Abstract
Microfabricated regular sieving structures hold great promise as an alternative to gels to improve biomolecule separation speed and resolution. In contrast to disordered gel porous networks, these regular structures also provide well-defined environments ideal for study of molecular dynamics in confining spaces. However, previous regular sieving structures have been limited for separation of long DNA molecules, and separation of smaller, physiologically-relevant macromolecules, such as proteins, still remains as a challenge. Here we report a microfabricated anisotropic sieving structure consisting of a two-dimensional periodic nanofluidic filter array (Anisotropic Nanofilter Array: ANA). The designed structural anisotropy in the ANA causes different-sized or -charged biomolecules to follow distinct trajectories, leading to efficient separation. Continuous-flow size-based separation of DNA and proteins as well as electrostatic separation of proteins were achieved, thus demonstrating the potential of the ANA as a generic molecular sieving structure for an integrated biomolecule sample preparation and analysis system.
Efficient methods of separating and purifying biomolecules from a complex mixture are of utmost importance in biology and biomedical engineering. Currently, nucleic acids and proteins are routinely separated based on size by gel filtration chromatography or by gel electrophoresis1,2. Both techniques use gelatinous materials that consist of a cross-linked, three-dimensional pore network, where the sieving interaction with the migrating macromolecules determines the separation efficiency3,4. Both gel-based techniques represent the current standard for size-based macromolecule separation. However poor separation resolution in gel filtration chromatography and difficult sample recovery with gel electrophoresis make neither method optimal in separating complex mixtures for downstream analysis1. Liquid and solid gelatinous materials have also been integrated in microchip-based systems for rapid separation of biomolecules (e.g., DNA, proteins and carbohydrates) with high resolution5–7. However, the foreign sieving matrices pose intrinsic difficulties for the integration of automated multi-step bioanalysis microsystems. Furthermore, these microchip-based systems are limited to analytical separation of biomolecules, due to the difficulty of harvesting purified biomolecules for downstream analysis.
Recently, there has been great interest in switching from disordered porous gel media to patterned regular sieving structures, either by colloidal templating of self-assembled bead arrays8,9 or by microfabrication techniques10–15. While significantly more efficient than gels in terms of separation speed and resolution, these regular sieving structures still largely resemble gels in the sense that separation is achieved by repeated sieving through multiple, identical “pores”. More recently, microfabricated asymmetric obstacle courses were used to continuously separate macromolecules either by diffusion16,17 or by asymmetric bifurcation of laminar flow18. This later work took advantage of asymmetric interaction of macromolecules with the device geometries, which enabled these novel separation mechanisms. However, the regular sieving structures discussed so far have proven efficacious only for separation of long DNA and microspheres, and their applicability to smaller, physiologically-relevant macromolecules remains questionable19,20, which clearly limits progress on a future integrated bioanalysis system.
Here we report a unique molecular sieving structure design, named “anisotropic nanofilter array” (ANA)21, and its implementation for continuous-flow separation of DNA and proteins based on either size or charge. The designed structural anisotropy of the ANA is critical to the continuous-flow separation, which is not readily possible with a random isotropic sieving medium (e.g., gel, liquid gel, and ampholytes). Moreover, the ANA allows for various sieving mechanisms (e.g., Ogston sieving22–24, entropic trapping25–28, and electrostatic sieving29,30) to take effect for separation of biomolecules of very broad biological size scales and based on different molecular properties, which is a clear advantage over the conventional and nonconventional techniques discussed above. The continuous-flow operation of the ANA further permits continuous-harvesting of purified biomolecules for downstream analysis, rendering the ANA a promising generic sieving structure for an integrated biomolecule sample preparation and analysis system.
RESULTS
Design of anisotropic nanofilter array
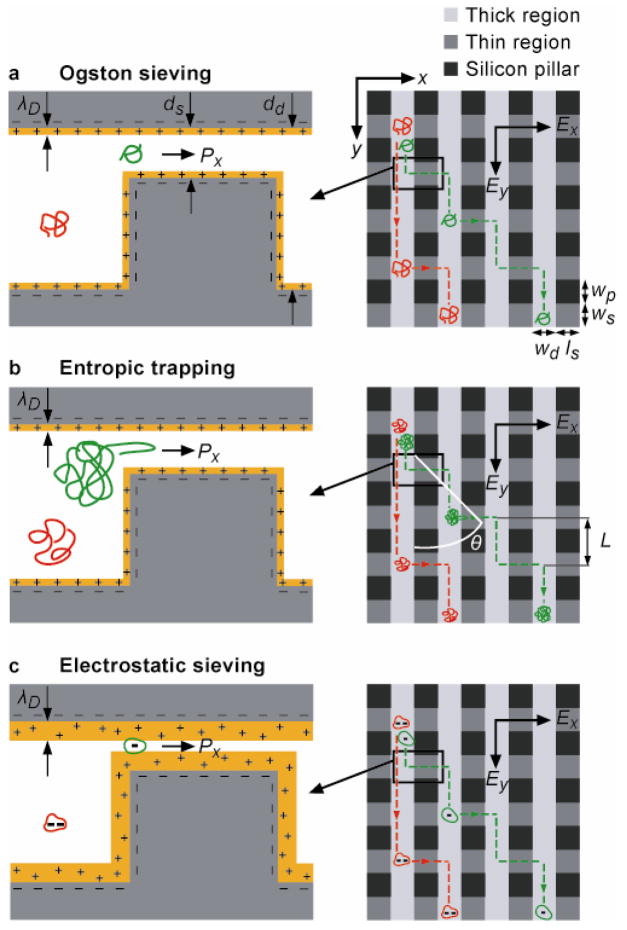
The design of the ANA consists of a two-dimensional (2D) periodic nanofilter array (Fig. 1). The separation mechanism of the ANA relies on different sieving characteristics along two orthogonal directions within the ANA, which are set perpendicular and parallel to the nanofilter rows (indicated as x- and y-axis, respectively, in Fig. 1). Upon application of an electric field Ey along the y-axis, negatively charged macromolecules are driven into the nanofilter array following the direction of either electrophoretic force (electrophoresis) or hydrodynamic drag force from electroosmotic flow (electroosmosis) (see Method). An orthogonal electric field Ex is superimposed along the x-axis across the nanofilters, and this field selectively drives the drifting macromolecules in the deep channel to jump across the nanofilter in the positive x-direction to the adjacent deep channel. Molecular crossings of the nanofilter under the influence of Ex can be described as biased thermally activated jumps across free energy barriers at the nanofilter threshold28,31, and these free energy barriers depend on both steric and electrostatic interactions between charged macromolecules and charged nanofilter walls30. At high ionic strength where the Debye length λD is negligible compared to the nanofilter shallow region depth ds, electrostatic interactions between charged macromolecules and charged nanofilter walls are largely screened. The energy barriers are therefore solely determined by the configurational or conformational entropy loss within the constriction due to steric exclusion (a purely steric limit)28,31,32. For biomolecules with diameters smaller than the nanofilter constriction (i.e., Ogston sieving), the steric energy barrier has been shown to favor DNA and proteins with a smaller size for passage24 (Fig. 1a), resulting in a greater jump passage rate Px for smaller molecules. Therefore, in Ogston sieving, smaller molecules exercise a shorter mean characteristic drift distance L in the deep channels between two consecutive nanofilter crossings, leading to a larger stream deflection angle θ. For macromolecules with diameters greater than the nanofilter constriction size, passage requires the molecules to deform and form hernias at the cost of their internal conformational entropy (i.e., entropic trapping)9,12. A previous study on long DNA molecules trapped at a similar nanofluidic constriction showed that the steric energy barrier for DNA escape depends solely on the inverse of the electric field strength (~1/Ex)28. Further, longer molecules have a larger surface area contacting the constriction and thus have a greater probability to form hernias that initiate the escape process (i.e., a higher escape attempt frequency)33 (Fig. 1b). Therefore, in entropic trapping, longer molecules assume a greater jump passage rate Px, resulting in a larger deflection angle θ.
Figure 1. O Schematic showing negatively charged macromolecules assuming bidirectional motion in the ANA under the influence of two orthogonal electric fields Ex and Ey.
Nanofilters (width: ws, length: ls, depth: ds) arranged in rows are separated by deep channels (width: ld, depth: dd). Rectangular pillars (width: wp, length: ls) serve as supporting structures to prevent collapse of top ceiling. When the Debye length λD = ds (Debye layer highlighted in yellow) (a, b), steric exclusion effect dictates jump dynamics. For Ogston sieving (a), smaller-sized molecules (green) are preferred for nanofilter passage, resulting in a greater nanofilter jump passage rate Px. For entropic trapping (b), longer linear molecules (green) assume a greater probability for hernia formation and thus a greater passage rate Px. Electrostatic sieving becomes dominant when λD~ds (c). Similar sized globular molecules with a lower negative net charge (green) experiences smaller electrostatic repulsion when crossing negatively-charged nanofilter, resulting in a greater passage rate Px. The mean drift distance L between two consecutive nanofilter crossings plays a determinant role for the migration trajectory, with a shorter L leading to a larger stream deflection angle θ where θ is defined with respect to the positive y-axis.
For low ionic strength solutions where the Debye length λD becomes comparable to the nanofilter shallow region depth ds, repulsive electrostatic interactions between negatively charged biomolecules and like charged nanofilter walls become prominent and start to dictate jump dynamics across the nanofilter (Fig. 1c)29,30. The electric potential remains negative in the whole nanochannel, resulting in an electrostatic exclusion of negatively charged molecules. Such electrostatic effects on the partitioning of macromolecules through nanopores have been well studied in membrane science29,30, and recently have been applied for pH-controlled diffusion of proteins across a nanochannel34. Therefore, similar sized biomolecules bearing a lower net charge are energetically favored for passage through the nanofilter, resulting in a greater jump passage rate Px and a larger deflection angle θ.
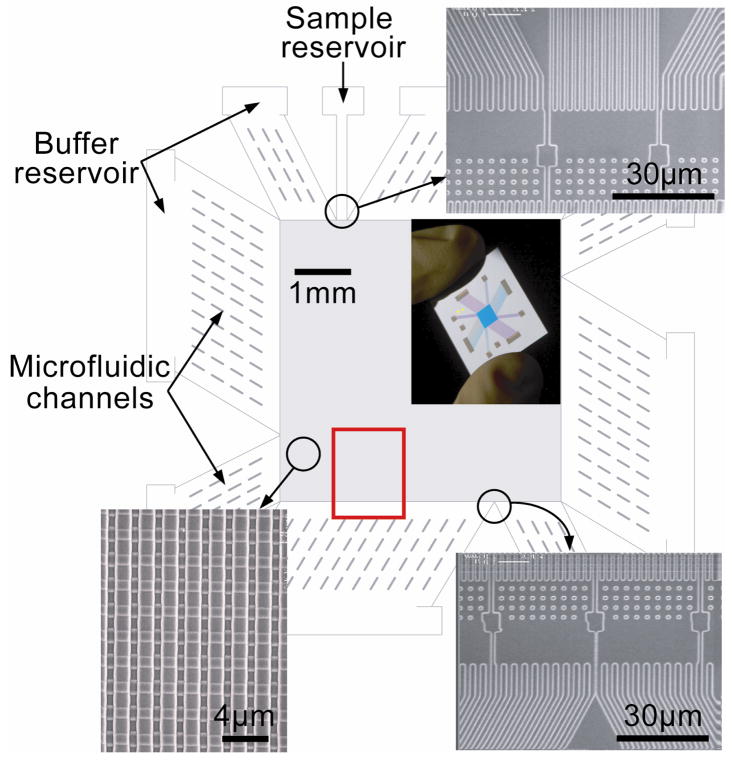
We fabricated a silicon-based device that incorporates the ANA as the sieving structure (Fig. 2). The ANA contains nanofilters with a constriction size of 55 nm (ds) and a width of 1 μm (ws). Deep channels separating the nanofilter rows are 1 μm wide (wd) and 300 nm deep (dd). The initial biomolecule stream is continuously injected into the deep channels on the top left of the device. The fractionated biomolecule streams are collected at intervals along the opposite edge. Microfluidic channels surrounding the ANA connect to fluid reservoirs, where voltages are applied. The microfluidic channels provide sample loading and collection ports, and further act as electric-current injectors to create uniform electric fields Ex and Ey over the ANA structure13,35.
Figure 2. O Structure of the microfabricated device incorporating the ANA.
Scanning electron microscopy images show details of different device regions (clockwise from top right: sample injection channels, sample collection channels, and ANA). The inset shows a photograph of the thumbnail-sized device. The rectangular ANA is 5 mm × 5 mm, and nanofilters (ws=1 gm, ls=1 gm and ds=55 nm) are spaced by 1 μm × 1 μm square-shaped silicon pillars. Deep channels are 1 μm wide (wd) and 300 nm deep (dd). Injection channels connecting sample reservoir (1 mm from the ANA top left corner) inject biomolecule samples as a 30 μm wide stream. The red rectangle highlights the area in which fluorescence photographs in Fig. 3 were taken.
Ogston sieving of short DNA molecules
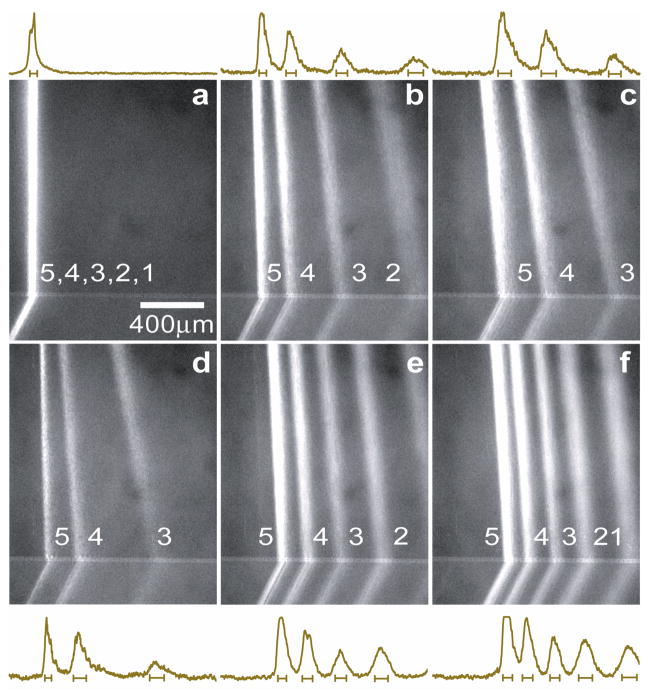
To explicitly demonstrate the steric sieving effect of the ANA, we first injected a low molecular weight (MW) DNA ladder sample (the PCR marker) at Tris-borate-EDTA (TBE) 5× buffer (0.445 M Tris-Borate, 10 mM EDTA, pH≈8.3) under a broad range of field conditions (Fig. 3). Since TBE 5× buffer has an equivalent ionic strength about 130 mM36 with a corresponding Debye length λD of about 0.84 nm (= ds=55 nm), steric interactions dominate jump dynamics. The PCR marker contains 5 different DNA fragments of sizes ranging from 50- to 766-base pairs (bp). Since the persistence length of DNA is about 50 nm (about the contour length of 150-bp DNA)37, the PCR marker fragments appear relatively straight, and recognizable as rigid, rod-like molecules with an end-to-end distance of about 16 nm to 150 nm31,38. The entry into the confining nanofilter can only be realized if the rod-like DNA molecules are properly positioned and oriented without overlapping the wall, which limits the configurational freedom and creates an entropic barrier (i.e., Ogston sieving)2,31,39. Figure 3a–3f show 6 fluorescence photographs of the PCR marker stream pattern in the ANA when horizontal and vertical fields of different values were applied. The horizontal field Ex quickly deflected DNA fragments according to their molecular weights (size), with the stream deflection angle and the stream width depending on the exact field conditions (Supplementary Video 1). Increasing the horizontal field Ex resulted in larger deflection angles as well as wider spreading of the streams.
Figure 3. O Ogston sieving of short DNA (the PCR marker) through the ANA.
Fluorescent photographs of the PCR marker stream pattern were taken in the area highlighted by the red rectangle in Fig. 2. For a, only Ey applied and Ey=25 V/cm; for b, Ex=35 V/cm, Ey=25 V/cm; for c, Ex=60 V/cm, Ey=25 V/cm; for d, Ex=35 V/cm, Ey=12.5 V/cm; for e, Ex=35 V/cm, Ey=50 V/cm; for f, Ex=35 V/cm, Ey=75 V/cm. Band assignment: (1) 50-bp; (2) 150-bp; (3) 300-bp; (4) 500-bp; (5) 766-bp. Fluorescence intensity profiles (of arbitrary units) were measured at the ANA bottom edge. The bars underneath the peaks are centered at the means and label the stream widths (±s.d.).
In Ogston sieving, the nanofilter jump passage rate Px for short DNA of a bp number N can be calculated based on the equilibrium partitioning theory and the Kramer’s rate theory31. In the limit of low field, the passage rate Px is proportional to , where K is the DNA equilibrium partitioning coefficient across the nanofilter31. Therefore, increasing Ex enhances the jump passage rate Px, leading to a shorter drift distance L and thus a larger deflection angle θ. Based on the calculation of Px, we have constructed a course-grained kinetic model to explain the field-dependent stream deflection angle θ (see Supplementary Text and Supplementary Figure 1).
The vertical electric field Ey also affects the deflection angle θ. As Ey was raised from 25 V/cm to 75 V/cm at fixed Ex=35 V/cm (Fig. 3d–3f), the DNA stream pattern became more focused with shorter DNA fragments (50-bp, 150-bp) shifting towards the negative x-direction and longer DNA fragments (300-bp, 500-bp, 766-bp) shifting towards the positive. A greater vertical field Ey shortens the time for DNA to explore the transition through a nanofilter threshold, which explains the behavior of short DNA with increased Ey. The long DNA fragments shifted with Ey in ways not yet fully understood, although the changes were reproducible with Ey up to 125 V/cm. We suspect this phenomenon might be due to the slight non-uniformity of Ex and Ey over the ANA.
Entropic trapping of long DNA molecules
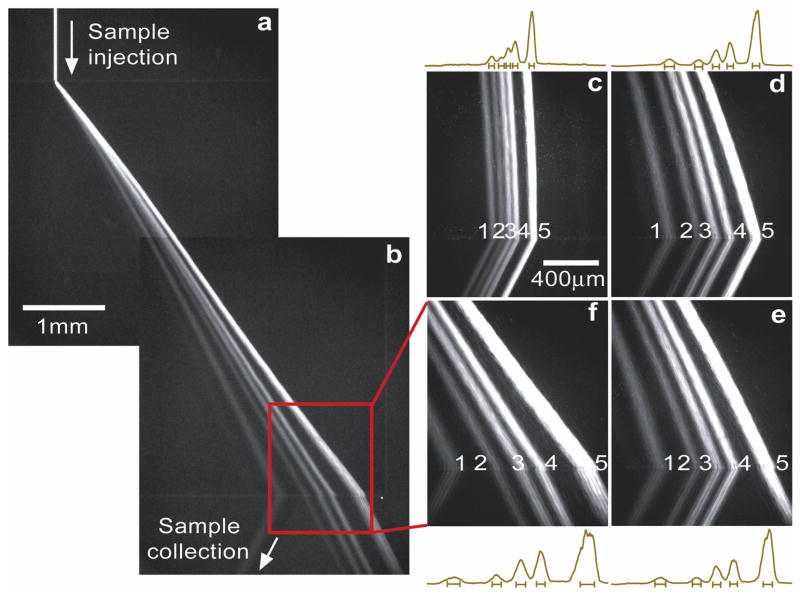
The ANA can separate long DNA molecules based on the entropic trapping mechanism. We prepared a mixture of long DNA molecules (the λ DNA–Hind III digest) in TBE 5× buffer, which contains 6 DNA fragments with sizes ranging from 2027- to 23130-bp and corresponding equilibrium (unconfined) radii of gyration Rg of about 140 nm to 520 nm40. These equilibrium radii of gyration are useful estimates of the spherical DNA size, and they are all greater than the nanofilter constriction depth ds. Therefore the nanofilter jump dynamics involves necessarily the deformation and hernia nucleation (i.e., entropic trapping). With application of the horizontal field Ex=185 V/cm and the vertical field Ey=100 V/cm, λ DNA–Hind III digest was separated in less than 1 min with base-line resolution (Fig. 4a–4b and Supplementary Video 2; note the shortest 2027-bp fragment was too dim for clear visualization in Fig. 4, but with higher CCD gain setting and longer CCD exposure time, the 2027-bp fragment was identified to be base-line separated with the others). A closer look at the video revealed that, as expected, longer DNA fragments followed more deflected migration trajectories than shorter ones, a clear distinction of entropic trapping from Ogston sieving. The streams of λ DNA–Hind III digest followed more deflected and resolved trajectories as Ex was increased (Fig. 4c–4f). This observation is consistent with the argument that increased Ex lowers the activation energy barrier leading to a higher jump passage rate Px28.
Figure 4. O Entropic trapping of long DNA (the λ DNA–Hind III digest) through the ANA.
Fluorescent photographs show separation of λ DNA–Hind III digest with different electric field conditions. For a, b, f, Ex=185 V/cm, Ey=100 V/cm; for c, Ex=50 V/cm, Ey=100 V/cm; for d, Ex=145 V/cm, Ey=100 V/cm; for e, Ex=170 V/cm, Ey=100 V/cm. Band assignment: (1) 2322-bp; (2) 4361-bp; (3) 6557-bp; (4) 9416-bp; (5) 23130-bp. Fluorescence intensity profiles were measured at the ANA bottom edge. The bars underneath the peaks are centered at the means and label the stream widths (±s.d.).
Size- or charged-based separation of proteins with the ANA
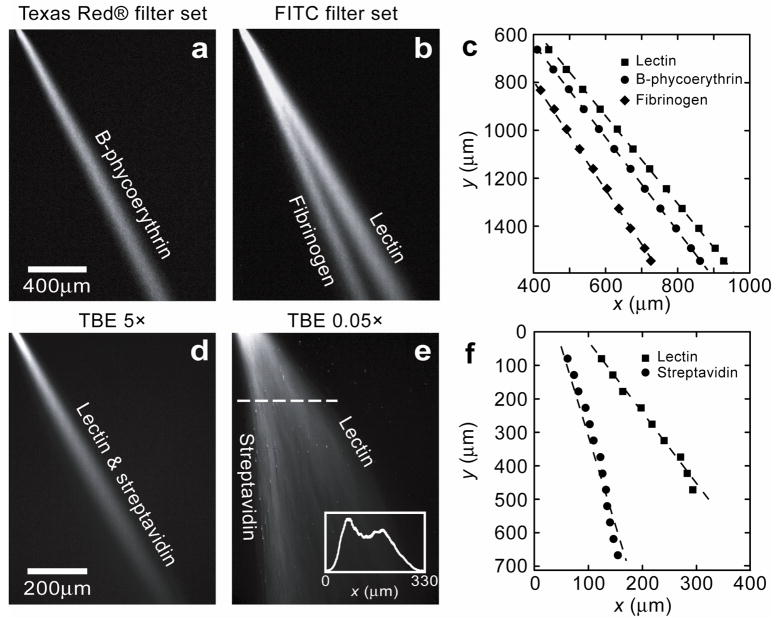
Mixtures of proteins under both native (Fig. 5) and denaturing conditions (Supplementary Figure 2) have been separated based on either size or charge with the ANA, depending on the buffer ionic strength. As proof of concept, we investigated various standard proteins dissolved in either TBE 5× or TBE 0.05× (4.45 mM Tris-Borate, 0.1 mM EDTA) buffer, both at pH=9.6: lectin from Lens culinaris (lentil) (MW~49-kDa, isoelectric point (pI, characteristic pH value at which proteins exhibit zero net charge)≈8.0–8.8), streptavidin (MW~52.8-kDa, pI≈5–6), B-phycoerythrin (MW~240-kDa, pI≈4.2–4.4), and fibrinogen (MW~340-kDa, pI≈5.5). Under the horizontal field Ex=100 V/cm and the vertical field Ey=50 V/cm, a mixture of lectin, B-phycoerythrin, and fibrinogen was driven through the ANA at TBE 5×. The three proteins were clearly separated into 3 distinct streams according to their molecular weight, and non-specific adsorption on the ANA was not significant, possibly due to electrostatic repulsion from the like charged hydrophilic ANA walls41 (Fig. 5a–5c). The stream deflection angles of lectin, B-phycoerythrin, and fibrinogen are about 30.21°, 27.88°, and 24.04°, respectively. The resolution Rs for lectin and B-phycoerythrin at 1.5 mm and 5 mm (extrapolated, see Method) from the injection point are 0.33 and 0.47, respectively, while for B-phycoerythrin and fibrinogen, Rs are 0.95 and 1.24, respectively. In all the experiments, lectin was deflected most followed by B-phycoerythrin and then fibrinogen, suggesting Ogston sieving to account for the jump dynamics. Further increasing Ex resulted in larger lateral separation and broader lateral diffusion of the streams. Similar separation experiments with sodium dodecyl sulfate (SDS)-protein complexes at TBE 5× also supported Ogston sieving for the differential jump dynamics across nanofilters (Supplementary Figure 2).
Figure 5. O Continuous-flow separation of proteins through the ANA.
Proteins are driven through the ANA following electroosmosis. With TBE 5×, separation time was within a few minutes; with TBE 0.05×, separation time was about tens of seconds. a–b, Fluorescent photographs show separation of lectin, B-phycoerythrin, and fibrinogen at TBE 5× with Ex=100 V/cm and Ey= 50 V/cm. Image a and b were taken for the same ANA area (a with a Texas Red® filter set, b with a FITC filter set). c, Maximum fluorescence intensity along the streams measured for both a and b as a function of x and y. d, No separation was observed for lectin and streptavidin at TBE 5× with Ex=150 V/cm and Ey= 75 V/cm. e, Fluorescent photograph shows separation of lectin and streptavidin at TBE 0.05× with Ex=250 V/cm and Ey= 75 V/cm. The inset shows fluorescence intensity profile scanned along the dashed line (at y=175 μm). Maximum fluorescence intensity along the streams measured for e as a function of x and y is shown in f.
Electrostatic sieving in the ANA was demonstrated at TBE 0.05× by separating two proteins, lectin and streptavidin, under native conditions. Both the proteins have similar molecular weight and a relatively large pI value difference. No separation of these two proteins was observed at TBE 5× (Fig. 5d), which excludes the possibility of size-based separation. However, at TBE 0.05× (equivalent ionic strength of 1.3 mM36 and a corresponding Debye length λD of 8.4 nm) where electrostatic interactions become dominant29,30,42, separation of lectin and streptavidin was clearly achieved with 2 distinct streams under the horizontal field Ex=250 V/cm and the vertical field Ey= 75 V/cm. Streptavidin is more negatively charged at pH=9.6 compared to lectin due to its lower pI value, and therefore streptavidin experiences greater repulsion during the jump across the nanofilter constriction29, leading to a lower jump passage rate Px and a smaller deflection angle θ (Fig. 5e). The stream deflection angles of streptavidin and lectin were about 7.44° and 28.50°, respectively, and the resolution Rs at 0.9 mm and 5 mm (extrapolated) from the injection point are 0.32 and 1.96, respectively, indicating base-line resolution at the bottom of ANA.
DISCUSSION
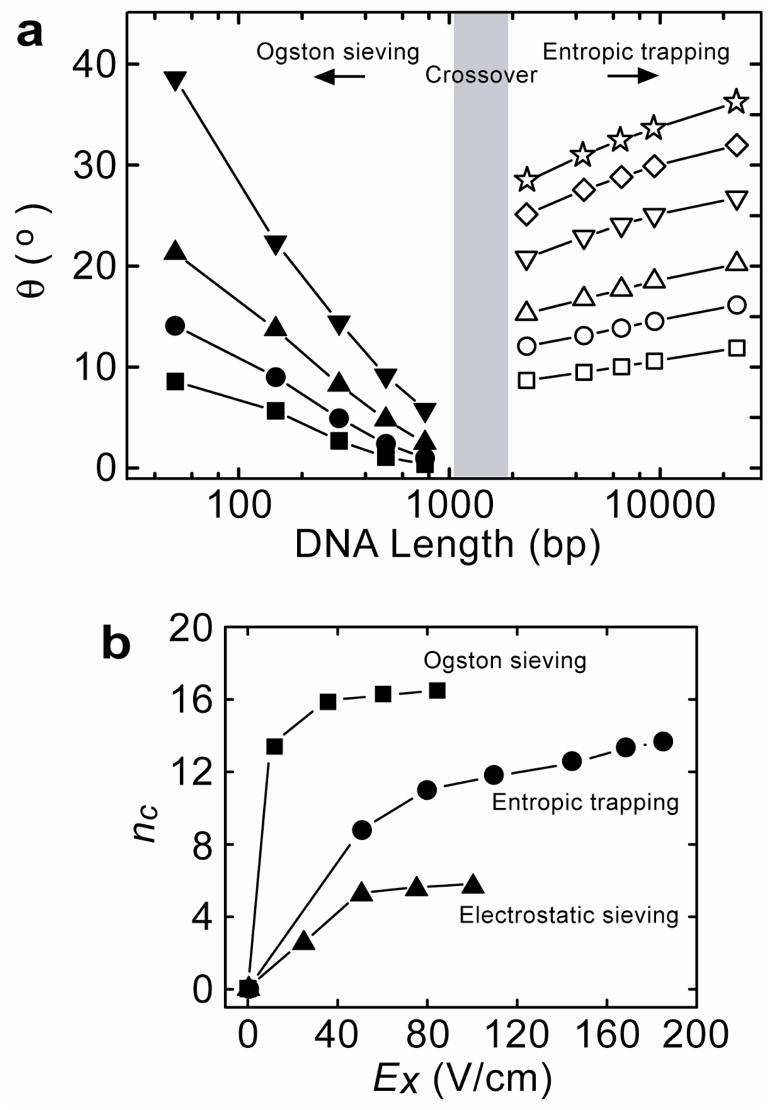
We have observed direct experimental evidence of an unambiguous transition between Ogston sieving and entropic trapping in the ANA. The trajectories of different-sized DNA molecules are consistent with either Ogston sieving or entropic trapping (Fig. 6a). Crossover from Ogston sieving to entropic trapping is between 1000-bp and 2000-bp, which is concurrently with the DNA rod-like conformation to coiled conformation transition and is consistent with observations in one-dimensional (1D) nanofilter arrays31. No saturation plateau was observed for entropic trapping in the ANA, in contrast to the 1D nanofilter array31, indicating possible separation of long DNA in the ANA over an even broader size range. This different observation might be attributed to the more complex structural geometry of the ANA. In addition, the 2D anisotropic energy landscapes of the ANA is modulated by the two independent orthogonal fields Ex and Ey; therefore, the local nanofilter jump dynamics of biomolecules is critically different from in the 1D nanofilter array. It is largely unknown how our understanding of the local nanofilter jump dynamics in 1D nanofilter array applies to the ANA. The effects of the two actively modulated orthogonal fields as well as all the ANA structural parameters need to be considered for a quantitative understanding of the different separation modes.
Figure 6. O Ogston sieving, entropic trapping and electrostatic sieving of DNA and proteins in the ANA.
a, Stream deflection angle θ as a function of DNA length. Left side (Ogston sieving), Ey=25 V/cm, and Ex: 10 V/cm (□), 35 V/cm (○), 60 V/cm (▲), 85 V/cm (▼). Right side (entropic trapping), Ey=100 V/cm and Ex: 50 V/cm (□), Ex: 80 V/cm (○), Ex: 110 V/cm (△), Ex: 145 V/cm (▽), Ex: 170 V/cm (◇), Ex: 185 V/cm (⋆). The ±s.d. of θ derived from the stream half-width are all less than 1°, so statistical error bars θ for are not plotted. b, Dependence of the effective peak capacity nc on Ex. For Ogston sieving (■), Ey=25 V/cm; for entropic trapping (●), Ey=100 V/cm; for electrostatic sieving (▲), Ey=50 V/cm.
From the fluorescence intensity profile of Fig. 3b, the coefficients of variation (CVs) for the 150-bp, 300-bp, and 500-bp DNA stream profiles are 8.6, 6.0, and 4.5%, respectively. Therefore, the size selectivity of the ANA in the Ogston sieving regime is about 5 nm (corresponding to the end-to-end distance of 20-bp DNA). The separation efficiency of the ANA can be further characterized by the effective peak capacity nc that defines the maximum number of separated streams that fit into the space provided by the separation39. Figure 6b shows the dependence of nc on the horizontal field Ex for the regimes of Ogston sieving, entropic trapping and electrostatic sieving. All the effective peak capacity curves initially increased quickly with Ex and then leveled off. This asymptotic behavior of nc can be largely attributed to the stream widening with increased Ex, which cancels out the increased lateral separation between the streams. The effective peak capacity nc can be improved by increasing separation distance. An optimized ANA structure with a gradient of decreasing constriction size along the positive x-direction (an equivalent “gradient gel”) should also provide better resolution and separate proteins over a wider molecular weight range, similar to the effect of gradient-SDS gels for protein separation1,43. Incorporating gate electrodes on the nanofilter walls can allow for additional active adjustment of the surface potential, thus introducing a new degree of control to enhance the electrostatic sieving across the nanofilter44,45.
Other regular sieving structures for continuous-flow sorting of long DNA molecules and microspheres have been reported recently (see Ref. 10–18); however, none of these techniques has demonstrated separation of smaller, physiologically-relevant macromolecules, such as proteins, as we report here. The ANA also represents a significant advance compared to our earlier work in 1D nanofilter arrays, since the continuous-flow operation of the ANA permits continuous-harvesting of the subset of biomolecules of interest to enhance the specificity and sensitivity for downstream biosensing and detection, which is highly desirable for integrated bioanalysis microsystems because of the low sample throughput. In addition, separation speed and resolution in 1D nanofilter arrays cannot be both enhanced without compromising each other24, while in the ANA, they are mainly modulated by the two independent fields Ex and Ey, respectively. Therefore, careful regulation of both Ex and Ey can always achieve rapid separation with high resolution at the same time.
The designed structural anisotropy of the ANA is essential for continuous-flow separation. The continuous-flow separation through the ANA should be applicable to any interaction mechanism (either size-, charge-, or hydrophobicity-based) along the orthogonal x-direction that can lead to differential transport across the nanofilters. The high-resolution separation and ease of sample collection may prove useful for preparative separation of complex biological samples, which has promising implications for proteomic research and biomarker discovery46,47. The sample throughput of the ANA can be further scaled up by parallelism with multi-device processing. We believe the ANA can be used as a generic sieving structure to separate other particles of interest with nanoscale dimensions, including nanoparticles and nanowires, viruses and cell organelles. In addition, we envisage it is possible to develop anisotropic gel- or membrane-based large-scale biomolecule separation systems operating in the continuous-flow mode by introducing structural anisotropy by either photo-patterning anisotropic gel structures or stacking membranes in layers48.
In summary, we have designed and implemented a novel anisotropic nanofluidic sieving structure (i.e., the ANA) that can efficiently separate biologically-relevant macromolecules of broad size scales. The designed structural anisotropy causes different-sized or -charged macromolecules to follow distinct trajectories consistent with either Ogston sieving, entropic trapping, or electrostatic sieving. We have successfully demonstrated high-resolution continuous-flow separation of a wide range of DNA fragments (50-bp – 23000-bp) and proteins (11 kDa – 400 kDa) within a few minutes. By virtue of its separation efficiency, ease of sample recovery and high throughput enabled by continuous-flow operation, the ANA holds great promise as an integrated biomolecule sample preparation and analysis system.
METHODS
Device design, fabrication and metrology
The shallow and deep regions of the ANA, as well as the microfluidic channels were defined and etched into a Si wafer using photolithography and reactive-ion etching techniques. A 5× reduction step-and-repeat projection stepper (Nikon NSR2005i9, Nikon Precision Inc., CA) was used for patterning. KOH etching was performed at 80°C to etch through the wafer for creating buffer access holes. A 500 nm thermal oxide layer was grown to provide an electrical isolation between the conductive Si substrate and buffer solution. Finally, the device was sealed by bonding a Pyrex wafer on the front surface of the silicon wafer. The bonded wafers were cut by diesaw into individual devices for channel filling and separation experiments. The depths of shallow and deep regions of the ANA were measured with a surface profilometer (Prometrix P-10, KLA-Tenco Co., CA) before the anodic bonding process. The depths and surface uniformity of the nanofilter shallow regions were further checked by imaging the cross-section of the nanofilter with scanning electron microscopy (JEOL6320FV, JEOL USA, Inc., Peabody, MA) after anodic bonding.
Sample preparation
The PCR marker as well as λ DNA–Hind III digest (New England BioLabs, Beverly, MA) were labeled with the intercalating fluorescence dye YOYO-1 (Molecular Probes, Eugene, OR) in TBE 5× buffer. The dye to DNA base pair ratio was about 1:2 and the final DNA concentration was about 42.18 μg/ml (PCR) and 104 μg/ml (λ DNA–Hind III digest). Proteins were dissolved in either TBE 5× or TBE 0.05× buffer, and the solution pH was further adjusted to 9.6 by addition of potassium hydroxide (Sigma-Aldrich, St. Louis, MO). The following commercially available proteins and protein-conjugates were investigated in this work: fluorescent B-phycoerythrin (Alexis Biochemicals, San Diego, CA), Alexa Fluor 488 conjugated fibrinogen and streptavidin (both from Molecular Probes, Eugene, OR), fluorescein isothiocyanate (FITC) conjugated lectin from Lens culinaris (lentil) (Sigma-Aldrich, St. Louis, MO). The final concentration of each protein sample in the mixture was about 0.1–0.2 mg/ml, except for lectin (0.2–0.4 mg/ml) due to its lower fluorescence signal. For SDS-protein experiments, Alexa Fluor 488 conjugated cholera toxin subunit B was purchased from Molecular Probes. β-galactosidase from E. coli was obtained from Sigma, and was custom labeled with Alexa Fluor 488 by Molecular Probes. The complete denaturation of both proteins was performed by adding SDS (Sigma-Aldrich, St. Louis, MO) and dithiothreitol (DTT, Sigma-Aldrich, St. Louis, MO). The SDS-DTT protein mixture contained 2 wt% SDS and 0.1M DDT and was treated in an 80°C water bath for 10 min. The resultant SDS-protein complex solutions were mixed and further diluted in TBE 5× buffer. The final SDS-protein complex sample solution contained 15.1 μg/ml cholera toxin subunit B, 90.9 μg/ml β-galactosidase, 0.1 wt% SDS, and 5 μM DTT.
Separation
The ANA was filled with TBE 5× buffer for size-based DNA and protein separation and TBE 0.05× for charged-based protein separation, respectively. Additional 0.1wt% SDS was added to TBE 5× buffer for SDS-protein experiments. In all experiments, DNA and SDS-protein complexes followed the direction of electrophoresis (due to diminished electroosmosis under high buffer ionic strength), while proteins under native conditions followed the direction of electroosmosis (presumably due to their lower net charge compared to DNA and therefore less experienced electrophoretic drag). An inverted epi-fluorescence microscope (IX-71, Olympus, Melville, NY) equipped with a thermoelectrically cooled CCD camera (Sensicam QE, Cooke Co., Auburn Hill, MI) was used for fluorescence imaging, and a 100W mercury lamp (Chiu Technical Corp., Kings Park, NY) was used for illumination. B-phycoerythrin was visualized with a Texas Red® filter set (exitation: 562 nm, emission: 624 nm, Semrock, Rochester, NY) whereas all the other biomolecules were observed using a FITC filter set (exitation: 482 nm, emission: 536 nm, Semrock, Rochester, NY). The images were analyzed with image processing software (IPLab, Scanalytics, BD Bioscience, Rockville, MD).
Size selectivity, resolution and peak capacity
The coefficient of variation (CV) of biomolecules with molecular weight m is defined as σm/m×100%, where σm is the standard deviation (±s.d.) of m. When used as a measure for size selectivity, CV is calculated according to , where θ is the measured deflection angle, as a function of m, and σθ is the ±s.d. of the deflection angle derived from the stream half-width. The effective peak capacity nc is calculated based on some specified separation resolution value Rs of adjacent streams. Separation resolution Rs between two streams (stream 1 and 2) is defined as Rs,12=0.5×Δx/(σ1+σ2), where Δx is the spatial distance between the two streams, and σ1 and σ2 are the ±s.d. of stream widths. In the ANA, the effective peak capacity nc for adjacent streams separated at Rs=1 is calculated as , where θ1 and θn denote the smallest and greatest stream deflection angles, respectively, σ̄W is the mean of ±s.d. of stream widths, and 5000 gm is the width of the rectangular ANA. For all the electropherograms measured, we have used Gaussian functions for fitting to determine the means (the maximum intensity) as well as the stream widths. Please note that, due to the low signal-to-background ratio, the separation resolution for electrostatic sieving of proteins at the bottom of ANA is calculated by linearly extrapolating the stream separation distance and stream width along the streams.
Supplementary Material
Acknowledgments
We acknowledge financial support from NIH (EB005743), DuPont-MIT Alliance, NSF (CTS-0347348) and Singapore-MIT Alliance (SMA-II, CE program). We thank P. Mao for helping take the scanning electron microscopy images and J. Yoo for contribution in the experimental setup. We acknowledge valuable comments and suggestions on the manuscript by P. Doyle, H. Bow and C. Rothman. The MIT Microsystems Technology Laboratories is acknowledged for support in fabrication.
Footnotes
AUTHOR CONTRIBUTION
J. F. and J. H. conceived and initiated ANA concept. J. F. designed and fabricated ANA device. J. F. conceived and performed experiments with short DNA, long DNA and denatured proteins. R.B.S. conceived and performed experiments with native proteins. J. F. and R. B. S. analyzed data. A. L. S. performed gel analysis. J. F. and J. H. developed theoretical model. J. F. and R. B. S. wrote manuscript. J. H. and S. R. T. supervised the project.
COMPETING INTERESTS STATEMENT
The authors declared no competing interests.
References
- 1.Scopes RK. Protein Purification, Principles and Practice. 3. Springer-Verlag; New York: 1993. [Google Scholar]
- 2.Giddings JC. Principles and Theory. Marcel Dekker; New York: 1965. Dynamics of Chromatography. Part 1. [Google Scholar]
- 3.Slater GW, Mayer P, Drouin G. Migration of DNA through gels. Methods Enzymol. 1996;270:272–295. doi: 10.1016/s0076-6879(96)70014-9. [DOI] [PubMed] [Google Scholar]
- 4.Viovy JL. Electrophoresis of DNA and other polyelectrolytes: physical mechanisms. Rev Mod Phys. 2000;72:813–872. [Google Scholar]
- 5.Woolley AT, Mathies RA. Ultra-high-speed DNA fragment separations using microfabricated capillary array electrophoresis chips. Proc Natl Acad Sci U S A. 1994;91:11348–11352. doi: 10.1073/pnas.91.24.11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao G, et al. SDS capillary gel electrophoresis of proteins in microfabricated channels. Proc Natl Acad Sci U S A. 1999;96:5372–5377. doi: 10.1073/pnas.96.10.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callewaert N, Contreras R, Mitnik-Gankin L, Carey L, Matsudaira P, Ehrlich D. Total serum protein N-glycome profiling on a capillary electrophoresis-microfluidics platform. Electrophoresis. 2004;25:3128–3131. doi: 10.1002/elps.200406020. [DOI] [PubMed] [Google Scholar]
- 8.Liu L, Li P, Asher SA. Entropic trapping of macromolecules by mesoscopic periodic voids in a polymer hydrogel. Nature. 1999;397:141–144. doi: 10.1038/16426. [DOI] [PubMed] [Google Scholar]
- 9.Nykypanchuk D, Strey HH, Hoagland DA. Brownian motion of DNA confined within a two-dimensional array. Science. 2002;297:987–990. doi: 10.1126/science.1073277. [DOI] [PubMed] [Google Scholar]
- 10.Volkmuth WD, Austin RH. DNA electrophoresis in microlithographic arrays. Nature. 1992;358:600–602. doi: 10.1038/358600a0. [DOI] [PubMed] [Google Scholar]
- 11.Turner SW, Perez AM, Lopez A, Craighead HG. Monolithic nanofluid sieving structures for DNA manipulation. J Vac Sci Technol B. 1998;16:3538–3840. [Google Scholar]
- 12.Han J, Craighead HG. Separation of long DNA molecules in a microfabricated entropic trap array. Science. 2000;288:1026–1029. doi: 10.1126/science.288.5468.1026. [DOI] [PubMed] [Google Scholar]
- 13.Huang LR, et al. A DNA prism: high speed continuous fractionation of large DNA molecules. Nature Biotech. 2002;20:1048–1051. doi: 10.1038/nbt733. [DOI] [PubMed] [Google Scholar]
- 14.Baba M, et al. DNA size separation using artificially nanostructured matrix. Appl Phys Lett. 2003;83:1468–1470. [Google Scholar]
- 15.Kaji N, et al. Separation of long DNA molecules by quartz nanopillar chips under a direct current electric field. Anal Chem. 2004;76:15–22. doi: 10.1021/ac030303m. [DOI] [PubMed] [Google Scholar]
- 16.Chou F, et al. Sorting by diffusion: an asymmetric obstacle course for continuous molecular separation. Proc Natl Acad Sci USA. 1999;96:13762–13765. doi: 10.1073/pnas.96.24.13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Oudenaarden A, Boxer SG. Brownian ratchets: molecular separation in lipid bilayers supported on patterned arrays. Science. 1999;285:1046–1048. doi: 10.1126/science.285.5430.1046. [DOI] [PubMed] [Google Scholar]
- 18.Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 19.Austin RH, et al. Ratchets: the problems with boundary conditions in insulating fluids. Appl Phys A. 2002;75:279–284. [Google Scholar]
- 20.Huang LR, et al. Role of molecular size in ratchet fractionation. Phys Rev Lett. 2002;89:art. no 178301. doi: 10.1103/PhysRevLett.89.178301. [DOI] [PubMed] [Google Scholar]
- 21.Fu J, Han J. Continuous-flow biomolecule separation through patterned anisotropic nanofluidic sieving structure. Proc Micro Total Analysis Systems. 2006;2006:519–521. [Google Scholar]
- 22.Ogston AG. The spaces in a uniform random suspension of fibres. Trans Faraday Soc. 1958;54:1754–1757. [Google Scholar]
- 23.Rodbard D, Chrambach A. Unified theory for gel electrophoresis and gel filtration. Proc Natl Acad Sci U S A. 1970;65:970–977. doi: 10.1073/pnas.65.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu J, Mao P, Han J. Nanofilter array chip for fast gel-free biomolecule separation. Appl Phys Lett. 2005;87:art no 263902. doi: 10.1063/1.2149979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muthukumar M, Baumgärtner A. Effects of entropic barriers on polymer dynamics. Macromolecules. 1989;22:1937–1941. [Google Scholar]
- 26.Smisek DL, Hoagland DA. Electrophoresis of flexible macromolecules: evidence for a new mode of transport in gels. Science. 1990;248:1221–1223. doi: 10.1126/science.2349481. [DOI] [PubMed] [Google Scholar]
- 27.Rousseau J, Drouin G, Slater GW. Entropic trapping of DNA during gel electrophoresis: effect of field intensity and gel concentration. Phys Rev Lett. 1997;79:1945–1948. [Google Scholar]
- 28.Han J, Turner SW, Craighead HG. Entropic trapping and escape of long DNA molecules at submicron size constriction. Phys Rev Lett. 1999;83:1688–1691. [Google Scholar]
- 29.Smith FG, Deen WM. Electrostatic effects on the partitioning of spherical colloids between dilute bulk solution and cylindrical pores. J Colloid Interface Sci. 1983;91:571–590. [Google Scholar]
- 30.Deen WM. Hindered transport of large molecules in liquid-filled pores. AIChE J. 1987;33:1409–1425. [Google Scholar]
- 31.Fu J, Yoo J, Han J. Molecular sieving in periodic free-energy landscapes created by patterned nanofilter arrays. Phys Rev Lett. 2006;97:art no 018103. doi: 10.1103/PhysRevLett.97.018103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giddings JC, Kucera E, Russell CP, Myers MN. Statistical theory for the equilibrium distribution of rigid molecules in inert porous networks. Exclusion chromatography. J Phys Chem. 1968;72:4397–4408. [Google Scholar]
- 33.Slater GW, Gratton Y, Kenward M, McCormick L, Tessier F. Deformation, stretching, and relaxation of single-polymer chains: fundamentals and examples. Soft Materials. 2003;1:365–391. [Google Scholar]
- 34.Schoch RB, Bertsch A, Renaud P. pH-controlled diffusion of proteins with different pI values across a nanochannel on a chip. Nano Lett. 2006;6:543–547. doi: 10.1021/nl052372h. [DOI] [PubMed] [Google Scholar]
- 35.Huang LR, et al. Generation of large-area tunable uniform electric fields in microfluid arrays for rapid DNA separation. Tech Dig Int Elect Dev Mtg. 2002:363–366. [Google Scholar]
- 36.Lide DR. CRC Handbook of Chemistry and Physics. 87. Taylor and Francis; Boca Raton, FL: 2007. [Google Scholar]
- 37.Hagerman PJ. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 38.Rubenstein M, Colby RH. Polymer Physics. Oxford; New York: 2003. [Google Scholar]
- 39.Giddings JC. Unified Separation Science. John Wiley & Sons; New York: 1991. [Google Scholar]
- 40.Smith DE, Perkins TT, Chu S. Dynamical scaling of DNA diffusion coefficients. Macromolecules. 1996;29:1372–1373. [Google Scholar]
- 41.Nakanishi K, Sakiyama T, Imamura K. On the adsorption of proteins on solid surfaces, a common but very complicated phenomenon. J Biosci Bioeng. 2001;91:233–244. doi: 10.1263/jbb.91.233. [DOI] [PubMed] [Google Scholar]
- 42.Schoch RB. PhD Thesis No 3538. EPFL, Lausanne: 2006. Transport Phenomena in Nanofluidics: From Ionic Studies to Proteomic Applications. [Google Scholar]
- 43.Margolis J, Kenrick KG. Polyacrylamide gel-electrophoresis across a molecular sieve gradient. Nature. 1967;214:1334–1336. doi: 10.1038/2141334a0. [DOI] [PubMed] [Google Scholar]
- 44.Karnik R, Castelino K, Majumdar A. Field-effect control of protein transport in a nanofluidic transistor circuit. Appl Phys Lett. 2006;88:art no 123114. [Google Scholar]
- 45.Eijkel JCT, van den Berg A. Nanotechnology for membranes, filters and sieves. Lab Chip. 2006;6:19–23. doi: 10.1039/b516903h. [DOI] [PubMed] [Google Scholar]
- 46.Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3:267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 47.Righetti PG, Castagna A, Herbert B, Reymond F, Rossier JS. Prefractionation techniques in proteome analysis. Proteomics. 2003;3:1397–1407. doi: 10.1002/pmic.200300472. [DOI] [PubMed] [Google Scholar]
- 48.Han J, Fu J. Continuous biomolecule separation in a nanofilter structure. United States Patent P-8195-USP. 5 October 2006
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.