autophagy [26, 29]. Under conditions in which one degradation system is compromised, enhanced degradation by an alternate pathway may become critical to maintaining pools of amino acids for protein synthesis and may protect against the accumulation of a toxic species. As mentioned above, dramatic illustration of the interrelatedness of the UPS and autophagy was provided by characterizations of mice with conditional knockout of the essential autophagy genes Atg5 or Atg7 in the central nervous system, which resulted in neurodegeneration with accumulation of ubiquitin-positive pathology [36, 37]. Given that these mice showed no observable defect in UPS function, these results suggest that some ubiquitin-tagged proteins may in fact normally be degraded by autophagy. This model is consistent with an older study showing that inactivation of the ubiquitin-activating enzyme E1 leads to a defect in autolysosomal degradation and to an absence of ubiquitin-positive proteins within lysosomes [68].
Further illustration of the relationship between the UPS and autophagy originated in a series of in vitro studies that examined the behavior of cells following challenge to the UPS. When cultured cells are challenged with excess misfolded protein that overwhelms the UPS, or treated with proteasome inhibitors, ubiquitinated misfolded proteins are actively transported to a cytoplasmic, juxtanuclear structure that has been termed an “aggresome” [69]. It has been inferred that aggresome formation in vitro is a cytoprotective response in cultured cells since their formation correlates inversely with cell death, whereas interventions that block aggresome formation enhance cytotoxicity and slow the rate of turnover of misfolded proteins [27, 64, 70–72]. While aggresomes superficially resemble the cytoplasmic inclusions present in some neurodegenerative diseases, evidence of aggresome formation in vivo is lacking, and most pathological inclusions found in neurodegenerative disease are clearly not aggresomes. Nevertheless, studying the phenomenon of aggresome formation in vitro has provided insight into the cellular management of misfolded proteins; for example, helping to identify molecular machinery that protects cells from misfolded protein stress. It has now been established that clearance of misfolded proteins from aggresomes is mediated at least in part by autophagy, implicating this pathway as a compensatory mechanism for degrading misfolded proteins when the proteasome is impaired [27, 64, 71, 73].
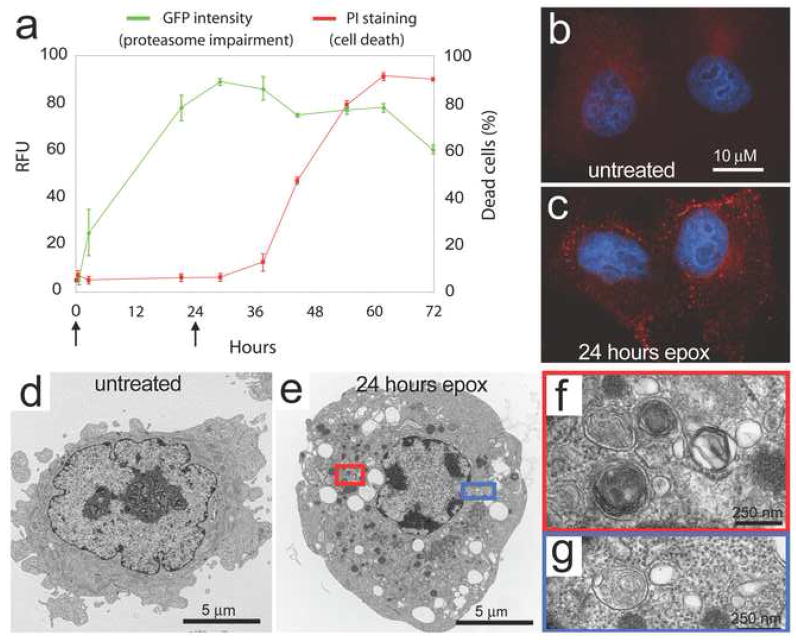
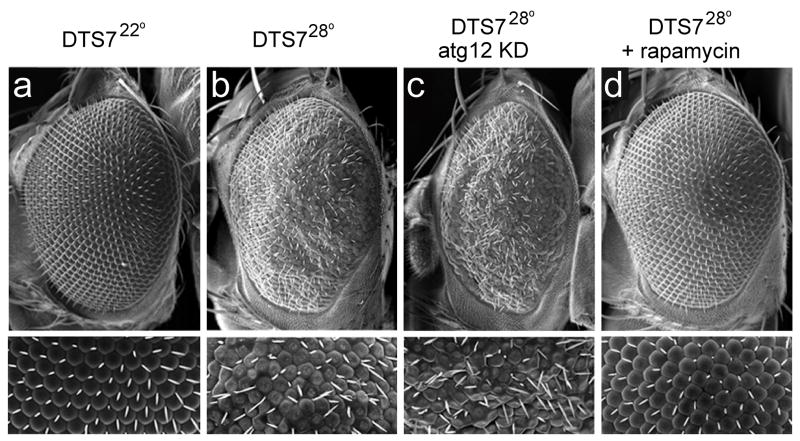
In addition to aggresome formation, impairment of the UPS in vitro has been found to induce autophagy [63, 64]. This is observed, for example, in HeLa cells after prolonged proteasomal inhibition as evidenced by redistribution of LC3 into numerous puncta (Figure 3a–c) and the accumulation of autophagic vacuoles based upon ultrastructural evaluation (Figure 3d–g). Similar induction of autophagy is observed in response to genetic impairment of the proteasome in Drosophila [50]. The role of autophagy induction in the setting of UPS impairment appears to be cytoprotective since degenerative phenotypes associated with proteasome impairment are enhanced in an autophagy-deficient background (Figure 4a–c), whereas this degeneration is suppressed when autophagy is induced with the TOR-inhibitor rapamycin (Figure 3d). Similar results have recently been observed in vitro using the proteasome inhibitor lactacystin, as pre-treatment with rapamycin attenuates lactacystin-induced apoptosis and reduces lactacystin-induced ubiquitinated protein aggregation [74].
Figure 3.
Proteasome impairment leads to upregulation of autophagic activity. (a) HeLa cells that stably express the UPS reporter UbG76V-GFP were treated with the irreversible proteasome inhibitor epoximicin for 72 hours and monitored for cell death. Increasing levels of the GFP substrate indicate impaired UPS function. Note the 24 hours time point used in (b)–(g) is within the window during which proteasome function is impaired, but the cells remain viable. (b–c) Images of LC3 staining (red) and DAPI (blue) show accumulation of LC3 puncta in epoximicin-treated cells. (d–e) Transmission electron microscopy images of cells reveal autophagic structures and prominent vacuolization in epoximicin-treated cells. (f–g) Increased magnification of structures in (e) reveal multi-membraned structures consistent with autophagic activity.
Figure 4.
A Drosophila model of proteasome impairment is modified by manipulation of autophagic activity. (a–b) The temperature-sensitive DTS7 mutant shows a normal eye phenotype at the permissive temperature of 22°C and a significant degenerative phenotype at the restrictive temperature of 28°C. (c) RNAi knockdown of the autophagy gene atg12 results in an enhancement of the DTS7 degenerative phenotype, suggesting that the autophagic activity that is induced in response to proteasome impairment is compensatory. (d) Treatment of DTS7 flies with rapamycin suppresses the degenerative phenotype, demonstrating that induction of autophagy can compensate for impaired proteasome function. Adapted from [50].
Although the mechanism whereby autophagy and UPS function are coordinated is little understood, several regulators have emerged as important players in mediating this crosstalk, including histone deacetylase 6 (HDAC6) [50, 64, 75], p62/sequestosome 1 (p62) [76], and the FYVE-domain containing protein Alfy [77]; notably, these proteins have all been found to regulate or be essential for aggresome formation. HDAC6 is a cytoplasmic microtubule-associated deacetylase whose targets include α-tubulin, Hsp90, and cortactin. HDAC6 interacts with polyubiquitinated proteins through a highly conserved Zn-finger ubiquitin-binding domain, and also interacts with dynein motors, suggesting that the molecule may provide a physical link between ubiquitinated cargo and transport machinery [70]. HDAC6 activity appears to be important for trafficking ubiquitinated proteins and lysosomes in vitro and this has led to the suggestion that HDAC6 coordinates delivery of substrates to autophagic machinery [64, 70, 78]. In Drosophila, HDAC6 overexpression was found to suppress degeneration associated with impaired UPS activity and also suppressed degeneration caused by toxic polyglutamine expression. In both cases, this rescue by HDAC6 was found to be autophagy dependent, consistent with a role for HDAC6 in linking the UPS and compensatory autophagy [50]. HDAC6 activity was also reported to regulate chaperone expression in response to heat shock by deacetylating Hsp90 leading to release and activation of the transcription factor HSF-1 [79].
p62 is another cytosolic protein whose structure suggests a function as an adaptor molecule linking ubiquitinated proteins to autophagic machinery. The C-terminal portion of p62 harbors both a ubiquitin-associated (UBA) domain which interacts non-covalently with ubiquitinated proteins [80, 81] as well as an LC3-interacting region (LIR) [82]. Cellular stresses such as polyQ expression, proteasome impairment, oxidative stress, and increased misfolded protein burden activate transcription and translation of p62, suggesting that it functions broadly in stress situations [83, 84]. p62 localizes to a variety of ubiquitin-positive neuropathological inclusions including Lewy bodies in Parkinson’s disease, neurofibrillary tangles in tauopathies, polyglutamine-expanded huntingtin aggregates in Huntington’s disease, and aggregates of mutant SOD1 in familial amyotrophic lateral sclerosis [85–87]. A role for p62 in protecting against misfolded protein stress is supported by the observation that RNAi-mediated knockdown of p62 exacerbates polyglutamine toxicity in vitro and diminishes the formation of ubiquitin-positive inclusions in response to misfolded protein stress [76] while reducing the ability of LC3 to co-precipitate ubiquitinated proteins [82]. Very recently, p62 was found to contain an LC3 recognition sequence that, when mutated, resulted in ubiquitin- and p62- positive inclusion formation [88]. Thus, it has been suggested that p62 provides a key link between autophagy and the UPS by facilitating autophagic degradation of ubiquitinated proteins. As predicted by this model, p62 null mice fail to form ubiquitin-positive protein aggregates in response to misfolded protein stress [89] and show age-related neurodegeneration [90]. Consistent results were obtained in studies of Drosophila deficient in Ref(2)p, the Drosophila homologue of p62 [91]. Recent models propose that p62 and HDAC6 function analogously to facilitate autophagic degradation of proteins that display specific polyubiquitin topology. Specifically, it is suggested that K63-linked polyubiquitin chains recruit p62 and HDAC6 providing a signal for autophagic degradation [92, 93].
Alfy (autophagy-linked FYVE protein) is a third possible molecular link between autophagy and the UPS. Alfy is a member of the FYVE-domain family of proteins. In cells that are exposed to stressors such as starvation or UPS inhibition, Alfy relocalizes from the nuclear envelope to filamentous cytoplasmic structures that are near autophagic membranes and ubiquitinated protein inclusions, as well as within autophagosomes [77]. Mutations in blue cheese, the Drosophila homology of human Alfy, lead to reduced longevity and the accumulation of ubiquitinated neural aggregates, suggesting that its role in autophagic degradation may be involved in the clearance of ubiquitin aggregates [77, 94].
Summary and unresolved questions
The last few years have led to substantial insight into the relationship between autophagy and the UPS. It has become apparent that there is significant similarity, and in some cases overlap, in the regulation of these catabolic pathways by UBL modification, leading to the suggestion that they evolved from a common biological origin[95]. Further, it has become evident that the function of autophagy and the UPS are coordinated. For example, impairment of the UPS results in upregulation of autophagy ([50, 64] and Figure 3), and in some contexts this upregulation of autophagy can compensate for impaired UPS function [50, 74]. However, it is not known whether this compensatory relationship is reciprocal, as few reagents exist to upregulate the UPS. One study found that upregulation of UPS may afford neuroprotection from toxicity caused by disease proteins, though the authors did not examine the effects of UPS upregulation in autophagy-deficient cells [58]. The mechanism and the molecular players that regulate the relationship between autophagy and the UPS are beginning to be elucidated and, perhaps not surprisingly, recognition of UBL modification is emerging as a consistent theme. CMA is also clearly involved in the coordinated functioning of proteolytic pathways. CMA can selectively degrade some subunits of the proteasome, highlighting a relationship between CMA and the UPS [96]. In addition, acute blockage of CMA results in short-term impairment of both the UPS and macroautophagy, followed by a recovery of these catabolic systems as CMA blockade persists [97]. Chronic blockage of CMA results in constitutive activation of macroautophagy, which appears to be compensatory [59]. These interrelationships suggest a model in which the preferred route of degradation for a particular substrate may be linked to which system is most capable of efficiently degrading it.
While much has been revealed in recent years, substantial questions remain. Most notable, it is largely unknown how the decision is made between degradative routes for any particular protein substrate when more than one pathway is available. HDAC6, p62, and Alfy have been implicated in directing ubiquitinated proteins for autophagic degradation, but the mechanisms whereby these proteins identify their targets and influence their degradation are still unknown. Tantalizing recent evidence suggests that different classes of substrates may be identified by polyubiquitin chains of differing topology, providing the signal for degradation by one proteolytic system or the other [93, 98]. More specifically, it has been suggested that polyubiquitin chains with K48-linked chains are primarily degraded by the UPS, whereas those with K63-linked chains are directed to autophagy. Indeed, one might envision a “ubiquitin code” that translates into interaction with specific ubiquitin binding proteins, including HDAC6, Alfy, p62 or other members of the UBA family [99], which may in turn determine the fate of the substrate. However, experimental limitations in distinguishing between K48-, K63- and mixed-linkage ubiquitin chains must be overcome in order to answer this question effectively, and many of these relationships remain largely unexplored. This concept of different classes of substrates that are destined for degradation by differing pathways is consistent with one recent report in which disease-associated proteins were found to partition into two distinct intracellular compartments, with soluble ubiquitinated proteins accumulating in a proteasome-rich juxtanuclear region, and insoluble aggregated proteins accumulating in perivacuolar inclusions that colocalize with Atg8 [100]. It would be interesting to determine whether these proteins could be distinguished by differing UBL modifications.
The mechanism by which upregulation of autophagy mitigates neurotoxicity associated with UPS impairment is also unresolved. It is unlikely that autophagy is able to compensate for the role of the UPS in fine-tuning the steady-state levels of short-lived regulatory proteins. More likely, augmentation of autophagy is neuroprotective by 1) maintaining the overall rate of catabolism, “freeing” amino acids that would otherwise lie useless in aggregated, nonfunctioning proteins, 2) eliminating specific protein substrates that would otherwise accumulate, aggregate and acquire toxic properties, or 3) a combination of these.
Further illumination of the relationship between the UPS, autophagy and the relationship to human disease is vitally important and could lead to harnessing intrinsic catabolic pathways for therapeutic benefit.
Acknowledgments
We thank Brett McCray and Mondira Kundu for helpful comments and critical review of the manuscript. This work was supported by a grant from the Muscular Dystrophy Association and NIH grant NS053825 to JPT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wheatley DN, Inglis MS. An intracellular perfusion system linking pools and protein synthesis. J Theor Biol. 1980;83:437–445. doi: 10.1016/0022-5193(80)90050-8. [DOI] [PubMed] [Google Scholar]
- 2.Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 3.Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 4.Yewdell JW. Serendipity strikes twice: the discovery and rediscovery of defective ribosomal products (DRiPS) Cell Mol Biol (Noisy-le-grand) 2005;51:635–641. [PubMed] [Google Scholar]
- 5.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 6.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 7.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 8.Abeliovich H, Klionsky DJ. Autophagy in yeast: mechanistic insights and physiological function. Microbiol Mol Biol Rev. 2001;65:463–479. doi: 10.1128/MMBR.65.3.463-479.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 10.Ahlberg J, Marzella L, Glaumann H. Uptake and degradation of proteins by isolated rat liver lysosomes. Suggestion of a microautophagic pathway of proteolysis. Lab Invest. 1982;47:523–532. [PubMed] [Google Scholar]
- 11.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 12.Arstila AU, Trump BF. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol. 1968;53:687–733. [PMC free article] [PubMed] [Google Scholar]
- 13.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 14.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 15.Reggiori F, Klionsky DJ. Autophagosomes: biogenesis from scratch? Curr Opin Cell Biol. 2005;17:415–422. doi: 10.1016/j.ceb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 17.Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 18.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 20.McCray BA, Taylor JP. The role of autophagy in age-related neurodegeneration. Neurosignals. 2008;16:75–84. doi: 10.1159/000109761. [DOI] [PubMed] [Google Scholar]
- 21.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 22.Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, Mouatt-Prigent A, Ruberg M, Hirsch EC, Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 23.Sikorska B, Liberski PP, Giraud P, Kopp N, Brown P. Autophagy is a part of ultrastructural synaptic pathology in Creutzfeldt-Jakob disease: a brain biopsy study. Int J Biochem Cell Biol. 2004;36:2563–2573. doi: 10.1016/j.biocel.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Sapp E, Schwarz C, Chase K, Bhide PG, Young AB, Penney J, Vonsattel JP, Aronin N, DiFiglia M. Huntingtin localization in brains of normal and Huntington’s disease patients. Ann Neurol. 1997;42:604–612. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- 25.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 26.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 27.Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, Markovic-Plese S, Fischbeck KH. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet. 2003;12:749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 28.Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Aparicio E, Yamamoto A, Hernandez F, Hen R, Avila J, Lucas JJ. Proteasomal-dependent aggregate reversal and absence of cell death in a conditional mouse model of Huntington’s disease. J Neurosci. 2001;21:8772–8781. doi: 10.1523/JNEUROSCI.21-22-08772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett MC, Bishop JF, Leng Y, Chock PB, Chase TN, Mouradian MM. Degradation of alpha-synuclein by proteasome. J Biol Chem. 1999;274:33855–33858. doi: 10.1074/jbc.274.48.33855. [DOI] [PubMed] [Google Scholar]
- 32.Cummings CJ, Reinstein E, Sun Y, Antalffy B, Jiang Y, Ciechanover A, Orr HT, Beaudet AL, Zoghbi HY. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 33.Koike M, Nakanishi H, Saftig P, Ezaki J, Isahara K, Ohsawa Y, Schulz-Schaeffer W, Watanabe T, Waguri S, Kametaka S, Shibata M, Yamamoto K, Kominami E, Peters C, von Figura K, Uchiyama Y. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci. 2000;20:6898–6906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shacka JJ, Klocke BJ, Young C, Shibata M, Olney JW, Uchiyama Y, Saftig P, Roth KA. Cathepsin D deficiency induces persistent neurodegeneration in the absence of Bax-dependent apoptosis. J Neurosci. 2007;27:2081–2090. doi: 10.1523/JNEUROSCI.5577-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myllykangas L, Tyynela J, Page-McCaw A, Rubin GM, Haltia MJ, Feany MB. Cathepsin D-deficient Drosophila recapitulate the key features of neuronal ceroid lipofuscinoses. Neurobiol Dis. 2005;19:194–199. doi: 10.1016/j.nbd.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 37.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 38.Nixon RA, Yang DS, Lee JH. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 2008;4:590–599. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- 39.Ning YP, Kanai K, Tomiyama H, Li Y, Funayama M, Yoshino H, Sato S, Asahina M, Kuwabara S, Takeda A, Hattori T, Mizuno Y, Hattori N. PARK9-linked parkinsonism in eastern Asia: mutation detection in ATP13A2 and clinical phenotype. Neurology. 2008;70:1491–1493. doi: 10.1212/01.wnl.0000310427.72236.68. [DOI] [PubMed] [Google Scholar]
- 40.Verhoeven K, De Jonghe P, Coen K, Verpoorten N, Auer-Grumbach M, Kwon JM, FitzPatrick D, Schmedding E, De Vriendt E, Jacobs A, Van Gerwen V, Wagner K, Hartung HP, Timmerman V. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet. 2003;72:722–727. doi: 10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puls I, Oh SJ, Sumner CJ, Wallace KE, Floeter MK, Mann EA, Kennedy WR, Wendelschafer-Crabb G, Vortmeyer A, Powers R, Finnegan K, Holzbaur EL, Fischbeck KH, Ludlow CL. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann Neurol. 2005;57:687–694. doi: 10.1002/ana.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, Cotman SL. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem. 2006;281:20483–20493. doi: 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- 43.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 44.Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 45.Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 46.Kimura S, Noda T, Yoshimori T. Dynein-dependent Movement of Autophagosomes Mediates Efficient Encounters with Lysosomes. Cell Struct Funct. 2008;33:109–122. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- 47.Fader CM, Sanchez D, Furlan M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 48.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 49.Laird FM, Farah MH, Ackerley S, Hoke A, Maragakis N, Rothstein JD, Griffin J, Price DL, Martin LJ, Wong PC. Motor neuron disease occurring in a mutant dynactin mouse model is characterized by defects in vesicular trafficking. J Neurosci. 2008;28:1997–2005. doi: 10.1523/JNEUROSCI.4231-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 51.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, Rubinsztein DC. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 53.Sarkar S, Krishna G, Imarisio S, Saiki S, O’Kane CJ, Rubinsztein DC. A rational mechanism for combination treatment of Huntington’s disease using lithium and rapamycin. Hum Mol Genet. 2008;17:170–178. doi: 10.1093/hmg/ddm294. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, Kurosawa M, Nekooki M, Nukina N. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med. 2004;10:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 55.Davies JE, Sarkar S, Rubinsztein DC. Trehalose reduces aggregate formation and delays pathology in a transgenic mouse model of oculopharyngeal muscular dystrophy. Hum Mol Genet. 2006;15:23–31. doi: 10.1093/hmg/ddi422. [DOI] [PubMed] [Google Scholar]
- 56.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 57.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2008 doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 58.Seo H, Sonntag KC, Kim W, Cattaneo E, Isacson O. Proteasome activator enhances survival of huntington’s disease neuronal model cells. PLoS ONE. 2007;2:e238. doi: 10.1371/journal.pone.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 61.Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee JH, Mohan PS, Mercken M, Farmery MR, Tjernberg LO, Jiang Y, Duff K, Uchiyama Y, Naslund J, Mathews PM, Cataldo AM, Nixon RA. Macroautophagy--a novel Beta-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ross CA, Pickart CM. The ubiquitin-proteasome pathway in Parkinson’s disease and other neurodegenerative diseases. Trends Cell Biol. 2004;14:703–711. doi: 10.1016/j.tcb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 63.Rideout HJ, Lang-Rollin I, Stefanis L. Involvement of macroautophagy in the dissolution of neuronal inclusions. Int J Biochem Cell Biol. 2004;36:2551–2562. doi: 10.1016/j.biocel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 64.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 65.Fuertes G, Villarroya A, Knecht E. Role of proteasomes in the degradation of short-lived proteins in human fibroblasts under various growth conditions. Int J Biochem Cell Biol. 2003;35:651–664. doi: 10.1016/s1357-2725(02)00382-5. [DOI] [PubMed] [Google Scholar]
- 66.Li D. Selective degradation of the IkappaB kinase (IKK) by autophagy. Cell Res. 2006;16:855–856. doi: 10.1038/sj.cr.7310110. [DOI] [PubMed] [Google Scholar]
- 67.Fuertes G, Martin De Llano JJ, Villarroya A, Rivett AJ, Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lenk SE, Dunn WA, Jr, Trausch JS, Ciechanover A, Schwartz AL. Ubiquitin-activating enzyme, E1, is associated with maturation of autophagic vacuoles. J Cell Biol. 1992;118:301–308. doi: 10.1083/jcb.118.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 73.Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, Kopito RR. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci U S A. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pan T, Kondo S, Zhu W, Xie W, Jankovic J, Le W. Neuroprotection of rapamycin in lactacystin-induced neurodegeneration via autophagy enhancement. Neurobiol Dis. 2008;32:16–25. doi: 10.1016/j.nbd.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 75.Pandey UB, Batlevi Y, Baehrecke EH, Taylor JP. HDAC6 at the intersection of autophagy, the ubiquitin-proteasome system and neurodegeneration. Autophagy. 2007;3:643–645. doi: 10.4161/auto.5050. [DOI] [PubMed] [Google Scholar]
- 76.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 78.Kopito RR. The missing linker: an unexpected role for a histone deacetylase. Mol Cell. 2003;12:1349–1351. doi: 10.1016/s1097-2765(03)00498-2. [DOI] [PubMed] [Google Scholar]
- 79.Boyault C, Zhang Y, Fritah S, Caron C, Gilquin B, Kwon SH, Garrido C, Yao TP, Vourc’h C, Matthias P, Khochbin S. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geetha T, Wooten MW. Structure and functional properties of the ubiquitin binding protein p62. FEBS Lett. 2002;512:19–24. doi: 10.1016/s0014-5793(02)02286-x. [DOI] [PubMed] [Google Scholar]
- 81.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 83.Kuusisto E, Suuronen T, Salminen A. Ubiquitin-binding protein p62 expression is induced during apoptosis and proteasomal inhibition in neuronal cells. Biochem Biophys Res Commun. 2001;280:223–228. doi: 10.1006/bbrc.2000.4107. [DOI] [PubMed] [Google Scholar]
- 84.Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, Oyama F, Nukina N. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem. 2004;91:57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- 85.Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 86.Kuusisto E, Salminen A, Alafuzoff I. Early accumulation of p62 in neurofibrillary tangles in Alzheimer’s disease: possible role in tangle formation. Neuropathol Appl Neurobiol. 2002;28:228–237. doi: 10.1046/j.1365-2990.2002.00394.x. [DOI] [PubMed] [Google Scholar]
- 87.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 89.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy–deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 90.Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom AL, Kemppainen R, Cox N, Zhu H, Wooten MC, Diaz-Meco MT, Moscat J, Wooten MW. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 91.Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, Brech A. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tan JM, Wong ES, Dawson VL, Dawson TM, Lim KL. Lysine 63-linked polyubiquitin potentially partners with p62 to promote the clearance of protein inclusions by autophagy. Autophagy. 2007;4:251–253. [Google Scholar]
- 93.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, Rodriguez MH, Hwang CE, Benedetti M, McKeown M. blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J Neurosci. 2003;23:1254–1264. doi: 10.1523/JNEUROSCI.23-04-01254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hughes T, Rusten TE. Origin and evolution of self-consumption: autophagy. Adv Exp Med Biol. 2007;607:111–118. doi: 10.1007/978-0-387-74021-8_9. [DOI] [PubMed] [Google Scholar]
- 96.Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- 97.Massey AC, Follenzi A, Kiffin R, Zhang C, Cuervo AM. Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy. 2008;4:442–456. doi: 10.4161/auto.5654. [DOI] [PubMed] [Google Scholar]
- 98.Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, Dawson VL, Dawson TM. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 100.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]