Abstract
Neuregulin 1 (Nrg1) and ErbB receptor tyrosine kinase signalling is essential for the formation and proper functioning of multiple organ systems and inappropriate Nrg1/ErbB signalling severely compromises health, contributing to such diverse pathologies as cancer and neuropsychiatric disorders. Numerous genetic modelling studies in mice demonstrate that Nrg1 signalling is important in the development of normal neuronal connectivity. Recent studies have identified novel signalling mechanisms and revealed unexpected roles of Nrg1 isoforms in both the developing and adult nervous system. Of particular interest to this discussion are findings linking deficits in Nrg1—ErbB4 signalling to perturbations of synaptic transmission, myelination, and the survival of particular sets of neurons and glia.
Genetic association of neuregulin 1 and schizophrenia
The Nrg1 gene is a schizophrenia susceptibility gene. This was initially reported by Decode in 2002 in a study in which a seven-marker haplotype (five single nucleotide polymorphisms [SNPs] and two microsatellites) at the extreme 5′ end was identified as significantly associated with disease diagnosis in an Icelandic population (Stefansson et al 2002). The association of these, and/or additional sequence polymorphisms in the Nrg1 gene with disease has been reproduced in numerous studies in multiple populations. A recent count of the literature indicates that there have been at least 30 studies of which about two or three show association between Nrg1 and schizophrenia and the rest do not. Both positive and negative findings have been reported in case-control and family based studies and in numerous geographically distinct populations (for recent update, see: http://www.schizophreniaforum.org/res/sczgene/geneoverview.asp?geneid=311). Several recent studies have extended the Nrg1 connection beyond schizophrenia raising the possibility that dysregulation of Nrg1–ErbB signalling contributes to other disorders such as bipolar (Thomson et al 2007) and Alzheimer’s disease with psychosis (Go et al 2005). In addition there are data emerging that support additional links between ErbB4 and schizophrenia susceptibility (Silberberg et al 2006, Law et al 2007).
Over 80 SNPs have been described in the association studies between the Nrg1 gene and risk of schizophrenia and disease associated SNPs are grouped into several ‘at risk’ haplotypes that span much of the 1–1.5 million base pairs encoding Nrg1 proteins. Of these 80, only four result in changes in the predicted coding sequence of the protein. The significance of these changes is not known but the most recent, a valine to leucine substitution in the transmembrane domain will be discussed below (Walss-Bass et al 2006). The absence of clear mutations in protein coding sequences that are associated with disease risk, are consistent with the conclusion that changes in the level of expression of one or more isoforms of Nrg1 contribute to the aetiology of this disorder (Law et al 2006).
Neuregulin 1 functions: why are there so many isoforms, and does this diversity provide insight into pathology?
We face two major challenges to bridging the gap between results of human genetic studies and understanding how Nrg1–ErbB signalling contributes to psychiatric disease. First, we need to identify the relevant DNA sequence polymorphisms and gain insight into how these polymorphisms affect expression of the Nrg1 gene. Second, we need to acquire a deep enough understanding of how Nrg1 functions in the CNS to allow us to make the leap from DNA sequences to changes in function that might explain the cell and neuronal circuit based changes that contribute to disease.
In the following sections I will (1) review the functional importance of different exons/isoforms and (2) present data supporting the use of genetically modified mice to better understand the role of Nrg1 in establishing and maintaining important (with respect to this disease) circuits in the mouse brain.
Neuregulin 1 is one of about 20 vertebrate genes encoding ligands for the four ErbB receptors (Stein & Staros 2006). In general, ErbB1 (=EGFR) and ErbB4 are the key targets—seven genes encode ligands for ErbB1 and nine encode ligands for ErbB4 (with three, HB-EGF, epiregulin and betacellulin, shared). ErbB3 is more selective; to date only Nrg1 and Nrg2 and neuroglycan have been shown to bind directly to ErbB3. ErbB2 (also called Her2/neu) is a co-receptor that does not bind ligand on its own, but forms heterodimers with the other ErbBs altering their signalling properties.
Nrg1 is a large and complex gene that encodes 15–20 different proteins. These isoforms are generated following transcription from six promoters coupled with a significant degree of alternative splicing of primary transcripts (Fig. 1). Aspects of both promoter usage and splicing appear to be developmentally regulated and occur in tissue and cell type specific patterns. Until recently little attention has been paid to the factors that regulate the choice of these promoters and/or splice sites. This lack of data on the function of non-coding sequences in neurons and glia limit the ability do generate informed hypotheses linking disease associated polymorphisms with potential functional consequences. A very recent report from A. Law and colleagues provides the first experimental demonstration that a disease associated polymorphism alters promoter strength, a result that should pave the way for a more systematic assessment of the biological significance of non-coding SNPs in the Nrg1 gene (Tan et al 2007).
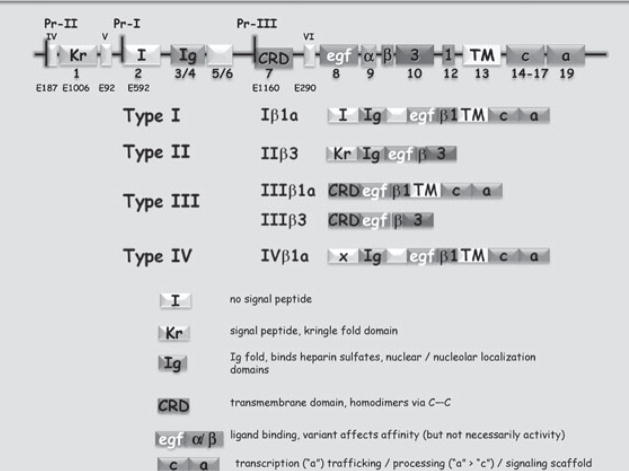
FIG. 1.
The Neuregulin 1 gene encodes an abundance of related but functionally distinct proteins. The Neuregulin 1 gene is schematized at the top. Boxes represent coding exons drawn approximately to scale. Introns are not shown to scale. In primates, six families of Nrg1 isoforms have been described and are thought to all be transcribed from distinct core promoters (I–VI) (Steinthorsdottir et al 2004). Only types I–III appear to be present in non-primates. Each of the six families of isoforms has a unique N-terminal sequence that is encoded by 6 different 5′ exons (I = E592, II= E1006, III = E1160, IV = E187, V = E92 and VI = E290). No clear signal sequence is present in E592, E187, E92 or E290. E1006 encodes a classic signal peptide and E1160 enocodes a transmembrane domain that anchors the Type III isoforms in the membrane. Other domains of interest include the Ig domain (encoded by exons 3 and 4), the EGF domain encoded by exon 8, the C-terminal EGF-like domain encoded by combinations of exons 9–12, the common transmembrane domain encoded by exon 13 and the intracellular domain, encoded by exons 14–19. The functions associated with these domains are described in the text. What is clear from this diagram is that combinations of differential promoter usage with differential splicing gives rise to families of proteins with distinct signalling strategies.
To date Nrg1 isoforms have been divided into three major families (types I, II and III) present in all vertebrates examined and three minor families (types IV, V and VI) which appear to be restricted to primates. Although post-mortem studies have focused attention on type I Nrg1 and type IV Nrg1 (Law et al 2006), the fact that SNPs and haplotypes spanning nearly the entire 1.5 Mbp have been linked to disease risk, reinforces the need to understand the functional contribution of all of the isoforms to the development and maintenance of the CNS.
In contrast to what we know about non-coding regions, we know a lot more about the functions of discrete regions of the encoded proteins, both those that are common to all known functional isoforms and those that are isoforms or family specific. In sum: the unique N-termini associated with types I, II and III all have functional implications. The N-terminus of the type I isoforms (and presumably types IV–VI) is notable for what it lacks: a signal sequence or alternative mechanism for entering the secretory pathway. Type I Nrg1 isoforms depend on the central transmembrane domain and portions of the intracellular domain to enter the secretory pathway, reach the cell surface and then undergo proteolytic release of the growth factor domain (Liu et al 1998, Montero et al 2000, Wang et al 2001). Without a transmembrane domain, type 1 Nrg1 does not reach the cell surface, and experimentally, are expressed as soluble, cytosolic proteins that are targeted to the nucleolus (Golding et al 2004).
The type II isoforms contain N-terminal domains blessed with both a classic signal peptide and a Kringle-like domain. These isoforms readily enter the secretory pathway and are released from cells. Kringle domains in other proteins mediate protein–protein interactions, but no partners for type II Nrg1 have been reported.
Types I, II, IV and V contain an immunoglobulin-like domain near their N-termini. In peripheral nerves and tissues outside of the nervous system, this Ig-domain is thought to contribute to specific interactions with extracellular matrix components and modify (a) the distance over which these growth factors act and (b) the local concentration of growth factor (Li & Loeb 2001).
Type III Nrg1 is unique in a number of ways. First, type III expression is mostly limited to neurons; no other isoforms displays this degree of restricted expression (Yang et al 1998, Wolpowitz et al 2000). Second, the cysteine-rich domain encoded by the type III-specific N-terminal exon forms a transmembrane domain (Wang et al 2001). As a result, type III Nrg1 proteins have cytosolic N-termini, and have membrane-tethered EGF-like domains. This topology (a) restricts type III Nrg1 signalling to cell–cell interactions (i.e. juxtacrine signalling) and (b) allows type III Nrg1 to act as a receptor. Indeed we have demonstrated that type III Nrg1 is a bidirectional signalling protein that functions as both a ligand that activates ErbB receptors and as a receptor that modulates the behaviour of type III Nrg1-expressing neurons (Bao et al 2003).
All known functional variants of Nrg1 contain an EGF-like domain essential for interaction with ErbB receptors. The EGF-like domain consists of three regions; the N-terminal part of the EGF domain is constant for all Nrg1 isoforms. This is followed by either an α type or a β type exon and a variable ‘stalk’ region (the stalk is the extracellular juxtamembrane segment separating the EGF-like domain from the transmembrane domain). The variability in the EGF-like domain has functional implications. Variations in this region affect the affinity of the Nrg1 ligand for receptors with the β isoforms having significantly greater affinity than α isoforms. The relatively subtle variations in the stalk region affect the efficiency by which the transmembrane precursors are processed by metalloproteases (which for non-type III Nrg1 isoforms is necessary for release of active growth factor), with longer ‘stalks’ being better substrates than shorter ones (Montero et al 2000). These relatively subtle differences have not been explored in depth in vitro or in vivo.
The C-terminal intracellular domain (ICD) has important functions. As noted above, in the case of the Type I Nrg1 isoforms, the intracellular domain is required for intra-cellular trafficking of the protein (Liu et al 1998). In addition, the longest variant of the intracellular domain (the ‘a’ form) functions as an inducible transcription factor (Bao et al 2003, 2004). Both ErbB binding and depolarization stimulate a γ-secretase catalysed intramembranous cleavage that releases the C-terminal ICD. Subsequent to cleavage, the ICD translocates to the nucleus where it contributes to the transcriptional regulation of genes involved in neuronal survival and in synaptic maturation and maintenance. In more recent studies we have uncovered additional signalling mechanisms that are engaged by Nrg1-ICD, and find that these signalling events regulate growth cone dynamics and trafficking of neurotransmitter receptors. These latter responses the Nrg1-ICD in which Nrg1 behaves like a receptor, require that the full length form of Nrg1 is presented at the cell surface. In our hands, only Type III Nrg1 isoforms elicit responses consistent with this receptor like function, but in the systems we have studied, type I/II isoforms are rapidly released from the cell surface and therefore are unlikely to contribute to bidirectional signalling. In addition, in recent studies we have found a number of amino acid residues within the transmembrane domain that are necessary for γ-secretase cleavage. The recent finding of a Val/Leu substitution within the transmembrane domain that is associated with risk of schizophrenia in some populations (Walss-Bass et al 2006) raises the possibility that alterations in bidirectional signalling could contribute to disease pathology.
Although the complexity of the Nrg1 gene has been known for 15 years, there is still relatively little information available about how the functional diversity of the Nrg1 isoforms influences the biology of the CNS. What information we do have, comes from two general types of experimental approaches; addition of recombinant neuregulin to neurons followed by measurements of various responses and studies on the effect of genetic disruption of one or more Nrg1 isoform in mice. The results of these studies provide compelling evidence for the biological plausibility of Nrg1 as a schizophrenia susceptibility gene. In particular these studies demonstrate that Nrg1 is critical for neuronal development (including neurogenesis, migration, axonal targeting, and neurotransmitter expression and targeting), the survival of neurons and the maintenance of circuit plasticity.
Using mouse genetic models to probe Nrg1 function in brain circuitry
In order to study aspects of the biological changes that underlie psychiatric diseases, one would ideally like to introduce disease associated mutations into model systems. Unfortunately most of the DNA sequence changes that link Nrg1 with schizophrenia occur in regions of the gene that do not encode protein and are believed to have subtle effects levels and/or patterns of expression. As a starting point, we and others have examined the effects of reducing Nrg1 expression by half on the behaviour of adult mice (homozygous mutations in Nrg1 result in embryonic or perinatal lethality) (Meyer & Birchmeier 1995, Wolpowitz et al 2000). Mutations that have been examined include those that eliminate all Nrg1 expression as well as ones that eliminate Ig-Nrg1 expression (types I and II) (Gerlai et al 2000, Bjarnadottir et al 2007), type III Nrg1 expression or all Nrg1 isoforms that contain the common transmembrane domain (all type I and subsets of types II and III) (Stefansson et al 2002, O’Tuathaigh et al 2006, Bjarnadottir et al 2007, Boucher et al 2007, Karl et al 2007). Not surprisingly, the results are complicated. Some behaviours are altered in all mutants examined whereas others are associated with the disruption of specific Nrg1 isoforms. In addition, behaviours affected include those that are thought to correspond to endophenotypes associated with schizophrenia as well as behaviours that do not model schizophrenia phenotypes. For example, Nrg1 mutant mice show elevated levels of activity in a novel open environment (with the exception of the type III Nrg1 mutants), have deficits in pre-pulse inhibition of an acoustic startle reflex (a model of sensorimotor gating), alterations in anxiety related behaviours, and altered responses to drugs include THC and nicotine (Boucher et al 2007). Similar results have been noted in mice bearing mutations in ErbB receptors, although the effect of mutation tends to be less pronounced in ErbB mutant mice than in Nrg1 mutant mice (Gerlai et al 2000, Stefansson et al 2002). We are tempted to speculate that the picture will clarify once more precise mutations are generated, both in terms of specific brain regions affected as well as studying more ‘schizophrenia-like’ alterations in Nrg1.
Neuregulin 1 signalling in synaptic development and maintenance: building the case for biological plausibility
The pathology of schizophrenia has been associated with underlying deficits in a number of cellular and molecular systems in the brain, including those involving oligodendrocytes, prefrontal and striatal dopamine signalling, cortical GABAergic interneurons and diffuse NMDA mediated glutamatergic transmission (Lewis 2000, Hof et al 2002, Coyle & Tsai 2004, Harrison & Weinberger 2005). In vivo studies in rodents, as well as in vitro studies using tissue from a variety of species including human post-mortem samples, have provided evidence that Nrg1–ErbB signalling can affect each of these targets. For example, Nrg1 is essential for directing myelination of peripheral nerves (Michailov et al 2004, Taveggia et al 2005) and disruption of oligodendroglial ErbB signalling alters myelination of central axons and widely dysregulates dopamine signalling (Roy et al 2007). Nrg1–ErbB signalling is an essential regulator of tangential migration of cortical GABAergic interneurons during early brain development and adult mice lacking neuronal ErbB4 expression or heterozygous for disruption of Type III Nrg1 expression have deficits in subpopulations of these cortical interneurons (Flames et al 2004). In vitro studies show that short-term responses of cerebellar and cortical neurons to the application of recombinant Nrg1 include transcriptional regulation of GABA-A receptor expression (Rieff et al 1999, Okada & Corfas 2004) and presynaptic modulation of GABA release (Woo et al 2007). Thus Nrg1–ErbB signalling contributes to the migration, differentiation and modulation of a population of cortical interneurons that are affected in individuals with schizophrenia.
Nrg1–ErbB signalling also affects glutamatergic transmission, again at multiple levels. Early in development Nrg1–ErbB signalling is important for radial migration of differentiating pyramidal neurons and for targeting of glutamatergic projections from the dorsal thalamus to the neocortex (Anton et al 1997, Lopez-Bendito et al 2006). We have found that heterozygous type III Nrg1 mutant mice have alterations in the formation and function of glutamatergic ventral hippocampal synapses on striatal medium spiny neurons (unpublished observations). Altering Nrg1–ErbB signalling has complex effects on activity dependent synaptic plasticity in hippocampal and cortical slice cultures, altering both NMDA receptor levels and phosphorylation and AMPA receptor trafficking. Although these studies point to complex, and sometimes contradictory effects of Nrg1-ErbB signalling at glutamatergic synapses, in sum they point to the need for a fine balance of Nrg1–ErbB signalling with both deficient and excessive signalling interfering with synaptic plasticity (Gu et al 2005, Kwon et al 2005, Hahn et al 2006, Bjarnadottir et al 2007, Li et al 2007, Role & Talmage 2007). It is likely that there is a similar requirement for balanced signalling at other synapses and at extrasynaptic sites (Fig. 2). Given the apparent sensitivity of the Nrg–ErbB signalling network to perturbations, it is becoming clearer how subtle changes in the levels and types of Nrg1–ErbB interactions could alter the ability of key brain circuits to withstand additional genetic and environmental insults and contribute to disease.
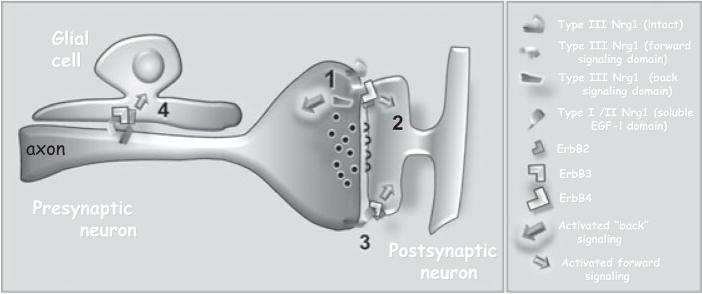
FIG. 2.
Differential axonal expression of Neuregulin 1 isoforms affects synaptic Nrg1-ErbB signalling strategies. At left is a cartoon depicting an axon terminal segment interacting with a postsynaptic spine and a glial cell (1). At the synapse, presynaptic Type III Nrg1 interacts with post-synaptic ErbB4 dimers (shown as an ErbB4:ErbB2 heterodimer, but could also be an ErbB4 homodimer). This interaction activates ErbB tyrosine kinase activity eliciting both local changes in the spine and transcriptional responses in the post-synaptic neuron (2). At glutamatergic synapses responses include regulation of NMDA receptor expression and regulation of both NMDA and AMPA receptor trafficking; responses that modulate synaptic plasticity. Type III Nrg1–ErbB interactions can also result in signalling by the Type III Nrg1 in the pre-synaptic cell. One type of signalling that we have described (Bao et al 2003) involves the γ-secretase dependent cleavage of type III Nrg1 releasing the C-terminal intracellular domain which has transcriptional regulatory activity. ‘Back-signalling’ contributes to the survival of pre-synaptic neurons during development and regulates the targeting of neurotransmitter receptors to presynaptic sites where these receptors modulate synaptic activity. Non-type III Nrg1 isoforms also signal at synapses, but are likely to do so in predominantly a unidirectional manner (3). Altering the ratio of type III Nrg1 to non-type III Nrg1 levels is predicted to change the balance of unidirectional vs. bidirectional signalling. Axonal Nrg1 also interacts with ErbB3:ErbB2 heterodimers on glia (4), a signalling partnership that is essential for proper myelination.
Acknowledgments
All work, published and unpublished from the Talmage lab has been carried out in collaboration with Dr Lorna W. Role and was funded by NS29071.
DISCUSSION
Chao: In the heterozygotes for neuregulin, what is the level of the type III proteins?
Talmage: It is close to 50% of wild-type levels in systems where we can get clean tissue and good clean blots.
Chao: Are the other isoforms of neuregulin affected?
Talmage: They are unaffected at the RNA level. There aren’t good antibody probes to look at protein expression levels, but we have done RNase protection assays for various isoforms, and they are unaffected by the full type III knockout.
Akil: Do you have any inducibles?
Talmage: Not at this point.
Akil: I am basing my comment on my thinking on fi broblast growth factor (FGF). There is one programme during development, and quite a different programme during adulthood. It might be worth sorting out which one we are talking about when we are thinking about animal models.
Talmage: We want to do those experiments. As an example, the interneuron migration story involves neuregulins. The migrating interneurons don’t express neuregulin, but they express ErbB4. At that point in development in the mouse, the E12/E14 window, there is very little neuregulin and type III is not expressed in the neocortex. It is expressed in regions of the developing striatum where it interacts with the migrating neurons and the axons that have been targeted. By E18 there is type III expression in the cortex. In adults there is clear expression in both pyramidal and non-pyramidal cortical neurons. There is a window where it is important in setting up the structure of the neocortex but it is not expressed in the cells that get there. Yet it is turned on later. One would expect it would then have another function distinct from its role in early development. We think there are probably both embryonic and adult phenotypes.
Lu: The functional studies you did were in cultures of extracellular domain (ECD) of the ErbB4 receptor. Let’s think in the physiological context: how would the ECD be generated and is it regulated? Is it regulated at the synapse?
Talmage: One possibility is that the ErbB4 ECD is released from cells in a regulated fashion.
Lu: Has that been demonstrated?
Talmage: In vitro, yes. There are splice variants of ErbB4 that undergo ectodomain shedding and γ-secretase-mediated processing (e.g. Rio et al 2000, Ni et al 2000). We think there is direct interaction between type III neuregulin and ErbB4 (or ErbB3). Neuregulin is shipped to axons that go to target fi elds where ErbB4 is expressed. Everyone is in the right place, but we don’t know at the electron microscopy (EM) level if they are really in the right place. Conceptually, both proteins are where they need to be to do what we think they are doing. The γ-secretase processing of type III neuregulin can be stimulated by activity as well.
Lu: I have another question about the acute versus long-term effects. The long-term regulation is a nuclear event. How long does it take for the intracellular fragment to go into the nucleus, and do you know about the targets?
Talmage: It takes 12–15 minutes to appear in the nucleus after stimulus. It physically interacts with Eos, a member of the Ikaros transcription family (Bao et al 2004). We have genes whose transcription is regulated up and down as a consequence of stimulating the back signalling event. We get slightly different transcriptional profi les if we stimulate with Erb4 than we do following depolarization. There may be additional subtleties. We haven’t shown binding to promoter for those target genes yet.
Bothwell: What do Alzheimer disease mutations of amyloid precursor protein (APP) or presenilins do to neuregulin signalling?
Talmage: I don’t know. Processing of type III neuregulin does not occur in cells from presenilin knockout mice
Buonanno: Is the back signalling of type I Neuregulin also included in this? Are type I and ErbB4 in those axons that you were talking about?
Talmage: We don’t detect ErbB4 in those axons. If we inhibit ErbB4 kinase activity it affects nothing. If we add soluble neuregulin we don’t get any of the same responses. Type I and type II neuregulin isoforms are probably expressed but in these neurons type I (or type II) seems to be constitutively released from the cell surface, so we can’t ask whether it participates in back-signaling. There are other cells where type I neuregulin stays on the surface much longer and its release is regulated by other events. It probably can function as a receptor in those contexts.
Buonanno: So if you use an antibody that is specifi c for type III and run this on a Western blot, do you see that the proportion of unprocessed type III is large compared with type I?
Talmage: Yes, and we are also collecting type I from the conditioning medium, showing that it is there. We have stained neurons from type III knockout animals with the ErbB4 extracellular domain so we can look where it is binding. Without type III expression we don’t detect any ErbB4 binding.
References
- Bao J, Lin H, Ouyang Y, et al. Activity-dependent transcription regulation of PSD-95 by neuregulin-1 and Eos. Nat Neurosci. 2004;7:1250–1258. doi: 10.1038/nn1342. [DOI] [PubMed] [Google Scholar]
- Ni CY, Murphy MP, Golde TE, Carpenter G. γ-Secretase Cleavage and Nuclear Localization of ErbB-4 Receptor Tyrosine Kinase. Science. 2000;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- Rio C, Buxbaum JD, Peschon JJ, Corfas G. Tumor Necrosis Factor-alpha-converting Enzyme is Required for Cleavage of erbB4/HER4. J Biol Chem. 2000;275:10379–10387. doi: 10.1074/jbc.275.14.10379. [DOI] [PubMed] [Google Scholar]
References
- Anton ES, Marchionni MA, Lee KF, Rakic P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development. 1997;124:3501–3510. doi: 10.1242/dev.124.18.3501. [DOI] [PubMed] [Google Scholar]
- Bao J, Wolpowitz D, Role LW, Talmage DA. Back signaling by the Nrg-1 intracellular domain. J Cell Biol. 2003;161:1133–1141. doi: 10.1083/jcb.200212085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Lin H, Ouyang Y, et al. Activity-dependent transcription regulation of PSD-95 by neuregulin-1 and Eos. Nat Neurosci. 2004;7:1250–1258. doi: 10.1038/nn1342. [DOI] [PubMed] [Google Scholar]
- Bjarnadottir M, Misner DL, Haverfield-Gross S, et al. Neuregulin 1 (NRG1) signaling through Fyn modulates NMDA receptor phosphorylation: differential synaptic function in NRG1+/− knock-outs compared with wild-type mice. J Neurosci. 2007;27:4519–4529. doi: 10.1523/JNEUROSCI.4314-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher AA, Arnold JC, Duffy L, Schofield PR, Micheau J, Karl T. Heterozygous neuregulin 1 mice are more sensitive to the behavioural effects of Delta9-tetrahydrocannabinol. Psychopharmacology (Berl) 2007;192:325–336. doi: 10.1007/s00213-007-0721-3. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Tsai G. NMDA receptor function, neuroplasticity, and the pathophysiology of schizophrenia. Int Rev Neurobiol. 2004;59:491–515. doi: 10.1016/S0074-7742(04)59019-0. [DOI] [PubMed] [Google Scholar]
- Flames N, Long JE, Garratt AN, et al. Short- and long-range attraction of cortical GAB-Aergic interneurons by neuregulin-1. Neuron. 2004;44:251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Gerlai R, Pisacane P, Erickson S. Heregulin, but not ErbB2 or ErbB3, heterozygous mutant mice exhibit hyperactivity in multiple behavioral tasks. Behav Brain Res. 2000;109:219–227. doi: 10.1016/s0166-4328(99)00175-8. [DOI] [PubMed] [Google Scholar]
- Go RC, Perry RT, Wiener H, et al. Neuregulin-1 polymorphism in late onset Alzheimer’s disease families with psychoses. Am J Med Genet B Neuropsychiatr Genet. 2005;139:28–32. doi: 10.1002/ajmg.b.30219. [DOI] [PubMed] [Google Scholar]
- Golding M, Ruhrberg C, Sandle J, Gullick WJ. Mapping nucleolar and spliceosome localization sequences of neuregulin1-beta3. Exp Cell Res. 2004;299:110–118. doi: 10.1016/j.yexcr.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Gu Z, Jiang Q, Fu AK, Ip NY, Yan Z. Regulation of NMDA receptors by neuregulin signaling in prefrontal cortex. J Neurosci. 2005;25:4974–4984. doi: 10.1523/JNEUROSCI.1086-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CG, Wang HY, Cho DS, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12:824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- Hof PR, Haroutunian V, Copland C, Davis KL, Buxbaum JD. Molecular and cellular evidence for an oligodendrocyte abnormality in schizophrenia. Neurochem Res. 2002;27:1193–1200. doi: 10.1023/a:1020981510759. [DOI] [PubMed] [Google Scholar]
- Karl T, Duffy L, Scimone A, Harvey RP, Schofield PR. Altered motor activity, exploration and anxiety in heterozygous neuregulin 1 mutant mice: implications for understanding schizophrenia. Genes Brain Behav. 2007 doi: 10.1111/j.1601-183X.2006.00298.x. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Kwon OB, Longart M, Vullhorst D, Hoffman DA, Buonanno A. Neuregulin-1 reverses long-term potentiation at CA1 hippocampal synapses. J Neurosci. 2005;25:9378–9383. doi: 10.1523/JNEUROSCI.2100-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AJ, Lipska BK, Weickert CS, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16:129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- Lewis DA. GABAergic local circuit neurons and prefrontal cortical dysfunction in schizophrenia. Brain Res Brain Res Rev. 2000;31:270–276. doi: 10.1016/s0165-0173(99)00042-9. [DOI] [PubMed] [Google Scholar]
- Li Q, Loeb JA. Neuregulin-heparan-sulfate proteoglycan interactions produce sustained erbB receptor activation required for the induction of acetylcholine receptors in muscle. J Biol Chem. 2001;276:38068–38075. doi: 10.1074/jbc.M104485200. [DOI] [PubMed] [Google Scholar]
- Li B, Woo RS, Malinow R. The neuregulin-1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron. 2007;54:583–597. doi: 10.1016/j.neuron.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hwang H, Cao L, et al. Domain-specific gene disruption reveals critical regulation of neuregulin signaling by its cytoplasmic tail. Proc Natl Acad Sci USA. 1998;95:13024–13029. doi: 10.1073/pnas.95.22.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bendito G, Cautinat A, Sánchez JA, et al. Tangential neuronal migration controls axon guidance: a role for neuregulin-1 in thalamocortical axon navigation. Cell. 2006;125:127–142. doi: 10.1016/j.cell.2006.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- Michailov GV, Sereda MW, Brinkmann BG, et al. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- Montero JC, Yuste L, Diaz-Rodriguez E, Esparis-Ogando A, Pandiella A. Differential shedding of transmembrane neuregulin isoforms by the tumor necrosis factor-alpha-converting enzyme. Mol Cell Neurosci. 2000;16:631–648. doi: 10.1006/mcne.2000.0896. [DOI] [PubMed] [Google Scholar]
- Okada M, Corfas G. Neuregulin1 downregulates postsynaptic GABAA receptors at the hippocampal inhibitory synapse. Hippocampus. 2004;14:337–344. doi: 10.1002/hipo.10185. [DOI] [PubMed] [Google Scholar]
- O’Tuathaigh CM, O’Sullivan GJ, Kinsella A, et al. Sexually dimorphic changes in the exploratory and habituation profiles of heterozygous neuregulin-1 knockout mice. Neurore-port. 2006;17:79–83. doi: 10.1097/01.wnr.0000192738.31029.0a. [DOI] [PubMed] [Google Scholar]
- Rieff HI, Raetzman LT, Sapp DW, Yeh HH, Siegel RE, Corfas G. Neuregulin induces GABA(A) receptor subunit expression and neurite outgrowth in cerebellar granule cells. J Neurosci. 1999;19:10757–10766. doi: 10.1523/JNEUROSCI.19-24-10757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Role LW, Talmage DA. Neurobiology: new order for thought disorders. Nature. 2007;448:263–265. doi: 10.1038/448263a. [DOI] [PubMed] [Google Scholar]
- Roy K, Murtie JC, El-Khodor BF, et al. Loss of erbB signaling in oligodendrocytes alters myelin and dopaminergic function, a potential mechanism for neuropsychiatric disorders. Proc Natl Acad Sci USA. 2007;104:8131–8136. doi: 10.1073/pnas.0702157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg G, Darvasi A, Pinkas-Kramarski R, Navon R. The involvement of ErbB4 with schizophrenia: association and expression studies. Am J Med Genet B Neuropsychiatr Genet. 2006;141:142–148. doi: 10.1002/ajmg.b.30275. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RA, Staros JV. Insights into the evolution of the ErbB receptor family and their ligands from sequence analysis. BMC Evol Biol. 2006;6:79. doi: 10.1186/1471-2148-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinthorsdottir V, Stefansson H, Ghosh S, et al. Multiple novel transcription initiation sites for NRG1. Gene. 2004;342:97–105. doi: 10.1016/j.gene.2004.07.029. [DOI] [PubMed] [Google Scholar]
- Tan W, Wang Y, Gold B, et al. Molecular cloning of a brain-specific, developmentally regulated Neuregulin 1 (NRG1) isoform and identification of a functional promoter variant associated with schizophrenia. J Biol Chem. 2007 doi: 10.1074/jbc.M702953200. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Taveggia C, Zanazzi G, Petrylak A, et al. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. doi: 10.1016/j.neuron.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson PA, Christoforou A, Morris SW, et al. Association of Neuregulin 1 with schizophrenia and bipolar disorder in a second cohort from the Scottish population. Mol Psychiatry. 2007;12:94–104. doi: 10.1038/sj.mp.4001889. [DOI] [PubMed] [Google Scholar]
- Walss-Bass C, Liu W, Lew DF, et al. A novel missense mutation in the transmembrane domain of neuregulin 1 is associated with schizophrenia. Biol Psychiatry. 2006;60:548–553. doi: 10.1016/j.biopsych.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Wang JY, Miller SJ, Falls DL. The N-terminal region of neuregulin isoforms determines the accumulation of cell surface and released neuregulin ectodomain. J Biol Chem. 2001;276:2841–2851. doi: 10.1074/jbc.M005700200. [DOI] [PubMed] [Google Scholar]
- Wolpowitz D, Mason TB, Dietrich P, Mendelsohn M, Talmage DA, Role LW. Cysteine-rich domain isoforms of the neuregulin-1 gene are required for maintenance of peripheral synapses. Neuron. 2000;25:79–91. doi: 10.1016/s0896-6273(00)80873-9. [DOI] [PubMed] [Google Scholar]
- Woo RS, Li XM, Tao Y, et al. Neuregulin-1 enhances depolarization-induced GABA release. Neuron. 2007;54:599–610. doi: 10.1016/j.neuron.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Yang X, Kuo Y, Devay P, Yu C, Role L. A cysteine-rich isoform of neuregulin controls the level of expression of neuronal nicotinic receptor channels during synaptogenesis. Neuron. 1998;20:255–270. doi: 10.1016/s0896-6273(00)80454-7. [DOI] [PubMed] [Google Scholar]