Summary
VHL tumor suppressor loss results in hypoxia inducible factor-alpha (HIF-α) stabilization, and occurs in 70% of sporadic clear cell renal carcinomas (ccRCCs). To determine whether opposing influences of HIF-1α and HIF-2α on c-Myc activity regulate human ccRCC progression, we analyzed VHL genotype and HIF-α expression in 160 primary tumors, which segregated into three groups with distinct molecular characteristics. Interestingly, ccRCCs with intact VHL, as well as pVHL-deficient, HIF-1α/HIF-2α expressing ccRCCs, exhibited enhanced Akt/mTOR and ERK/MAPK signaling. In contrast, pVHL-deficient ccRCCs expressing only HIF-2α displayed elevated c-Myc activity, resulting in enhanced proliferation and resistance to replication stress. These reproducible distinctions in ccRCC behavior delineate HIF-α effects on c-Myc in vivo and suggest molecular criteria for selecting targeted therapies.
Significance
Constitutive HIF activity is clearly associated with ccRCC tumorigenesis; however, the influence of individual HIF-α subunits on cell growth mechanisms in vivo is unknown. Few dominant oncogenic pathways have been identified within ccRCC, making it difficult to select optimal targeted therapies for patients, or to predict disease outcome, except by grade and stage. Cell culture experiments indicate that HIF-1α inhibits the c-Myc oncoprotein, whereas HIF-2α potentiates c-Myc transcriptional activity and cellular proliferation. The findings reported here indicate that HIF-1α and HIF-2α promote distinct oncogene activation in human ccRCCs, and reveal a critical role for HIF-2α and c-Myc in promoting genomic integrity. These results suggest that evaluating pVHL status and HIF-α expression may aid targeted therapy selection for human ccRCCs.
Introduction
Mutation or silencing of the von Hippel-Lindau (VHL) tumor suppressor gene occurs in a majority of inherited and sporadic clear cell renal cancers (ccRCC) (Banks et al., 2006; Kim and Kaelin, 2004; Kondo et al., 2002b; Lonser et al., 2003). VHL encodes pVHL, a critical regulator of the hypoxia inducible factor (HIF) transcriptional activators (Kim and Kaelin, 2004). HIFs are heterodimeric bHLH-PAS proteins, consisting of an α subunit (HIF-1α or HIF-2α) and a β-subunit (HIF-1β or ARNT [for aryl hydrocarbon receptor nuclear translocator]). HIFs mediate cellular adaptation to low O2 by activating the transcription of target genes involved in metabolism, angiogenesis, and extracellular matrix (ECM) remodeling (Gordan and Simon, 2007; Pugh and Ratcliffe, 2003). pVHL is part of the recognition component of a ubiquitin ligase complex that targets HIF-α subunits for normoxic degradation, a process that is inhibited under hypoxic conditions (typically <5% O2) by several mechanisms. Although pVHL has additional molecular functions affecting fibronectin assembly (Kim and Kaelin, 2004), microtubule stability (Hergovich et al., 2003), and atypical Protein Kinase C activity (Lee et al., 2005), results from ccRCC xenograft experiments indicate that HIF-α regulation is critical for VHL tumor suppressor function.
HIF-1α and HIF-2α have overlapping effects on aspects of angiogenesis and ECM remodeling; however, they also exhibit distinct effects on cell metabolism and proliferation (Gordan et al., 2007b). For example, HIF-1α is uniquely able to stimulate glycolytic enzyme expression (Hu et al., 2003; Wang et al., 2005) while blocking anabolic biosynthesis by limiting mitochondrial pyruvate consumption (Kim et al., 2006; Lum et al., 2007; Papandreou et al., 2006). HIF-1α also opposes cell cycle progression in vitro by post-translationally inhibiting the c-Myc oncoprotein (Koshiji et al., 2004). In direct contrast, HIF-2α does not regulate glycolytic gene expression (Hu et al., 2003), but uniquely stimulates expression of the stem cell factor Oct-4 (Covello et al., 2006), and promotes cell cycle progression by enhancing c-Myc-mediated activation of cyclin D2 and E2F1, and repression of p21 and p27 (Gordan et al., 2007a). Intriguingly, independent reports demonstrated that the HIF-2α subunit is primarily responsible for the growth of pVHL-deficient human ccRCC xenografts (Kondo et al., 2003; Kondo et al., 2002a; Maranchie et al., 2002). Moreover, in VHL disease, the cancer susceptibility syndrome associated with germline VHL mutation, HIF-1α expression gradually decreases whereas HIF-2α expression increases as ccRCCs develop (Mandriota et al., 2002; Raval et al., 2005). The differential effects of HIF-1α and HIF-2α on c-Myc provide an appealing mechanistic explanation for the activity of HIF-2α in VHL disease-associated ccRCC (Mandriota et al., 2002; Raval et al., 2005). However, no studies have directly assessed the differential effects of HIF-1α and HIF-2α on c-Myc or other oncogenic pathways in human ccRCC.
In addition to driving cellular proliferation, oncogenes can also contribute to genomic instability by disrupting cell cycle controls and cellular metabolism (Halazonetis et al., 2008). Cell cycle checkpoint inactivation allows DNA replication in aneuploid cells, and may favor oncogenic genomic amplifications by repetitively triggering replication origins during a single S-phase (Hook et al., 2007). Furthermore, many oncogenes alter cellular metabolism, leading to the production of reactive oxygen species that directly modify DNA (Lee et al., 1999). Both processes result in disrupted chromosomal structures and replication fork stalling or collapse, triggering a cell-intrinsic DNA damage response (Bartkova et al., 2005; Gorgoulis et al., 2005). In low stage tumors, this damage causes a G0-like phenotype following incomplete DNA replication (Bartkova et al., 2006; Di Micco et al., 2006), while its impact on more advanced tumors is less clear. High levels of c-Myc overexpression can result in accumulation of significant DNA damage (Ray et al., 2006; Vafa et al., 2002). In contrast, intermediate c-Myc expression levels may actually promote genomic stability, consistent with the observation that DNA damage is reduced when c-Myc is inhibited by HIF-1α (Huang et al., 2007; Koshiji et al., 2005). Therefore, we also examined the possibility that activation of c-Myc by HIF-2α might enhance genomic stability in low stage ccRCCs, thus enhancing tumor cell proliferation by limiting DNA damage checkpoint activation.
In this report, we describe the characterization of more than 160 sporadic human ccRCC tumor samples based on VHL genotype and HIF-α expression patterns. Our initial investigations identified three distinct ccRCC groups: (1) tumors with wild type VHL alleles and undetectable HIF-α protein expression (designated “VHL WT”), (2) VHL-deficient tumors expressing detectable HIF-1α and HIF-2α proteins (designated “H1H2”), and (3) VHL-deficient tumors expressing HIF-2α exclusively (designated “H2”). VHL WT, H1H2, and H2 tumors were extensively characterized for differences in cell proliferation, patterns of oncogene activation, and genomic integrity. These analyses were extended to include two independent renal cancer sample sets and HIF-1α or HIF-2α deficient tumor cell lines. We wished to determine if human ccRCCs can be subclassified into distinct groups based on VHL status and HIF-α expression patterns, which in turn correspond to the activation of distinct oncogenic signaling and DNA repair pathways. The overall objective of these studies is to ultimately provide a strategy to stratify patients for targeted therapies.
Results
Identifying three groups of ccRCC
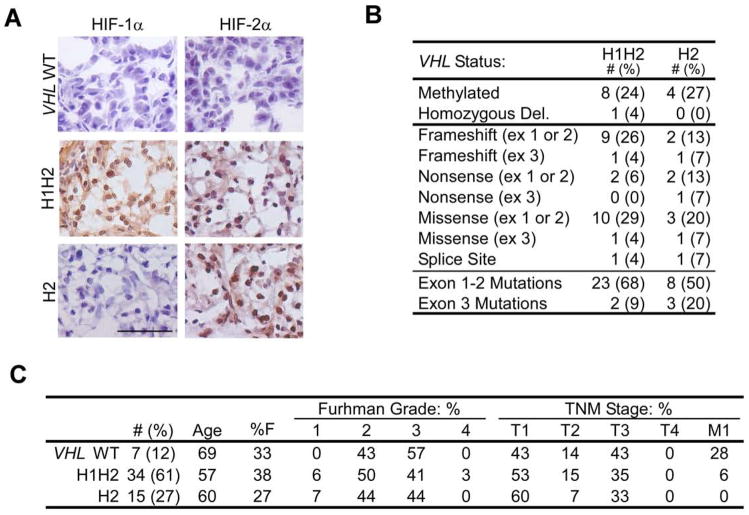
We determined the HIF-α expression patterns and VHL status of 57 independent sporadic human ccRCCs obtained from the Collaborative Human Tissue Network (CHTN). HIF immunostaining patterns separated the tumors into three distinct groups: no HIF-α protein detected (“VHL WT,” 12%), HIF-1α and HIF-2α detected (“H1H2,” 61%), or HIF-2α detected exclusively (“H2,” 27%). Classification was based on HIF-α expression solely in tumor cells. Interestingly, none of the sporadic tumors we evaluated expressed only HIF-1α, strongly suggesting that HIF-2α is critical for development of pVHL-deficient human ccRCCs. HIF-α protein staining in H1H2 and H2 specimens was predominantly nuclear and appeared in >75% of tumor cells, whereas VHL WT tumors exhibited HIF-α staining in <5% of nuclei (Fig. 1A). VHL sequencing, copy number and methylation analysis confirmed that all HIF-α negative tumors (7/7) had wild type VHL alleles, whereas all but one of the HIF-α positive tumors (49/50) harbored bi-allelic VHL inactivation by mutation, deletion or methylation (Fig. 1B and Supplemental Table 1). Most tumors displayed single allele point or frameshift VHL mutations, accompanied by deletion of the remaining allele. The spectrum and relative frequencies of VHL mutations were consistent with published analyses of sporadic ccRCC (Banks et al., 2006; Kondo et al., 2002b). Similar to previous observations (Banks et al., 2006; Kondo et al., 2002b), we found two tumors with one deleted and one intact VHL allele: of these, one tumor had no HIF-α staining and was classified VHL WT, whereas the other exhibited faint, but distinct, HIF-2α staining and was excluded from further analysis.
Figure 1. Categorization of ccRCCs by HIF-α expression and VHL status.
A. Representative HIF-1α and HIF-2α staining of fresh frozen tumors categorized as VHL WT, H1H2 and H2. Scale bar is 1 μM. B. Summary of VHL disruption in H1H2 and H2 tumors. Mutations are separated into those occuring in the first two exons or the last. C. Summary of clinical parameters for each tumor group. In addition to summarized patient characteristics, histological grade by Fuhrman score and clinical stage by the American Joint Committee on Cancer Tumor Node Metastasis (TNM) system are shown. Tumors scored as T1 or T2 (confined to the kidney) were considered low stage, while T3, T4, and M1 (invasive or metastatic tumors) were considered advanced.
Multiple controls were performed to substantiate the immunohistochemistry (IHC) and genotyping. As measured by QRT-PCR, VHL mRNA levels were reduced to <50% and <20% of normal renal epithelium in VHL-mutant tumors and VHL-methylated tumors, respectively (data not shown). HIF-α protein was measured by western blot assays, and protein levels in the three subgroups were consistent with IHC results. Immunoblots showed low levels of HIF-1α expression in H2 tumors (less than 10% the level in H1H2 tumors), which likely reflects HIF-1α protein in endothelial cells (Supplemental Figure 1). By comparing tumor protein extracts to known quantities of epitope-tagged HIF-α protein, we determined that HIF-1α and HIF-2α are expressed at essentially equivalent levels in H1H2 tumors (Supplemental Figure 1 and data not shown). There was no clear difference in HIF-1α mRNA levels between H1H2 and H2 tumors, although slightly increased HIF-2α mRNA levels were noted in H2 tumors (data not shown). The apparent discrepancy between HIF-1α mRNA and protein expression in H2 tumors can be explained by multiple mechanisms, including mutant pVHL proteins that selectively promote HIF-1α degradation (Rathmell et al., 2004) and direct effects of HIF-2α on HIF-1α mRNA translation (Raval et al., 2005). Finally, available clinical information was segregated by group and no significant differences were observed between VHL WT, H1H2, or H2 tumors in either grade or stage of disease at surgery, although VHL WT tumors showed a trend toward more frequent metastasis (Fig. 1C).
HIF-α expression modulates oncogene activation
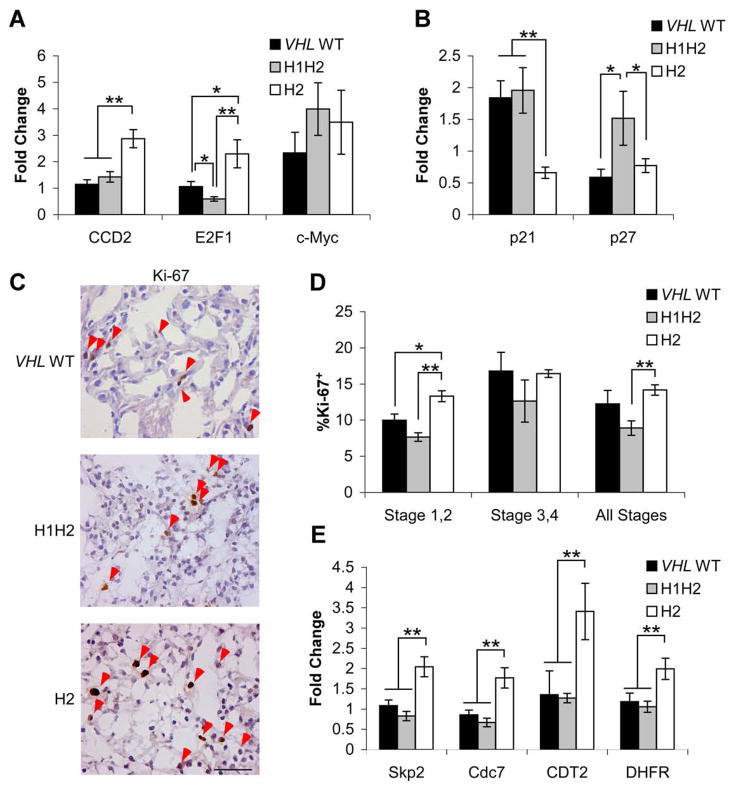
Based on results from cell culture models (Gordan et al., 2007a), we hypothesized that H2 tumors would display evidence of increased c-Myc activity. Indeed, expression of c-Myc-activated targets Cyclin D2 and E2F1 was 2–3 fold higher in H2 tumors than H1H2 or VHL WT tumors when measured by QRT-PCR (Fig. 2A). Similarly, c-Myc-repressed targets p21 and p27 were decreased in H2 tumors (Fig. 2B). To rule out alterations in overall c-Myc expression levels as an explanation for these results, c-Myc mRNA and protein abundance were measured, and no consistent difference was found between groups (Fig. 2A and data not shown), nor were significant c-Myc amplifications detected with Q-PCR (Supplemental Table 1). Ki-67 staining was performed to quantify cell proliferation (Fig. 2C), and a 55% increase in Ki-67+ nuclei was observed in H2 relative to H1H2 tumors (Fig 2D). The clearest difference was between low stage tumors, which displayed approximately 60% more Ki-67+ nuclei in H2 tumors than in VHL WT or H1H2 tumors, although the analysis of high stage tumors was limited by the smaller number in this sample set (Fig. 2D). Overall, these data support our hypothesis that HIF-2α activation of c-Myc occurs in human tumor specimens, and correlates with increased tumor cell proliferation. Although HIF-2α is expressed in H1H2 tumors, our previous data show that HIF-1α-mediated c-Myc inhibition directly opposes HIF-2α-mediated c-Myc activation (Gordan et al., 2007a). Consistent with these data, H1H2 tumors displayed levels of c-Myc target gene expression similar to that in VHL WT tumors.
Figure 2. Upregulation of c-Myc cell cycle targets and proliferation in H2 tumors.
A. Expression of c-Myc and c-Myc activated targets measured by QRT-PCR. Cyclin D2, E2F1 and c-Myc in VHL WT (n=5), H1H2 (n=8) and H2 (n=8) tumors. Data are shown as a fold change relative to pooled normal renal epithelium, ±1 SEM. * p<.05, **p<.01. c-Myc expression was tested with two independent primer sets, as it showed a trend towards upregulation (FDR =.2) in VHL-deficient vs. VHL WT tumors by microarray analysis. c-Myc expression was highly variable within tumor groups, and not found significant by QRT-PCR (p>.25). B. Expression of c-Myc repressed targets p21 and p27, shown as in part A. C. Representative Ki-67 staining in fresh frozen tumors, with positive nuclei indicated with red arrows. Scale bar is 0.5 μM. D. Summary of Ki-67 staining from Stage 1,2 (n= 4 VHL WT, 9 H1H2 and 8 H2) Stage 3,4 (n= 2 VHL WT, 3 H1H2 and 3 H2) or both combined. Data are shown as mean percentage Ki-67 positive ±1 SEM. *p<.05, **p<.01. E. Expression of c-Myc activated targets and G1-S transition mediators Skp2, CDC7, CDT2 and DHFR, analyzed as above.
To extend these analyses in an unbiased manner, mRNA expression profiling was performed to identify global transcriptional effects of differential HIF-α expression. VHL WT tumors were included, both as a comparison group and to identify unique markers of ccRCCs lacking HIF-α expression. Five VHL WT, 8 H1H2 and 8 H2 tumors were analyzed. These tumors were selected to represent a similar range of early and advanced lesions in each group, and for optimal RNA quality. Comparisons were performed between VHL WT and VHL-deficient tumors, and between H1H2 and H2 tumors, identifying a large number of differentially expressed genes (Supplemental Table 2), a subset of which are discussed further here. These comparisons revealed that H2 tumors express elevated levels of additional c-Myc responsive transcripts involved in the G1-S phase cell cycle transition (Coller et al., 2000; O’Connell et al., 2003), including Skp2, Cdc7, CDT2, and dihydrofolate reductase (DHFR) (Fig. 2E and Supplemental Table 2). In contrast, VHL WT and H1H2 tumors expressed lower, but relatively similar, levels of these targets, as discussed above. IHC for Skp2 (Supplemental Fig. 2A,2B) and immunoblotting for Skp2 and Cdc7 (Supplemental Fig. 2C) confirmed that changes in mRNA levels were reflected in protein expression. The c-Myc antagonist Mxi was upregulated in VHL-deficient tumors (Corn et al., 2005; Zhang et al., 2007), consistent with decreased expression of c-Myc regulated mitochondrial enzyme gene expression relative to VHL WT tumors (Supplemental Table 2). However, Mxi appeared to have limited impact on other c-Myc targets, as c-Myc regulated cell cycle genes were induced in H2 tumors, but not VHL WT tumors, despite increased Mxi expression.
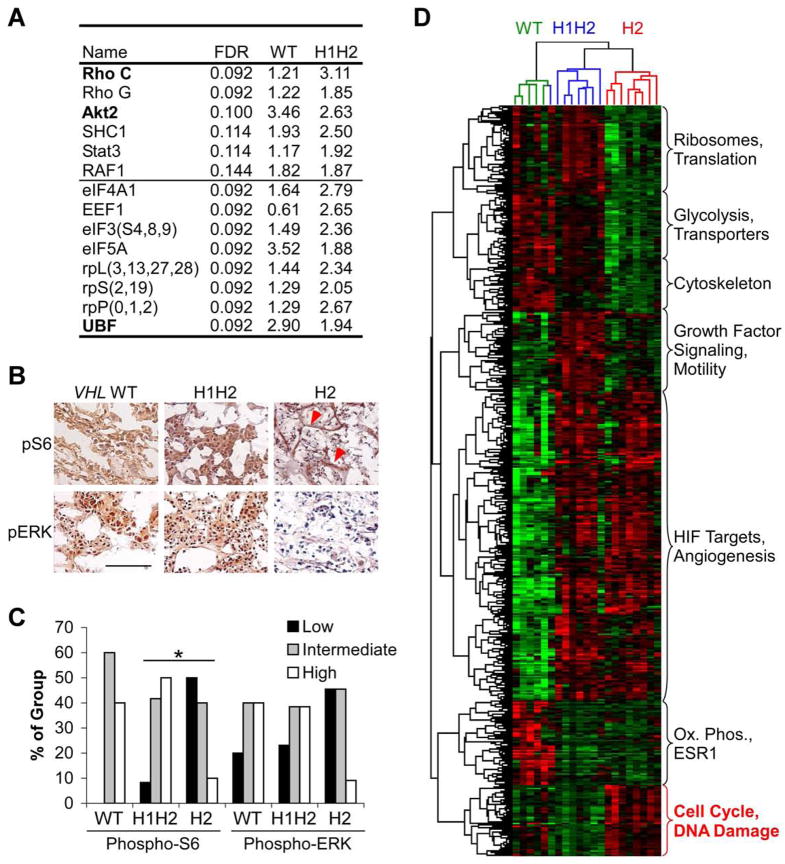
In striking contrast to c-Myc activation in H2 tumors, H1H2 tumors displayed increased expression of the growth factor signaling molecules Akt2 and RhoC, as well as ribosomal L, S and P proteins and the rRNA transcriptional regulator upstream binding factor (UBF). Several of these genes were also expressed at higher levels in VHL WT tumors relative to H2 samples (Fig. 3A). Intriguingly, we also noted increased expression of genes encoding cytoskeletal proteins in VHL WT and H1H2 tumors. This may be a result of enhanced growth factor signaling, given the impact of growth factor responsive proteins such as Rho C on cell motility. These changes in mRNA levels could represent an adaptive mechanism in H1H2 tumors, as VHL loss is associated with decreased matrix deposition and loss of cell polarity (Kim and Kaelin, 2004). To assess growth factor signaling more directly, we performed IHC for phosphorylated extracellular regulated kinase (ERK, also referred to as mitogen-actived protein kinase 1, or MAPK1) and ribosomal protein S6, a target of the mTOR substrate p70 S6 Kinase (Fig. 3B). These proteins are critical signaling molecules whose phosphorylation is induced by multiple growth factor signal transduction pathways. Increased phospho-S6 and phospho-ERK staining was observed in VHL WT and H1H2 compared to H2 tumors (p<.05 for H1H2 vs. H2, Fig. 3C). In H2 tumors, detectable phospho-S6 immunostaining was not observed in tumor cells, but rather restricted to stromal cells associated with blood vessels (Fig. 3B, red arrows). These findings define a functional distinction between ccRCC subsets: whereas c-Myc appears to drive proliferation in H2 tumors, VHL WT and H1H2 tumors utilize growth factor signaling pathways acting on ERK and S6.
Figure 3. Microarray analysis defines separable phenotypes in VHL WT, H1H2 and H2 tumors.
A. Genes associated with growth factor signaling and protein translation significantly upregulated in VHL WT and H1H2 tumors relative to H2 tumors. Statistical significance was measured by false discovery rate (FDR), and fold differences relative to H2 are shown. Gene names shown in bold have been confirmed by QRT-PCR in VHL WT (n=5), H1H2 (n=8) and H2 (n=8) tumors. Results for eIF3, and ribosomal protein S, L and P subunits are the average of genes shown. B. Representative IHC on fresh frozen VHL WT, H1H2 and H2 tumors for phospho-ERK and phospho-S6. Scale bar is 5 μM. C. Summary of phospho-ERK and phospho-S6 levels in VHL WT, H1H2 and H2 tumors, scored as weak, intermediate or strong based on intensity of staining. n=5, 12, and 11 respectively, p<.05. D. Two-way complete linkage clustering of significantly altered genes between each subgroup identifies both tumors and genes that show similar patterns of expression. Tumors are identified by color, with VHL WT highlighted in green, H1H2 in blue, and H2 in red. Red indicates higher levels of expression. The gene set involved in cell cycle and DNA damage responses are highlighted as this group was selected for further study.
As expected, the expression profiling experiments also revealed increased expression of multiple shared HIF-1α and HIF-2α targets (vascular endothelial growth factor, andrenomedullin, etc.) in all VHL-deficient tumors. Similarly, H1H2 tumors (but not H2 tumors) expressed elevated levels of HIF-1α specific target genes, such as those encoding glycolytic enzymes (Hu et al., 2003) (Supplemental Table 2). When two-way clustering was performed on the microarray data, tumors largely segregated into the same groups defined prospectively by HIF-α expression (Fig. 3D). Gene ontology analysis (Ficenec et al., 2003) also identified classes of differentially expressed genes in addition to those discussed here (Fig. 3D). Thus, the key features of each tumor subtype were consistently observed within groups, with activation of c-Myc targets detected exclusively in H2 tumors, in contrast to upregulated growth factor signaling components in VHL WT and H1H2 tumors.
HIF-2α limits DNA damage accumulation
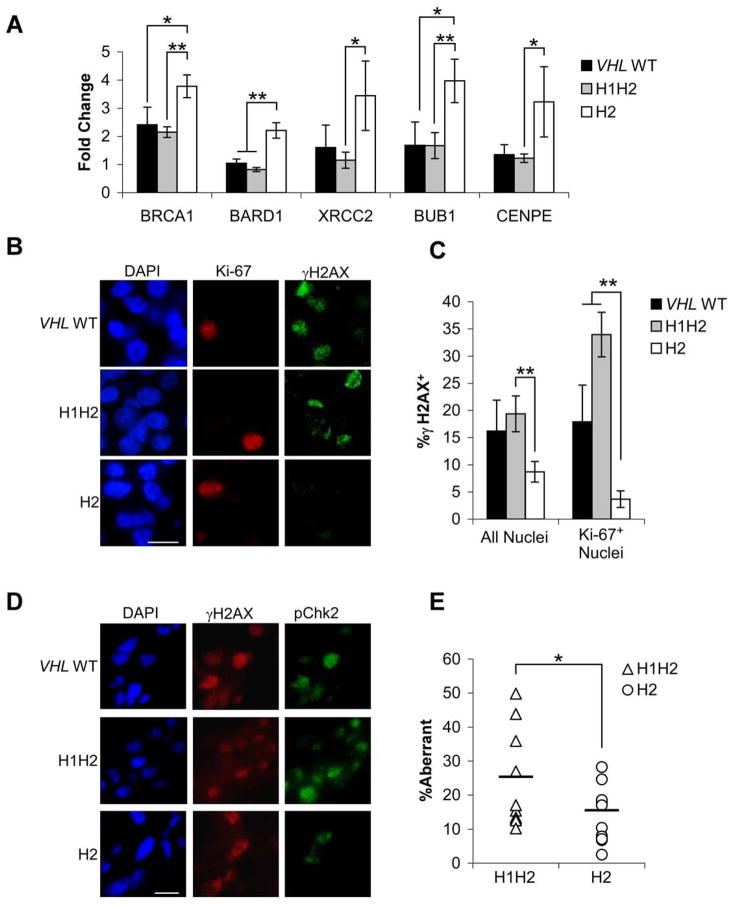
DNA damage from replication stress is commonly observed in hyperproliferative tumor cells, and can trigger a signaling pathway including ATR, ATM, phospho-Chk1, phospho-Chk2 and phospho-histone H2AX (γH2AX) (Bartkova et al., 2005; Gorgoulis et al., 2005), potentially resulting in cell cycle checkpoint activation. The ability to repair DNA damage through homologous recombination (HR) and non-homologous end joining (NHEJ) overcomes these responses and promotes cellular proliferation. In our expression profiling studies, we observed enhanced expression of the HR effectors BRCA1, BARD1, and XRCC2, as well as spindle assembly checkpoint genes BUB1 and CENPE, in H2 tumors compared to VHL WT and H1H2 tumors (Supplemental Table 2). We confirmed these findings with QRT-PCR (Fig. 4A). Immunofluorescence for γH2AX and Ki-67 revealed that H2 tumors contained 50% fewer strongly γH2AX+ nuclei than VHL WT or H1H2 tumors, with a clear difference in γH2AX+/Ki-67+ nuclei (H2 tumors <20% of VHL WT or H1H2) (Fig. 4B,C). Phospho-Chk2 staining also correlated to γH2AX staining patterns (Fig. 4D). The dramatic decrease in γH2AX+/Ki-67+ nuclei in H2 tumors strongly suggests that they accumulate less DNA damage during replication. DNA damage was directly measured by assessing the percentage of the genome that was aberrant (amplified or deleted) in 10 H1H2 and 11 H2 tumors by array-based Comparative Genomic Hybridization. Of note, a statistically significant 40% reduction was noted in H2 tumors (Figure 4E).
Figure 4. H2 tumors exhibit decreased accumulation of γH2AX and genomic aberrancy.
A. Enhanced expression of genes associated with HR and the spindle assembly checkpoint in H2 tumors. Data from VHL WT (n=5), H1H2 (n=8) and H2 (n=8) tumors are shown as a fold change relative to pooled normal renal epithelium, ±1 SEM. * p<.05, **p<.01. B. Representative γH2AX/Ki-67 co-staining in fresh frozen VHL WT, H1H2 and H2 only tumors. DAPI was used to identify nuclei; scale bar is 0.2 μM. C. Quantification of γH2AX staining in all cells or Ki-67+ cells from VHL WT, H1H2 and H2 tumors (n= 6, 12, and 11 respectively), ±1 SEM. * p<.05, **p<.01. Significant differences also were observed for H1H2 and H2 between all nuclei and Ki-67+ nuclei (p<0.05). D. Characteristic phospho-Chk2/γH2AX co-staining in a VHL WT, H1H2 and H2 tumor; scale bar is 0.2 μM. E. Measurement of copy number by Illumina SNP arrays in H1H2 and H2 tumors shows a significantly lower percentage of the genome is aberrant in H2 tumors (p<.04).
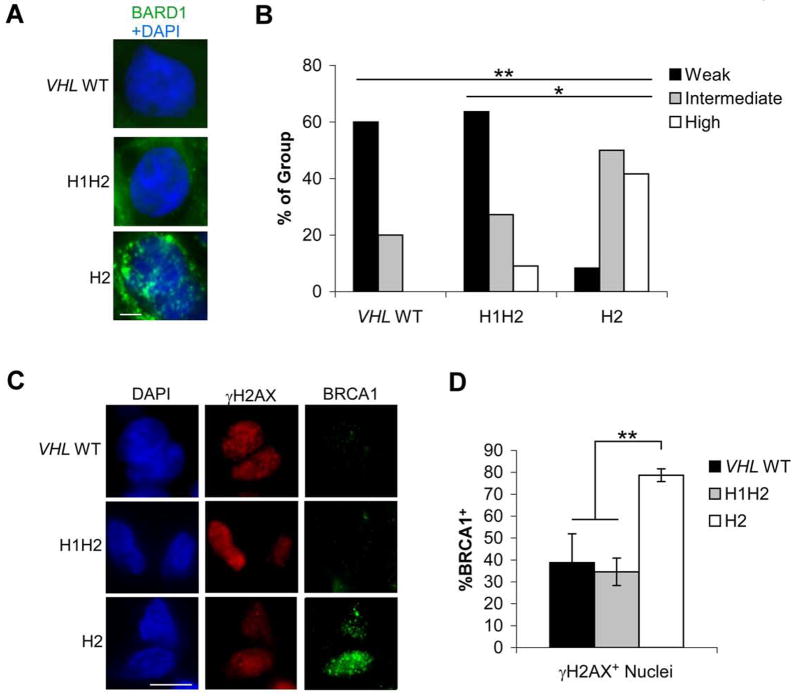
As our expression data implicated HR effectors in DNA damage responses in H2 tumors, we assessed protein abundance and function by immunofluorescence. When tumor sections were stained for BARD1, expression was detected at homogeneous levels within each section, but differences were noted in the number of foci between tumors (Fig. 5A). There was a statistically significant increase in the number of H2 tumors with moderate to high numbers of BARD1 foci (defined as 15–30 and >30 foci/nucleus respectively) compared to H1H2 tumors. This difference was also significant when H2 tumors were compared to H1H2 and VHL WT tumors grouped together (Fig. 5B). Whereas BARD1 staining tended to be homogeneous within tumor sections, BRCA1 staining (Fig. 5C) was more variable between nuclei for a given tumor. BRCA1 was present in a higher proportion of all nuclei in H2 tumors, including those that were γH2AX+ (Fig. 5D), suggesting that HR activity reduces the accumulation of stalled and collapsed replication forks in H2 tumors.
Figure 5. H2 tumors show enhanced signs of HR-mediated repair.
A. Representative BARD1 staining with DAPI to indicate nuclei in VHL WT, H1H2 and H2 tumors. Scale bar is 0.1 μM B. Quantification of BARD1 nuclear staining in 4, 11, and 12 VHL WT, H1H2 and H2 tumors. Weak staining was scored as 0–15, intermediate as 15–30 and strong as >30 foci/nucleus. C. Representative BRCA1/γH2AX co-staining in VHL WT, H1H2 and H2 tumors; scale bar is 0.1 μM. D. Quantification of BRCA1 foci in γH2AX+ nuclei from VHL WT, H1H2 and H2 tumors, ±1 SEM. * p<.05, **p<.01.
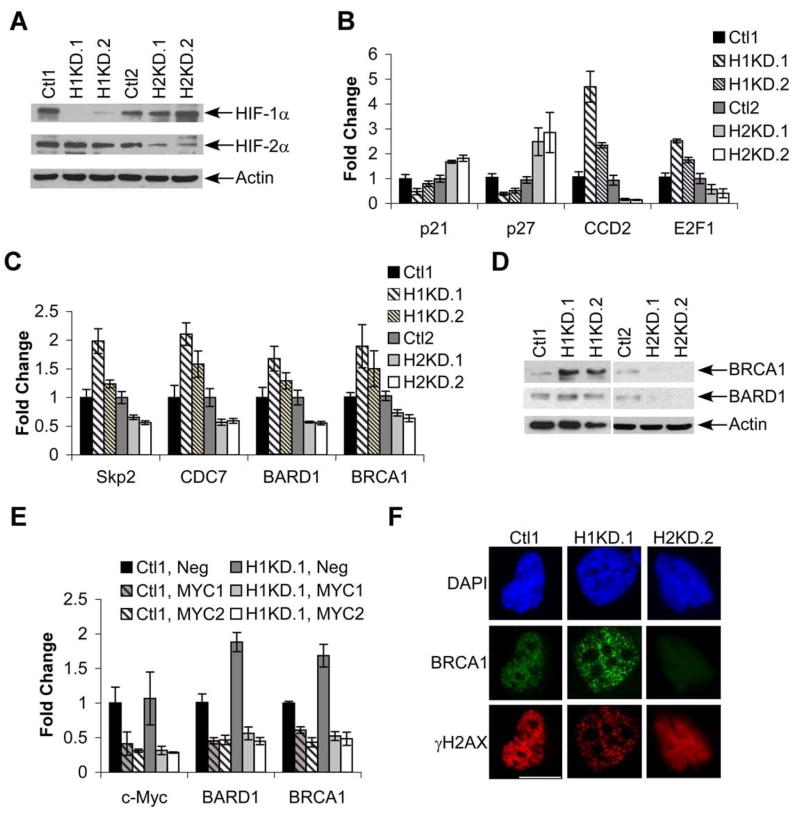
To test the role of HIF-α subunits in HR effector expression directly, we employed a VHL-mutant ccRCC cell line (RCC4) that expresses both HIF-1α and HIF-2α. Vector-transduced control cells were compared to HIF-1α knockdown (H1KD) cells (“H1KD.1” and “H1KD.2”) or HIF-2α knockdown (H2KD) cells (“H2KD.1” and “H2KD.2”) (Fig. 6A and Supplemental Figure 3A). Expression of a common HIF-1α and HIF-2α target gene (VEGF), a HIF-1α specific target gene (PGK), and a HIF-2α specific target gene (Oct-4) (Covello et al., 2006; Hu et al., 2003) was assessed to confirm functional HIF-α knockdown in these cells (Supplemental Fig. 3B). H1KD cells retain HIF-2α activity, and displayed enhanced c-Myc activity with elevated cyclin D2 and E2F1 expression, and decreased p21 and p27 expression. In contrast, decreased c-Myc activity was observed in H2KD cells, which retain HIF-1α expression (Fig. 6B). Similarly, HIF-1α knockdown promoted S-phase entry, enhancing proliferation, whereas HIF-2α knockdown caused accumulation in G1 and limited proliferation (Supplemental Fig. 3C, 3D). Expression of other c-Myc targets, including Skp2, CDC7, BARD1, and BRCA1 was found to be stimulated by HIF-1α knockdown (Fig. 6C) and inhibited by HIF-2α knockdown, consistent with our observations using primary ccRCCs. Although these mRNA changes are modest, they are likely to have additive effects. In the case of BRCA1, they also correspond to more dramatic changes in protein expression (Fig. 6D). HIF-1α and HIF-2α effects on BRCA1 and BARD1 mRNA expression were c-Myc-dependent, based on siRNA knockdown of c-Myc expression (Fig. 6E), consistent with previous studies showing that c-Myc stimulates BRCA1 expression (Bindra et al., 2005; Koshiji et al., 2004). Thus, manipulating the stoichiometry of HIF-1α and HIF-2α in RCC4 cells can modulate c-Myc activity, resulting in altered expression of HR mediators.
Figure 6. HIF-2α/c-Myc effects drive enhanced HR effector expression.
A. HIF-1α and HIF-2α expression in control and knockdown cell lines. “H1KD.1” and “H1KD.2” exhibit HIF-1α knockdown and “H2KD.1” and “H2KD.2” exhibit HIF-2α knockdown. Actin is shown as a loading control. See Supplemental Fig. 1A for changes in HIF-1α and HIF-2α mRNA levels in control and knockdown cells. B. Differential expression of c-Myc activated (Cyclin D2, E2F1) and repressed (p21, p27) targets in control and knockdown cell lines. Averages are shown from 4 experiments ±1 SEM. C. Expression of G1-S phase cell cycle targets and HR genes in knockdown cell lines, analyzed as in part B. D. Western blot analysis of BRCA1 and BARD1 in control and knockdown cell lines; actin is shown as a loading control. E. QRT-PCR for expression of c-Myc, BRCA1 and BARD1 in control and HIF-1α knockdown RCC4 clones transfected with control siRNA or two different siRNAs against c-Myc. Average values from 3 experiments, ±1 SEM. F. Representative images of BRCA1/γH2AX co-staining in control and HIF-α knockdown cell lines after 3 hr. treatment with 1.5 mM HU. Results from one control line are shown as both were equivalent results. Scale bar is 0.2 mM.
H1KD and H2KD cells were also evaluated for their response to hydroxyurea (HU)-induced replication stress. HU blocks nucleotide synthesis and inhibits DNA replication, resulting in replication fork stalling and ATR-mediated Chk1 phosphorylation. Failure to resolve stalled replication forks leads to their collapse and activation of ATM, Chk2 and p53, which can promote cell cycle arrest or senescence (Branzei and Foiani, 2007; Wang, 2007). Resolution of stalled forks requires HR, which is known to occur in BRCA1 nuclear foci (Wang, 2007). HU-treated H1KD (HIF-2α expressing) cells displayed >50 BRCA1 foci per nucleus, whereas <10 foci were detected in H2KD (HIF-1α expressing) cells. We also noted that H1KD cells exhibited less dramatic γH2AX staining when compared to control or H2KD cells (Fig. 6F). In each case, BRCA1 and γH2AX foci displayed overlapping nuclear distributions (Fig. 6F). These data are consistent with the large number of BRCA1 foci observed in H2 tumors, and suggest a specific role for HIF-2α in DNA damage responses in ccRCCs.
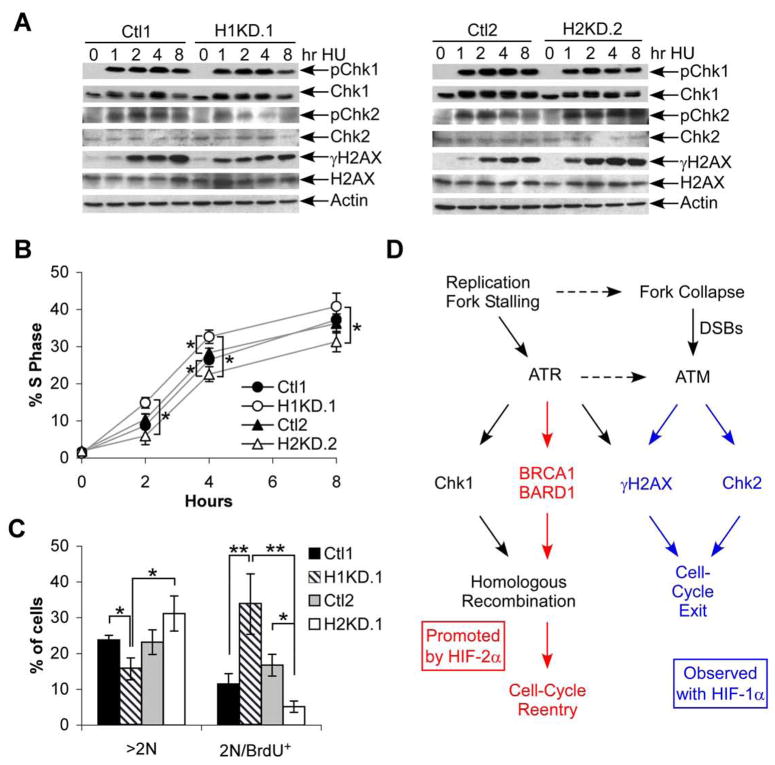
Functional consequences of differential DNA damage signaling in HIF-α knockdown cells were examined. HU treatment of cells synchronized in S-phase induced rapid Chk1 phosphorylation in all lines, suggesting equivalent activation of the intra-S-phase checkpoint (Fig. 7A). In contrast, Chk2 phosphorylation and γH2AX were decreased in H1KD cells compared to control cells, mirroring the differences observed between H2 and H1H2 tumor specimens (Fig. 7A, left panel), while the opposite result was observed in H2KD cells (Fig. 7A, right panel). Interestingly, H1KD cells returned more rapidly to cell cycle progression after HU treatment, whereas H2KD cells recovered more slowly (Fig. 7B). These data support the notion that HIF-2α, in contrast to HIF-1α, promotes more efficient HR-mediated repair and resolution of replication stress, reducing the accumulation of DNA damage and activation of checkpoint responses.
Figure 7. HIF-2α/c-Myc effects correlate with resistance to replication stress.
A. Western blot analysis of Chk1, Chk2 and γH2AX phosphorylation following 1.5 mM HU treatment of control and HIF-α knockdown cell lines synchronized in S-phase by timed release from serum withdrawal and confluency. Total Chk1, Chk2, H2AX and actin are shown as controls. B. Return to DNA replication following 1 hr. treatment of S-phase synchronized cells with 1.5 mM HU, measured by BRDU incorporation. Average percent BRDU+ are shown from 3 experiments ±1 SEM. Statistically significant differences between Ctl1 and H1.1, Ctl2 and H2.2, and H1.1and H2.2 were assessed, * p <.05. C. BRDU incorporation in cells grown for 20 hr. in the presence of 1 μg/ml Aphidicolin. Percentage of cells >2N DNA content is shown, as is the 2N/BRDU+ percentage. Data from one representative experiment, ±1 SD, * p<.05, **p<.01. D. Simplified model outlining responses to replication stress. Various stress inducers have been shown to activate ATR leading to Chk1 phosphorylation and DNA repair, with replication fork collapse activating ATM, enhancing γH2AX accumulation and promoting cell cycle exit. These data are consistent with a model where HIF-2α promotes the former pathway (highlighted in red) and HIF-1α expressing cells tend to exhibit the latter (highlighted in blue).
We used an independent experimental approach to evaluate the effect of HIF-α expression on cell cycle progression under conditions of replication stress. Cells were incubated in a low concentration of aphidicolin (1 μg/ml) for 20 hours, a concentration at which aphidicolin reduces DNA polymerase progression through complex genomic structures and can activate an intra-S-phase checkpoint characterized by cells with hyper-diploid DNA content (>2N). In RCC4 cells, HIF-1α knockdown promoted successful completion of S-phase, indicated by a significant decrease in the proportion of >2N cells compared to control lines (Fig. 7C), and consistent with the hypothesis that these cells respond more effectively to DNA replication stress. In contrast, H2KD cells showed a trend toward failure to complete S-phase (p=.06). Interestingly, cells completing G1 in the presence of low-dose aphidicolin treatment are capable of firing early origins of replication and incorporating BRDU, although DNA synthesis at newly fired origins is severely diminished (Dimitrova and Gilbert, 2000); consequently, these cells appear as BRDU-positive with 2N DNA content (2N/BRDU+). H1KD and H2KD cells were treated with aphidicolin as described above, and then pulsed with BRDU for 20 minutes to label newly fired origins in cells that had successfully completed G1. H1KD cells showed a significantly higher rate of replication initiation compared to controls, reflected as 2N/BRDU+ cells, whereas H2KD cells showed significantly lower rates (Fig. 7C). In summary, HIF-2α expression (in the absence of HIF-1α was correlated with increased BRCA1 expression and efficient S-phase completion, thereby permitting cell cycle progression in the face of replication stress while limiting phospho-Chk2 and γH2AX accumulation. Conversely, HIF-1α expression was associated with elevated markers of DNA damage and a limited ability to proliferate when challenged with replication stress, in keeping with the model presented in Figure 7D. Importantly, results from these functional in vitro studies using replication stress mimetics are consistent with our observations of DNA stress markers in ccRCC tumors.
Validation of expression and signaling changes in additional tumor samples
Having identified HIF-α-intrinsic effects on cell cycle and DNA repair pathways in human tumor specimens, we sought to confirm them in two independent sets of patient samples. Microarray analysis was performed on a second group of 12 ccRCCs, collected at the University of North Carolina. All tumors were scored as VHL-deficient because they showed strong HIF-α staining in >75% of cells, significant upregulation of multiple HIF targets, and decreased VHL expression compared to controls. Samples were separated into H1H2 and H2 subgroups (n=7 and 5, respectively), and differentially expressed genes analyzed. As described above, tumors clustered by HIF-α expression (Supplemental Fig. 4A), exhibiting gene expression consistent with increased glycolytic metabolism and growth factor signaling in H1H2 tumors, and enhanced cell cycle progression and HR gene expression in H2 tumors. Comparing these results to our primary analysis, we observed a 30–40% concordance between the data sets (p < 1×10−12), commensurate with published reports comparing arrays performed under similar conditions (Subramanian et al., 2005). Ki-67 staining (Supplemental Fig. 4B) was also consistent with data from our initial tumor set. Genes exhibiting increased expression in all H2 tumors are listed in Supplemental Figure 3C, and include Cyclin D2, E2F1, Skp2, DHFR, BARD1, and BRCA1.
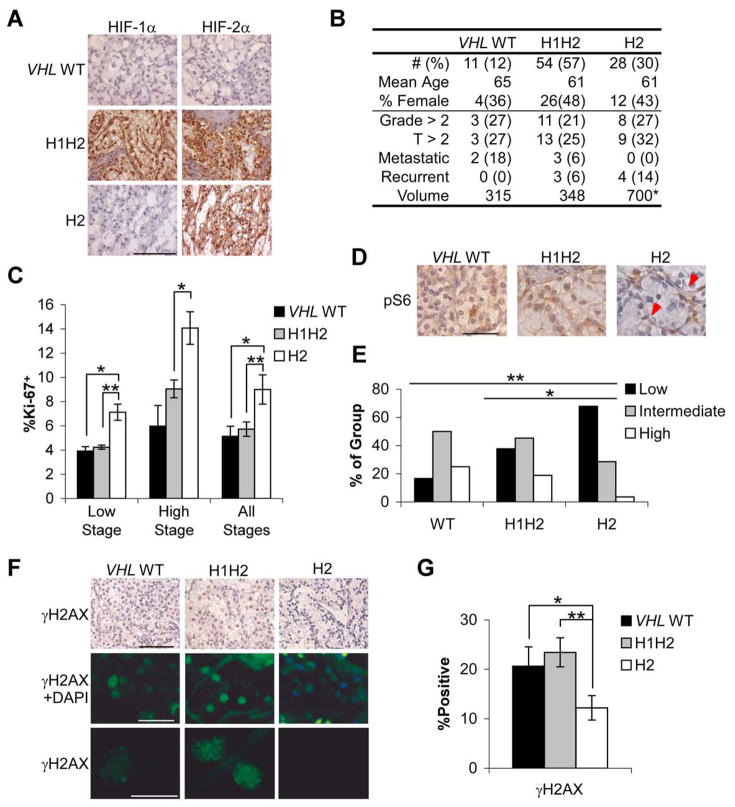
To confirm the biological phenotypes described above, and analyze a data set with information regarding clinical outcome, IHC was performed on a tissue microarray (TMA) containing triplicate samples from 93 randomly selected ccRCCs from the University of Pennsylvania. TMA slides were immunostained for HIF-1α and HIF-2α proteins (Fig. 8A) and a similar distribution of the three groups was observed (Fig. 8B). Tumors were categorized as VHL WT if neither HIF-1α nor HIF-2α staining was observed. The tumors in this data set tended to be of lower stage than the initial group, but a similar proportion was metastatic at presentation. Metastatic relapse was observed in H1H2 and H2 tumors over a median six years of follow up, with comparable overall rates between groups (Fig. 8B). Notably, H2 tumors were significantly larger in volume when compared to VHL WT or H1H2 (Fig. 8B, p <.008 and .04, respectively). Ki-67, phospho-S6 and γH2AX staining was performed, and H2 tumors displayed 60% more Ki-67+ cells relative to H1H2 and VHL WT tumors (Fig. 8C), whereas phospho-S6 (Fig. 8D,E) and γH2AX (Fig. 8F,G) staining was significantly higher in both VHL WT and H1H2 compared to H2 tumors. These data extended our analysis of HIF-α effects to advanced tumors, which had not previously been possible in the smaller initial tumor collection. These two data sets (105 additional samples) confirm and extend the phenotypic characterization of the initial ccRCC subgroups (Fig. 1), as well as the increased expression of specific gene targets in H2 tumors (Supplemental Table 2) associated with increased cell cycle progression and HR.
Figure 8. TMA analysis of biological parameters and patient outcome.
A. Representative HIF-1α and HIF-2α staining from paraffin-embedded TMA cores. Scale bar is 2 μM B. Summary of patient information and clinical outcomes across IHC determined groups. Volume was measured in CM3, * p<.05, **p<.01. C. Summary of Ki-67 staining from low and high stage tumors, or both combined. Data are shown as mean percentage Ki-67 positive ±1 SEM. * p<.05, **p<.01. D. Representative phospho-S6 staining. VHL WT and H1H2 sections shown were scored as intermediate, and the H2 section as negative, though endothelial cell phospho-S6 can be noted (red arrows). Scale bar is 0.5 μM E. Summary of phospho-S6 staining in VHL WT, H1H2 and H2 tumors, * p<.05, **p<.01. F. Representative γH2AX shown with DAB and fluorescent staining. Middle row also shows DAPI, so that negative nuclei can be appreciated, whereas lower row shows only γH2AX. Scale bars are 2, 0.5 and 0.2 mM G. Quantification of fluorescent γH2AX staining in all tumors, ±1 SEM. * p<.05, **p<.01.
Discussion
Human ccRCCs are typically subdivided into two clearly distinct groups based on VHL status. Approximately 70% of sporadic ccRCCs have lost pVHL expression through VHL deletion and/or silencing (Banks et al., 2006; Kim and Kaelin, 2004), but additional criteria by which pVHL-deficient tumors can be classified have not been described. Here, we demonstrate that VHL-deficient ccRCCs can be distinguished on the basis of differential HIF-α expression, and consequent HIF-dependent effects on c-Myc activity. Specifically, we found that H2 tumors displayed enhanced c-Myc activity, higher rates of proliferation, increased volume and lower levels of γH2AX accumulation than H1H2 or VHL WT tumors. In contrast, H1H2 and VHL WT tumors displayed increased activation of Akt/mTOR and ERK/MAPK1 growth factor signaling pathways and enhanced γH2AX accumulation. Although VHL WT and H1H2 tumors generally share these traits, prior work (Turner et al., 2002) and our expression profiling data strongly suggest that H1H2 tumors are more angiogenic than VHL WT tumors. This study presents evidence of differential HIF/c-Myc effects in patient-derived tumor samples, and describes a previously unappreciated interplay between HIF-2α, c-Myc and genome stability. Together, these data provide a mechanistic basis to subdivide ccRCC for molecularly targeted therapy.
The in vitro and in vivo findings presented here extend HIF-2α/c-Myc cell cycle effects to include promotion of efficient transit through S-phase by stimulating expression of HR mediators, as well as genes encoding proteins that limit replication stress by enhancing nucleotide pools (DHFR) (Milbrandt et al., 1981) or blocking re-replication (Skp2 and Cdt2) (Nishitani et al., 2006). As HIF-1α and HIF-2α have opposite effects on c-Myc, our findings are consistent with previous observations that HIF-1α inhibits mismatch repair by blocking c-Myc activity (Koshiji et al., 2005). It should be noted, however, that hypoxia directly promotes genomic instability in a HIF-1α independent fashion (Bristow and Hill, 2008). Thus, tumors expressing only HIF-2α may be able to compensate for hypoxia-induced genomic instability. Intriguingly, our findings suggest that Mxi upregulation by HIF-1α and HIF-2α (Zhang et al., 2007) exerts a specific effect on mitochondrial metabolism, without impacting several other c-Myc driven processes. These results delineate the in vivo impact of HIF/c-Myc effects in ccRCC, and highlight contrasting properties of HIF-1α and HIF-2α on c-Myc-regulated DNA damage repair.
The decreased levels of γH2AX and increased numbers of Ki-67+ nuclei observed in H2 tumors reveal a correlation between enhanced proliferation and decreased levels of DNA damage. Given the concomitant upregulation of HR effectors BRCA1 and BARD1, these findings implicate enhanced resolution of replication stress as a mechanism by which tumor cells escape the cell cycle block imposed by DNA damage-activated checkpoints. The fact that HIF-2α-expressing RCC4 cells, which display enhanced BRCA1 and BARD1 expression, recover more rapidly from HU and aphidicolin further supports the hypothesis that enhanced HR contributes to tumor cell proliferation in the context of replication stress. Similarly, a key role for HR in normal cell cycle progression has been suggested by targeted mutation of murine BRCA1 and BARD1, as disruption of either gene dramatically limits proliferation in embryonic cells (McCarthy et al., 2003; Xu et al., 1999). Differing levels of replication stress could underlie the variable activation of DNA damage response machinery observed in different tumor types. For example, whereas tumors of the lung, bladder, and skin appear dependent on stepwise dismantling of DNA damage responses for progression (Bartkova et al., 2005; Gorgoulis et al., 2005), recent studies suggest that stomach, colorectal and breast adenocarcinoma, as well as testicular germ cell tumors, have minimal DNA damage response activation at all stages (Bartkova et al., 2007; Nuciforo et al., 2007). More efficient progression through S-phase has not been previously described as a pathway for enhanced tumor cell proliferation, and is likely to be of particular importance in early stage tumor cells with intact DNA damage checkpoints.
The combination of VHL genotype and HIF-α expression allows the stratification of ccRCCs into biologically distinct groups, and suggests a framework for sub-classifying tumors for targeted therapies. Although several targeted drugs with distinct mechanisms of action are available to treat metastatic ccRCC, there are currently no parameters to select the optimal drug for each patient. Sunitinib and sorafenib each appear to inhibit tumor angiogenesis and tumor cell viability through their effects on VEGF signaling in endothelial cells and, potentially, tumor cells. These agents differ with respect to the potency of their interactions with VEGF and PDGF receptors and most notably with respect to the Raf/MAP kinase pathway, an exclusive target of sorafenib. We suggest that growth factor driven ccRCCs (VHL WT and H1H2) are more likely to respond to these drugs. Similarly, while treatment with erlotinib (targeting the epidermal growth factor receptor) and bevacizumab (inhibiting angiogenesis) conferred no benefit as compared to bevacizumab alone in unselected patients (Motzer and Bukowski, 2006), this combination might exclusively show increased efficacy against the H1H2 subset. Furthermore, whereas the anti-angiogenic effects of sorafenib and sunitinib should impact vascularity in H2 tumors, they are nevertheless likely to progress, as HIF-2α dependent c-Myc activation could promote growth in relatively well-oxygenated tumor subdomains. Expression of HIF-2α alone may therefore mark a subset of RCC that is uniquely resistant to the current targeted therapeutics. While retrospective and prospective studies with associated clinical trial data are needed to demonstrate a connection between HIF-α expression and therapeutic response, HIF-α analysis enables us to stratify ccRCC patients and thus examine predictors of outcome based on biological differences in clinical trials of targeted therapies.
Materials and Methods
Patient Material
Frozen material for the primary analysis was obtained through the Collaborative Human Tissue Network. OCT embedded blocks were sectioned until cut planes were >70% tumor. Sections were collected for DNA, RNA and protein extraction. Material for TMA analysis was obtained from archival specimens at the University of Pennsylvania. Both protocols were approved by the IRB at the University of Pennsylvania. Samples were catalogued, clinical information on cases was obtained through chart review, and patient identifiers were removed before analysis. Material for microarray validation was collected at the University of North Carolina from surgeries performed on site. An approval was given by the IRB at UNC.
VHL Sequence and Methylation Analysis
DNA was extracted from tumor samples using the EX-WAX DNA extraction kit (Chemicon). Mutation screening was performed on PCR amplified exons by direct sequencing with BigDyeTerminator Cycle kit on a 3130xl sequencer (Applied Biosystems). Primers and protocols were used as previously described (Stolle et al., 1998). Methylation studies were performed using the CpG Wiz Kit (Chemicon, Inc) and/or Not1 digestion (Herman et al., 1994).
Immunohistochemistry
For frozen specimens, 10 μM sections were fixed with 4% paraformaldehyde, permeabilized with PBS/.5% Triton X-100 and stained following standard protocols, detailed further in the Supplemental text. For HIF-α staining, slides were scored as positive if more than 40% of nuclei stained positive. Cytoplasmic staining was not considered. For Ki-67, Skp2, γH2AX, phospho-Chk2, and BRCA1, 3–4 high power images were counted for percentage positive, and then averaged to give a mean rate per tumor, except for Ki-67 on TMA cores, where the entire core was counted in each case. Only cells with clear tumor cell morphology were scored. For phospho-ERK, phospho-S6, and BARD1, samples were scored as negative/weak, intermediate or strong. For these and HIF-α staining, analysis was reviewed by multiple investigators, including J.D.G., P.L. and K.N.P.
TMA Design and Production
94 ccRCC samples were selected from tissue archives at University of Pennsylvania, representing 93 primary tumors and one metastasis. H&E stained sections were evaluated to confirm the initial diagnosis, and three 0.6 mm tissue cores were selected from different areas of each tumor. Matched normal renal epithelium was included for 2/3 of cases, and renal angiomyolipomas and spleen cores were also included as staining controls. Cores were assembled and cut into sections as previously described (Kononen et al., 1998).
Microarrays
Primary expression microarray analysis was performed at the University of Pennsylvania Microarray Core using the Affymetrix U133 Plus 2.0 array, with analysis using Gene Pattern (Broad Institute) (Reich et al., 2006). Array-based Comparative Genomic Hybridization (aCGH) was also performed at the University of Pennsylvania Microarray Core using the Illumina 550K-2 v3.1 array and analyzed with Partek Genomics Suite, while the validation set of expression arrays was hybridized at the University of North Carolina Genomics Core using the Agilent 4×44K array format, with a standard reference RNA used in the second channel and analyzed with Statistical Analysis of Microarrays (SAM). Analytical methods are described in the Supplemental Methods.
Statistical Analysis
Significance was typically evaluated by two-tailed student’s T test. When t tests were performed on data from multiple measurements (i.e., counting 4 images of one slide), values were averaged by tumor before inclusion in the test. For non-continuous measurements, Fisher’s Exact Test was used, with 2×2 tables except for phospho-S6 and phospho-ERK staining where 2×3 and 3×3 Chi-squared tables were used.
Cell Culture, Drug Treatments and Synchronization
RCC4 cells were cultured in DMEM with 10% FCS (Gemini), standard additives, and 1 ug/ml puromycin. Knockdown clones were generated by retroviral transduction using pBABE HIF-1α shRNA (Lum et al., 2007), and pRETROSUPER HIF-2α shRNA (Kondo et al., 2003) (generous gift of W.G. Kaelin). Hydroxyurea (Sigma) was prepared in water and used at 1.5 mM. Aphidicolin was prepared in DMSO and used at 1 μg/ml. Cells were synchronized in G1 by confluency and serum withdrawal for 24 hours, and released by trypsinization and replating. H1KD, control and H2KD cell lines were released 18, 20 and 22 hours respectively before HU treatment, ensuring 50–60% of cells in S Phase and the remainder in G1. All other in vitro methods are described in Supplemental Text.
Accession Numbers
The raw data for expression profiling and SNP based copy number data are available through the Gene Expression Omnibus (GEO) at http://www.ncbi.nlm.nih.gov/geo/. Expression profiling data is available at GSE11904 (UPenn) and GSE11985 (UNC) and SNP based copy number at GSE13282.
Supplementary Material
Acknowledgments
We are grateful to many members of the Abramson Family Cancer Research Institute, John Tomaszewski and Theresa Pasha of the University of Pennsylvania Pathology Department, and Don Baldwin and John Tobias of the Penn Center for Bioinformatics for advice and discussions. This work was supported by NIH Program Project Grant no. CA104838, the Howard Hughes Medical Institute and the DiBona Foundation. J.D.G. was supported by a Medical Scientist Training Grant and a NRSA T32 grant for Hemostasis and Thrombosis. W.K.R. was supported by R01 CA 01121781 and the V foundation. M.C.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banks RE, Tirukonda P, Taylor C, Hornigold N, Astuti D, Cohen D, Maher ER, Stanley AJ, Harnden P, Joyce A, et al. Genetic and epigenetic analysis of von Hippel-Lindau (VHL) gene alterations and relationship with clinical variables in sporadic renal cancer. Cancer Res. 2006;66:2000–2011. doi: 10.1158/0008-5472.CAN-05-3074. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Sehested M, Nesland JM, Rajpert-De Meyts E, Skakkebaek NE, Stucki M, Jackson S, Lukas J, Bartek J. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene. 2007;26:7414–7422. doi: 10.1038/sj.onc.1210553. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, Bristow RG, Classon MK, Glazer PM. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005;65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst) 2007;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corn PG, Ricci MS, Scata KA, Arsham AM, Simon MC, Dicker DT, El-Deiry WS. Mxi1 is induced by hypoxia in a HIF-1-dependent manner and protects cells from c-Myc-induced apoptosis. Cancer Biol Ther. 2005;4:1285–1294. doi: 10.4161/cbt.4.11.2299. [DOI] [PubMed] [Google Scholar]
- Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, Labosky PA, Simon MC, Keith B. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat Cell Biol. 2000;2:686–694. doi: 10.1038/35036309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficenec D, Osborne M, Pradines J, Richards D, Felciano R, Cho RJ, Chen RO, Liefeld T, Owen J, Ruttenberg A, et al. Computational knowledge integration in biopharmaceutical research. Brief Bioinform. 2003;4:260–278. doi: 10.1093/bib/4.3.260. [DOI] [PubMed] [Google Scholar]
- Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha Promotes Hypoxic Cell Proliferation by Enhancing c-Myc Transcriptional Activity. Cancer Cell. 2007a;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev. 2007;17:71–77. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: Sibling Rivals for Control of Cancer Cell Metabolism and Proliferation. Cancer Cell. 2007b;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Lisztwan J, Barry R, Ballschmieter P, Krek W. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat Cell Biol. 2003;5:64–70. doi: 10.1038/ncb899. [DOI] [PubMed] [Google Scholar]
- Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LE, Bindra RS, Glazer PM, Harris AL. Hypoxia-induced genetic instability--a calculated mechanism underlying tumor progression. J Mol Med. 2007;85:139–148. doi: 10.1007/s00109-006-0133-6. [DOI] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002a;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Kondo K, Yao M, Yoshida M, Kishida T, Shuin T, Miura T, Moriyama M, Kobayashi K, Sakai N, Kaneko S, et al. Comprehensive mutational analysis of the VHL gene in sporadic renal cell carcinoma: relationship to clinicopathological parameters. Genes Chromosomes Cancer. 2002b;34:58–68. doi: 10.1002/gcc.10054. [DOI] [PubMed] [Google Scholar]
- Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, Huang LE. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. Embo J. 2004;23:1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiji M, To KK, Hammer S, Kumamoto K, Harris AL, Modrich P, Huang LE. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Lee AC, Fenster BE, Ito H, Takeda K, Bae NS, Hirai T, Yu ZX, Ferrans VJ, Howard BH, Finkel T. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J Biol Chem. 1999;274:7936–7940. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- Lee S, Nakamura E, Yang H, Wei W, Linggi MS, Sajan MP, Farese RV, Freeman RS, Carter BD, Kaelin WG, Jr, Schlisio S. Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell. 2005;8:155–167. doi: 10.1016/j.ccr.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Lonser RR, Glenn GM, Walther M, Chew EY, Libutti SK, Linehan WM, Oldfield EH. von Hippel-Lindau disease. Lancet. 2003;361:2059–2067. doi: 10.1016/S0140-6736(03)13643-4. [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, Simon MC, Thompson CB. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21:1037–1049. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandriota SJ, Turner KJ, Davies DR, Murray PG, Morgan NV, Sowter HM, Wykoff CC, Maher ER, Harris AL, Ratcliffe PJ, Maxwell PH. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- Maranchie JK, Vasselli JR, Riss J, Bonifacino JS, Linehan WM, Klausner RD. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- McCarthy EE, Celebi JT, Baer R, Ludwig T. Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol Cell Biol. 2003;23:5056–5063. doi: 10.1128/MCB.23.14.5056-5063.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbrandt JD, Heintz NH, White WC, Rothman SM, Hamlin JL. Methotrexate-resistant Chinese hamster ovary cells have amplified a 135-kilobase-pair region that includes the dihydrofolate reductase gene. Proc Natl Acad Sci U S A. 1981;78:6043–6047. doi: 10.1073/pnas.78.10.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer RJ, Bukowski RM. Targeted therapy for metastatic renal cell carcinoma. J Clin Oncol. 2006;24:5601–5608. doi: 10.1200/JCO.2006.08.5415. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. Embo J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuciforo PG, Luise C, Capra M, Pelosi G, d’Adda di Fagagna F. Complex engagement of DNA damage response pathways in human cancer and in lung tumor progression. Carcinogenesis. 2007;28:2082–2088. doi: 10.1093/carcin/bgm108. [DOI] [PubMed] [Google Scholar]
- O’Connell BC, Cheung AF, Simkevich CP, Tam W, Ren X, Mateyak MK, Sedivy JM. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J Biol Chem. 2003;278:12563–12573. doi: 10.1074/jbc.M210462200. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- Rathmell WK, Hickey MM, Bezman NA, Chmielecki CA, Carraway NC, Simon MC. In vitro and in vivo models analyzing von Hippel-Lindau disease-specific mutations. Cancer Res. 2004;64:8595–8603. doi: 10.1158/0008-5472.CAN-04-1430. [DOI] [PubMed] [Google Scholar]
- Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Atkuri KR, Deb-Basu D, Adler AS, Chang HY, Herzenberg LA, Felsher DW. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Res. 2006;66:6598–6605. doi: 10.1158/0008-5472.CAN-05-3115. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Stolle C, Glenn G, Zbar B, Humphrey JS, Choyke P, Walther M, Pack S, Hurley K, Andrey C, Klausner R, Linehan WM. Improved detection of germline mutations in the von Hippel-Lindau disease tumor suppressor gene. Hum Mutat. 1998;12:417–423. doi: 10.1002/(SICI)1098-1004(1998)12:6<417::AID-HUMU8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KJ, Moore JW, Jones A, Taylor CF, Cuthbert-Heavens D, Han C, Leek RD, Gatter KC, Maxwell PH, Ratcliffe PJ, et al. Expression of hypoxia-inducible factors in human renal cancer: relationship to angiogenesis and to the von Hippel-Lindau gene mutation. Cancer Res. 2002;62:2957–2961. [PubMed] [Google Scholar]
- Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- Wang V, Davis DA, Haque M, Huang LE, Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res. 2005;65:3299–3306. doi: 10.1158/0008-5472.CAN-04-4130. [DOI] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 Inhibits Mitochondrial Biogenesis and Cellular Respiration in VHL-Deficient Renal Cell Carcinoma by Repression of C-MYC Activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.