Abstract
Endochondral bone formation is a highly orchestrated process involving coordination among cell-cell, cell-matrix and growth factor signaling that eventually results in the production of mineralized bone from a cartilage template. Chondrogenic and osteogenic differentiation occur in sequence during this process, and the temporospatial patterning clearly requires the activities of heparin binding growth factors and their receptors. Heparanase (HPSE) plays a role in osteogenesis, but the mechanism by which it does so is incompletely understood. We used a combination of ex vivo and in vitro approaches and a well described HPSE inhibitor, PI-88 to study HPSE in endochondral bone formation. In situ hybridization and immunolocalization with HPSE antibodies revealed that HPSE is expressed in the peri-chondrium, peri-osteum, and at the chondro-osseous junction, all sites of key signaling events and tissue morphogenesis. Transcripts encoding Hpse also were observed in the pre-hypertrophic zone. Addition of PI-88 to metatarsals in organ culture reduced growth and suggested that HPSE activity aids the transition from chondrogenic to osteogenic processes in growth of long bones. To study this, we used high density cultures of ATDC5 pre-chondrogenic cells grown under conditions favoring chondrogenesis or osteogenesis. Under chondrogenic conditions, HPSE/Hpse was expressed at high levels during the mid-culture period, at the onset of terminal chondrogenesis. PI-88 addition reduced chondrogenesis and accelerated osteogenesis, including a dramatic up-regulation of osteocalcin levels. In normal growth medium, addition of PI-88 reduced migration of ATDC-5 cells, suggesting that HPSE facilitates cartilage replacement by bone at the chondro-osseous junction by removing the HS component of proteoglycans, such as perlecan/HSPG2, that otherwise prevent osteogenic cells from remodeling hypertrophic cartilage.
Keywords: Heparanase, chondrogenesis, osteogenesis, bone, PI-88, migration
Introduction
Osteogenesis, the development of bone, is a complex process that requires coordination between cell-cell, cell-matrix, and growth factor mediated signaling events that conclude with the ossification and mineralization of bone [1]. In the craniofacial skeleton, mesenchymal cells differentiate into osteoblasts and form bone by intramembraneous ossification. In contrast, in the axial and limb skeleton, mesenchymal cells differentiate into chondrocytes and are later replaced by osteoblasts, a process known as endochondral ossification. Key events in this process include the commitment and differentiation of mesenchymal cells from the mesoderm into chondrocytes, condensation and proliferation of these committed mesenchymal cells, and subsequent invasion of blood vessels along with osteoblasts, bone forming cells, osteoclasts, bone resorbing cells, and hematopoietic cells to form the bone marrow [2]. Various studies have demonstrated that bioactive heparin binding factors such as Indian hedgehog (Ihh), Wnts, fibroblast growth factors (FGFs), and bone morphogenetic proteins (BMPs), in addition to components of the pericellular matrix in cartilage, are critical regulators of lineage commitment and cell proliferation and differentiation [3–9]. Dynamic paracrine interactions exist between the developing cartilage core of endochondral bone and the perichondrium (Pc) and support bone elongation and widening [10, 11]. A hallmark of developing endochondral bone is the chondro-osseous junction (COJ) the demarcation between terminally differentiated cartilage and mineralizing bone.
Investigations in our laboratory and others, using both human and animal models, revealed that heparan sulfate proteoglycan (HSPG) core proteins, including perlecan/HSPG2 [12] and their heparan sulfate (HS) chains influence the key events that occur during cartilage development [12–20] and fracture repair [21]. More specifically, it has been demonstrated that glycosaminoglycan (GAG)-bearing perlecan/HSPG2 domain I, but not perlecan/HSPG2 domain I lacking GAG chains, supports aggregation and expression of mature chondrogenic markers in C3H10T1/2 cells and assists growth factor delivery [22–25]. Accumulating evidence implicates a critical function for HSPGs in facilitating the interactions among HBGFs and their receptors [26]. Cleavage of HS chains facilitates formation of diffusible complexes of growth factors complexed with HS fragments which together form a trimolecular signaling complex [27]; however, the importance of HPSE as a modulator of growth factor bioavailability and HS catabolism during the transition from chondrogenesis to osteogenesis during endochondral ossification remains unclear.
HPSE, an endo-β-D-glucuronidase, is synthesized as an inactive 65kDa form (pro-HPSE) and is processed to an active heterodimer consisting of 50 and 8kDa subunits (active HPSE). As an active heterodimer, it cleaves HS chains attached to proteoglycans including perlecan/HSPG2 [28–30]. HPSE activity is inhibited by a structural mimic of HS, PI-88 [31]. HPSE also possesses non-enzymatic functions such as cell adhesion, proliferation, migration, and invasion [32]. HPSE has been identified in normal and malignant cells including skin fibroblasts, cytotrophoblasts, hepatocytes, Chinese hamster ovary (CHO) cells, and endothelial cells and tissues including skin, placenta, lung, and kidney [33–38] and is implicated in a number of normal and pathological conditions [31, 39–41]. Interestingly, HS chains have been identified in the pericellular matrix of both developing and mature cartilage and the Pc of developing growth plate. [12] Identifying the relationship between HPSE and HS expression will allow us to better understand the role of HPSE in growth plate development.
Most studies of HPSE have focused on its regulatory role in cancer progression. Constitutive overexpression of human HPSE in mice alters tissue architecture, vascularization, and metabolism [42]. In bone, HPSE overexpression creates a complex phenotype that favors osteogenesis, increases bone mass, but retards bone elongation in female transgenic mice [43]. Previous studies in our laboratory demonstrated a dramatic loss of HS at the COJ as endochondral bone formation progresses, suggesting that HS inhibits osteogenesis [12]. To expand on these observations, we sought to determine if HPSE influences the transition from chondrogenesis to osteogenesis during endochondral bone formation in mouse models. We designed our studies to identify HPSE/Hpse localization within the developing growth plate and to provide additional functional insight in the role of HPSE/Hpse during the process of endochondral bone formation.
Materials and Methods
Animals
All animal handling procedures were approved by the University of Delaware Institutional Animal Care and Use Committee. Long bones from C57/BL6 and ICR strain mice (Taconic, Germantown, NY) were dissected at various developmental stages and preserved in Tissue-Tek® Optimal Cutting Temperature (O.C.T.) (Sakura Finetechnical, Torrance, CA) at −80°C until cryosectioning.
Cell Culture
ATDC5 cells, a murine carcinoma-derived chondrogenic cell line, were obtained from Dr. Véronique Lefebvre (The Cleveland Clinic, Cleveland, OH) and maintained as monolayer cultures under conditions previously described [44]. Briefly, they were cultured at 37°C in air:CO2 [95:5% (v/v)] in regular growth media, Dulbecco’s Modified Eagle’s medium-Ham’s F12 (DMEM-F12) supplemented with 5% (v/v) fetal bovine serum (FBS), 100 units/ml pencillin G sodium and 100 µg/ml streptomycin sulfate in 0.085% (w/v) saline (PS). All cell culture reagents were purchased from Invitrogen (Carlsbad, CA) unless otherwise stated. For monolayer differentiation, cells were seeded into six-well tissue culture plates (Becton Dickinson Labware, Franklin Lakes, NJ) until they reached post-confluency at which culture medium was replaced with regular growth media (DMEM-F12 + 5% [v/v] FBS) containing 10 µg/ml bovine insulin (I), 10 µg/ml human transferrin (T), and 3 × 10−8 mol/L sodium selenite (S), (ITS). After 21 days in differentiation media, cells were transferred to 37°C in 97% air: 3% (v/v) CO2 and switched to α-MEM containing 5% (v/v) FBS, PS, and 1% (v/v) ITS for further differentiation into calcifying chondrocytes as described in [44].
In situ Hybridization
In situ hybridization followed a protocol previously described [31, 45]. Briefly, E18.5 mouse limbs were cryosectioned at 12µm thickness and mounted onto poly-L-lysine coated slides. Hybridization probes were generated as described in [31]. Sections hybridized with sense probes served as negative controls and indicated specificity of anti-sense probe. Sections were post-stained with hematoxylin and eosin to confirm orientation.
Immunohistochemistry
Frozen limb sections (10–12µm thickness) were fixed in methanol for 10 min at room temperature, rehydrated in PBS containing Ca2+ and Mg2+ and blocked with 1% (w/v) bovine serum albumin (BSA) (Sigma-Aldrich) in PBS for 30 min at room temperature. Sections were incubated at room temperature for 1 h with mouse monoclonal anti-human HPSE 1 antibody (Insight Biopharmaceuticals Limited, Rehevot, Israel) and Zenon™ reagent at 1:40 dilution and Draq5™ ( Biostatus Limited, Leicestershire, UK) at 1:2000 dilution. One µg of this antibody was used to prepare Zenon™ labeling complex using Zenon™ Technology (Invitrogen/Molecular Probes, Eugene, OR) according to the manufacturer’s protocol. Samples were rinsed three times with PBS containing Ca2+ and Mg2+ for 5 min at room temperature each. Next, sections and cells were post-fixed for 10 min at room temperature in 4% paraformaldehyde (w/v) in PBS. Finally, samples were mounted in Gel/Mount ™ (BioMeda Corp., Foster City, CA) to prevent fading. As a control for specificity, sections were incubated with mouse IgG2b. Samples were imaged with a Zeiss LSM 510 multi-photon confocal microscope at the Microscopy Core Facility of the University of Delaware.
Organ Culture
The three center metatarsals were microdissected from the feet of 2 day old ICR mice as described in [10, 46–48]. Metatarsals were placed in 24 well tissue culture treated plates (Corning Inc) containing 500µl of α-MEM (Gibco-BRL) supplemented with 0.05mg/ml ascorbic acid, 0.3mg/ml L-glutamine (SIGMA- Aldrich, St Louis, MO),1mM β-glycerophosphate (SIGMA- Aldrich), 250µg/ml of Amphotericin B (Invitrogen) (Fungizone Equivalent) and PS (Invitrogen). Explants were grown at 37°C in a humidified air:CO2 (95:5, v/v) incubator. Explants were cultured in media containing 0, 100, or 500µg/ml of PI-88. For PI-88 treated samples, the drug was generously provided by Progen Pharmaceuticals Ltd. (Toowong, Queensland, Australia). Media was changed every 2–3 days. Fresh PI-88 was added with each media change. Cultures were observed for contamination and photographed using a digital camera DXM1200F (Nikon, Japan) attached to phase-contrast microscope SMZ1500 (Nikon, Japan). Histomorphometric analyses of dissected metatarsals were performed as reported by Ueda et al., 2007 [49] with the relative contributions of the Rz, Pz, and Hz to the longitudinal length of the bone indicated. Images then were processed further and the length of the bones as well as the lengths of the growth plates and marrow cavities were measured on day 0 and after 5 days of culture using Adobe Photoshop® 7.0. The percentage of total bone growth and relative contribution of growth of each compartment was calculated from the measured values. The mean and standard deviation and significance were determined using GraphPad Software©. Groups with probability value less than 0.05 (p<0.05) were considered significantly different. To observe any morphological differences in the growth paltes that could result from treatment with PI-88, bones were sectioned on D5 of culture as described above and stained with hematoxylin and eosin. Bones were photographed at 10X.
Real Time PCR
Total RNA of differentiated ATDC5 cell was extracted using the RNeasy Mini Kit and QIAshredder column (Qiagen) and digested with DNAse I with the DNA free kit (Ambion) following the manufacturer’s instructions. Reverse transcription (RT) was performed with Omniscript Reverse Transcriptase (RT) kit (Qiagen) for conventional and real time quantitative (Q) polymerase chain reaction (PCR). Conventional PCR and Q-PCR were performed using the HotStar Taq DNA Polymerase kit (Qiagen) and the SYBR® green PCR master mix (Applied Biosystems, Foster City, CA). Oligonucleotide sequences were designed using Primer Express (Applied Biosystems, Foster City, CA) unless otherwise indicated. Oligonucleotide sequences used in PCR reactions are listed in Table 1. After 10 min of preincubation at 95°C, samples were incubated for 30 s at 55–65°C, and 60 s at 72°C for 35 cycles with a final extension for 10 min at 72°C. For Q-PCR, samples were cycled for 15 s at 95°C and 60 s at 60°C for 45 cycles. All data collected from real-time PCR was calculated using the ABI Prism® 7000 software (Applied Biosystems). The relative amounts of Hpse mRNA were identified using the comparative threshold cycle (Ct) method (User Bulletin No.2, ABI Prism 7700 Sequence Detection System). The Ct value of each target sequence was subtracted from the Ct value of ACTB, to derive ΔCt. The calculation of ΔΔCt involved subtraction of the ΔCt value of D0 differentiated ATDC5 cells. Similarity in amplifying efficiency (slope difference < 0.1) of the targets (Hpse) and reference (ActB) were validated. Each sample was assessed in triplicate. To confirm amplification specificity of the fluorescent detection system, Q-PCR products were analyzed in a 2% (w/v) agarose gel stained with ethidium bromide.
Table I.
PCR Primers Used In These Studies
| Gene | Forward Primer | Reverse Primer | Product Size (bp) | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|---|
| Aggrecan/Agg | 5’-CCAAGTTCCAGGGTCACTGT-3’ | 5’-TCTAGCATGCTCCACCACTG-3’ | 398 | 60 | [84] |
| Type II Collagen/ColII | 5’-TCATCGAGTACCGATCACAGA-3’ | 5’-GTTCGGGGGTTTTACAAGAAG-3’ | 130 | 55–60 | [56] |
| Type X Collagen/ColX | 5’-TAGGAGCTAAAGGAGTGCCT-3’ | 5’-TCACCCTTAGATCCAGGGAAT-3’ | 124 | 55–60 | [56] |
| Hpse | 5’-TTTGCAGCTGGCTTTATGTG-3’ | 5’-ACTGGTTTCCTGAACAACGG-3’ | 349 | 57.8 | new |
| Osteocalcin/Ocn | 5’-AAGTCCCACACAGCAGCTTG-3’ | 5’-AGCCGAGCTGCCAGAGTTTG-3’ | 400 | 65 | [85] |
| ActB | 5’-GTGGGCCGCTCTAGGCACCAA-3’ | 5’-CTCTTTGATGTCACGCACGATTTC-3’ | 502 | 55–60 | new |
| Hpse (for QPCR) | 5’-TGTCCTGAACCTCCATAATGTC-3’ | 5’-TACGTATCCACTGGTTTCCTGA-3’ | 79 | 60 | new |
| ActB (for QPPR) | 5’-AAATCGTGCGTGACATCAAAGA-3’ | 5’-GCCATCTCCTGCTTCGAAGTC-3’ | 80 | 60 | [56] |
Immunoblotting
For analysis of protein expression, total cell extract was used from ATDC5 cells that were lysed in buffer including 0.05M Tris pH 7.0, 8M urea, 1% (v/v) sodium dodecyl sulfate (SDS), 1% (v/v) β-mercaptoaethanol and 0.01% (v/v) phenylmethylsulphonyl fluoride (PMSF). Cell extracts were incubated for 5 min at 100°C with Laemmli Sample Buffer (BioRad Laboratories, Hercules, CA) containing 1% (v/v) β-mercaptoethanol and separated by SDS-polyacrylamide gel electrophoresis (PAGE) using a 4–12% (w/v) Bis-Tris gradient polyacrylamide gel with MES buffer (Invitrogen) or a 10% (w/v) polyacrylamide Porzio and Pearson SDS-PAGE gel [50, 51]. After electrophoresis, gels were transferred to Protran® Pure Nitrocellulose Transfer and Immobilization Membrane (Scheleicher & Schuell Bioscience Inc, Keene, NH) for immunoblotting with anti-heparanase polyclonal (1453) raised against the entire 65 kDa HPSE precursor which was kindly provided by Dr. Israel Vlodavsky (Cancer and Vascular Biology Research Center, The Bruce Rappaport Faculty of Medicine, Technion, Haifa 31096, Israel). Briefly, the blots were blocked for 3 h-overnight at 4°C with 5% (w/v) non-fat dry milk in phosphate buffer saline (PBS) plus 0.1% (v/v) Tween 20 (PBS-T) then incubated overnight at 4°C with the primary antibody at a final dilution of 1:2500 in 5% (w/v) non-fat dry milk/PBS-T. Blots were rinsed three times, 5 min each at room temperature with PBS-T to remove unbound antibody. Next, blots were incubated for 2 h at 4°C with anti-rabbit IgG peroxidase conjugate (Sigma, St Louis, MO) at a final dilution of 1:200,000 in 5% (w/v) non-fat dry milk/PBS-T. Finally, the blots were rinsed three times with PBS for 5 min each at room temperature and detected using the ECL system (Pierce, Rockford, IL) as described by the manufacturer. Blots were immunoblotted with anti-ACTB to determine equivalent loading. Statistical analyses were performed using one-way ANOVA and the Tukey-Kramer multiple comparisons test (GraphPad InStat program; GraphPad Software Inc., San Diego, CA).
Effects of PI-88 on HPSE Activity
Radiolabeled extracellular matrix-heparan sulfate proteoglycans (ECM-HSPGs) were prepared as described previously [31]. In summary, WiDR cells were cultured in Eagle’s Minimum Essential Medium (ATCC, Manassas, VA) supplemented with 10% (v/v) FBS, 2mM L-glutamine, 100U/ml penicillin and 100µg/ml streptomycin sulfate. Following the second passage, the cells were plated in a 24-well cell culture treated plate (Corning Inc) until they reached 80% confluency. The media was removed, cells were rinsed with low sulfate media containing RPMI-1640 (Invitrogen), 3.3mM magnesium chloride, 1.5mM HEPES, 1.2g/L sodium bicarbonate and 0.05% (v/v) penicillin/streptomycin solution. The pH of low sulfate media was adjusted to 7.3 before use. Cells were incubated in 1ml of low sulfate media containing 100µCi of Na2 35SO4 for 48h. After 48h, cells were rinsed, briefly, four times with PBS free of Ca2+ and Mg2+ to remove unincorporated Na2 35SO4. The wells were treated with 0.5% (v/v) Triton X-100/ 20mM ammonium hydroxide in PBS for 10min to solubilize the cell layer followed by four washes with PBS free of Ca2+ and Mg2+. The ECM-H(35S)PGs which remained intact and attached to the cell culture well were used immediately to test for HPSE activity as described [31, 52]. Total cellular extract from differentiated ATDC5 cells and B16BL6 cell was homogenized in a buffer containing 10mM TBS pH 7.2, 0.5% (v/v) Triton X-100, 0.1µg/ml (w/v) leupeptin, 0.1µg/ml (w/v) pepstatin and 0.2mM PMSF and prepared for HPSE activity assay as described in [31]. Prior to incubating samples with the ECM-H(35S)PGs, 50µg protein extracts were incubated with various concentrations of PI-88.
High Density Chondrogenic Culture
To prepare micromass cultures, ATDC5 cells were plated at a density of 1.0 × 105 cells in a total volume of 10µl of DMEM/F-12 supplemented with 5% (w/v) FBS in the center of four-well tissue culture treated plate (Nunc™, Rosklide, Denmark) as described in Denker et al. (1999) [53] . Cells were allowed to attach for 3 hr. After attachment, 0.5ml of regular growth media containing ITS and PS was added to each well. Media was changed every other day. For micromass cultures treated with PI-88, the procedure was as follows: a single cell suspension of cells was pretreated with 100µg/ml of PI-88 for 30 min at room temperature with occasional gentle resuspension prior to plating. Following 30 min incubation, 10µl of the pretreated cell suspension was plated at a density of 1.0 × 105 cells in DMEM/F-12 supplemented with 5% (w/v) FBS in the center of four-well tissue culture treated plate. Fresh PI-88 was added with each media change. For subsequent experiments, total RNA of differentiated ATDC5 micromasses were extracted using the RNeasy Mini Kit and QIAshredder column (Qiagen, Valencia, CA) and digested with DNAse I with the DNA free kit (Ambion, Austin, TX) following the manufacturer’s instructions. Reverse transcription (RT) was performed with Omniscript Reverse Transcriptase kit (Qiagen) for conventional and real time quantitative (Q) RT-polymerase chain reaction (PCR).
High Density Mineralization Culture
ATDC5 cells were cultured in micromass as described above with minimal modifications. Following attachment, 0.5ml of α-MEM containing 5% (v/v) FBS, 0.05mg/ml ascorbic acid, 10mM β-glycerophosphate (SIGMA- Aldrich), ITS, and PS. Media was changed every other day. For PI-88 treated samples, fresh PI-88 was added with each media change. Total RNA was extracted as described above. Whole-mount cell cultures were fixed and stained with Alcian Blue 8GX (Sigma-Aldrich) as described in [54–57] and Von Kossa stain as described in [46, 48] at the indicated time point(s).
Migration Assay
The effect of PI-88 on cell migration was determined using wound healing assay procedures as described [58]. Prior to plating cells, ATDC5 cell suspension was pre-incubated with 100µg/ml of PI-88 for 30 min at room temperature with occasional mixing. As a negative control single cell suspension was incubated with PBS alone. Immediately following incubation, cells were plated in six well tissue culture treated plates (Becton Dickinson) and grown to confluency in regular growth media. For cells treated with PI-88, fresh PI-88 was added to the regular growth media. Next, the cell monolayer was wounded by a sterile plastic micropipette tip and rinsed two times with PBS containing Ca2+ and Mg2+ to remove any unbound cells. The cells were imaged using a digital camera CoolPix 990 (Nikon Japan) attached to a phase contrast microscope (Nikon Japan) immediately following the creation of the wound. Cells were incubated at 37°C in a humidified incubator of air:CO2 (95:5). After 210 min, cells were re-imaged under phase-contrast microscopy. The percentage of cells repopulating the scratch zone was calculated by direct observation of the cell density in this area using Image J software (http://rsb.info.nih.gov/ij/).
Statistical Analysis
Statistical analyses were performed using one-way analysis of variance (ANOVA) and the Tukey-Kramer multiple comparisons test GraphPad Software©. Groups with probability value less than 0.05% (p<0.05) were considered significantly different.
Results
HPSE is present in the developing growth plate
In the first series of experiments, we determined the expression of Hpse mRNA and HPSE protein in developing mouse long bone and throughout the different regions of the growth plate. To determine Hpse mRNA localization in vivo, in situ hybridization was conducted using radioactively-labeled Hpse antisense probes on limbs obtained from day 18.5 mouse embryos, a time when all regional zones of endochondral bone are clearly evident. To confirm chondrogenic regions and orientation, cryosections were counterstained with hematoxylin and eosin (Figure 1A). The proliferative zone (Pz) is indicated by numerous closely packed round cells. Prehypertrophic (PHz) cells were identified by an increase in cell size and the initial formation of cells into a columnar organization flowing into the hypertrophic region (Hz) with large columnar cells. The chondro-osseous junction (COJ), the distinct area that lies between the Hz and mineralized tissue (MT), marks the initiation of large scale osteogenesis. In our in situ studies, the most intense signal for Hpse mRNA was observed in the Pc, Po, MT, and especially the COJ (Figure 1B), indicated by bright yellow hybridization signal (black arrows). Faint signal also was observed in the PHz and Hz as indicated by orange signal (arrowheads) (Figure 1B).
Figure 1. HPSE is present in the Pc and COJ of developing mouse limb.
Panel A shows hematoxylin and eosin counterstaining of the cryosection shown in (B). (B) Hpse mRNA (orange and yellow) was detected by in situ hybridization, in the periosteum (Po) and perichondrium (Pc), hypertrophic zone (Hz), and chondro-osseous junction (COJ) and mineralized tissue (MT) of day 18.5 embryos. (C) A cryosection of mouse hindlimb was analyzed by indirect immunofluorescence using a mouse monoclonal anti-human HPSE antibody as described in “Materials and Methods.” Positive HPSE immumostaining is indicated by green fluorescence in the Pc, Po, COJ, and MT. The section was double labeled with anti-perlecan/HSPG2, the major HSPG in the matrix of developing endochondral bone (red). (D) Close up view of HPSE immunostaining (green) in the Pc at the PHz/Hz boundary shown with perlecan/HSPG2 double stain (red). (E) HPSE staining of the COJ. No specific signal was observed using isoform matched normal mouse IgG under similar conditions used as a negative control (not shown). Arrows highlight areas of high HPSE/Hpse expression.
Next, we determined the localization of HPSE protein in 18.5 day mouse embryos using immunohistochemistry. An antibody against perlecan/HSPG2, the major HSPG in cartilage, was used to show the regions where the highest concentrations of HS are located (red in Figure 1C). The highest HPSE immunoreactivity was observed in the Pc surrounding the Hz region and Po along the edge of the developing long bone (white arrow). Expression also was evident in the COJ and MT as indicated by the asterisks. In contrast, expression throughout the Pz and PHz of cartilage was only moderately above background and did not appear to change throughout early differentiation. (Figure 1C) The expression of HPSE also was examined earlier in development. Figure 1D shows the very bright site of immunolocalization of HPSE in 16.5 day mouse embryos with intense staining in the Pc surrounding the transition to the Hz. In whole bone, this would appear as a “ring” of HPSE-expressing Pc demarcating the PHz-Hz boundary. It was of interest that very little overlap was observed for perlecan/HSPG2 and HPSE except for the Pc surrounding the PHz-HZ boundary, where a bright green/yellow signal (white arrow) was observed at high magnification (Figure 1D). HPSE staining also was evident in trabecular bone at the COJ (Figure 1E, white arrow), shown without the perlecan/HSPG2 staining.
PI-88 reduces growth plate expansion and increases bone marrow cavity proportion of Cultured metatarsals
To determine the effects of the HPSE inhibitor, PI-88, on long bone growth, metatarsal bones were dissected from 2–3 day old ICR mice and cultured ex vivo in the presence of 0, 100 or 500µg/ml of PI-88. Each study group consisted of five bones that were on average 3.53 mm in length at the initiation of the culture period. In the absence of PI-88, metatarsal bones increased in total length by approximately 21.5% when comparing D0 to D5 (Figure 2). There was no significant difference between untreated and metatarsal bones treated with 100µg/ml of PI-88 (Figure 2A). At 500µg/ml, the total mean increase in metatarsal bone length was only 15.4%, about a 6% reduction (Figure 2A, C). Similar results were seen when the data were analyzed as the median rather than the mean (Figure 2C). At D5 of culture, metatarsals were frozen in OCT and cryosectioned, then stained with hematoxylin and eosin. There was no indication of necrotic cell death as a result of PI-88 toxicity. Interestingly, bones from cultures treated with PI-88 (500 µg/ml) showed a significant reduction in cells in the Pz with a coordinate increase in the number of cells showing clear signs of hypertrophy (Figure 2B).
Figure 2. PI-88 reduces growth of cultured metatarsals.
Freshly dissected mouse metatarsals were cultured for 5 days as described in “Materials and Methods.” Cultures received either 100 or 500 µg/ml PI-88, or no treatment. Metatarsals were, on average, 3.53 mm in length at the initiation of the culture period. (Panels A, C) In the absence of PI-88, metatarsal bones increased in total length by 21.5% over the culture period. No change was observed at the lower dose (100 µg/ml) of PI-88. At the higher dose, a significant (p < 0.05) reduction in bone lengthening, expressed as a percent, was seen both by calculation of the mean and the median. (Panel B) Immunohistochemical analysis of sections of metatarsal bone treated with PI-88 (500 µsg/ml) showed a loss of cells in the proliferating zone (Pz) and a concomitant increase in cells in the hypertrophic zone (Hz). These features are consistent with limb shortening. (Panel C) Mean and median growth was assessed at two doses of PI-88 (n ≥ 8). The data shown is compiled from two independently performed experiments in which five bones were used in each treatment group (30 bones total).
To determine if there was a ratiometric change in the lengths of the growth plate relative to bone cavity, the length of each bone cavity was measured relative to total bone length (Table II) similarly to the method of Ueda et al. [49]. In freshly dissected bones, the growth plates occupied 62% of the total bone length, with the rest (38%) taken up by bone cavity. By D5, the growth plates occupied 69% of total bone length, with the remaining 31% occupied by cavity, reflecting the overall increase in bone volume occupied by expanding cartilage cells. The same was true for metatarsals treated with 100µg/ml PI-88. In contrast, metatarsals treated with 500µg/ml maintained dimensions of 60.2% growth plate and 39.8% bone cavity, similar to bones at the start of culture. Similarly, the bone cavity ratio of untreated metatarsal bones decreased by some 7% during the culture period compared to a nearly 2% increase in cavity ratio in metatarsals treated with PI-88 at 500µg/ml. These proportional changes in bone structure and increase in cells undergoing hypertrophy led us to examine HPSE/Hpse expression in an in vitro system where the processes of chondrocyte and osteoblast differentiation could be independently evaluated in the presence and absence of PI-88.
Table II.
Effects of PI-88 on metatarsal growth proportions.
| Fraction Occupied by Marrow Cavity (%) (D0) | Fraction Occupied by Marrow Cavity (%) (D5) | Fraction Occupied by Growth Plate (%) (D0) | Fraction Occupied by Growth Plate (%) (D5) | |
|---|---|---|---|---|
| Control (No Treatment) | 38** | 31 | 62** | 69 |
| 100µg/ml PI-88 | 38** | 30.8 | 62** | 69.2 |
| 500µg/ml PI-88 | 38** | 39.8 | 62** | 60.2 |
The average control bone was 3.53 mm in length at Day 0 (D0).
The D0 data were averaged for all bones prior to random division into treatment groups. See Materials and Methods for specifics and statistics.
HPSE/ Hpse is expressed in a differentiating chondrocytic cell line
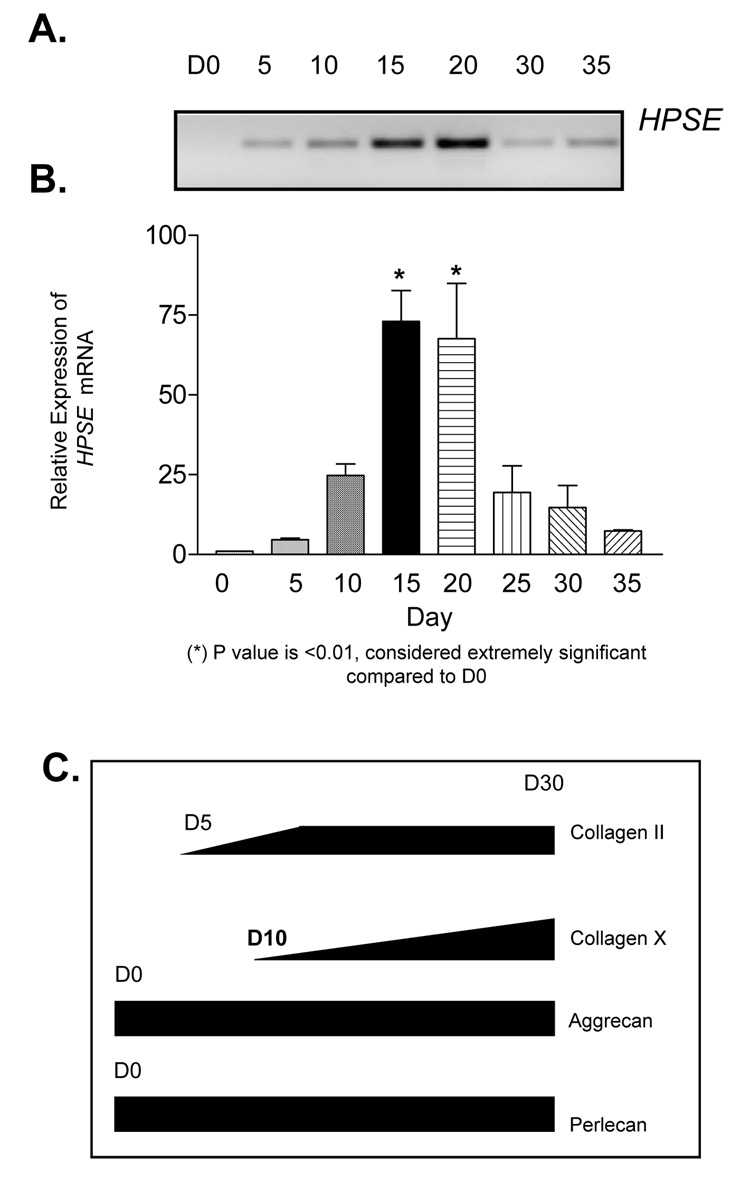
Using the ATDC5 in vitro model of chondrocyte differentiation, we examined Hpse mRNA expression as a function of differentiation in culture. By conventional PCR, a 349bp band appeared at D5 of culture revealing the first expression of Hpse mRNA. Sequencing confirmed the identity of this amplimer as a Hpse derived sequence. As assessed by the increase in intensity of the 349bp band, expression continued to increase through D15 and D20 of culture. The intensity of the band dramatically decreased by D30 and D35 of culture, approaching the levels observed at D5 and 10 of culture (Figure 3A). Q-PCR confirmed that Hpse mRNA increased significantly at D15 and 20 compared to D0, p<0.01 (Figure 3B). There was approximately a 60-fold increase in expression relative to D0.
Figure 3. Hpse mRNA is elevated as ATDC5 cells approach terminal differentiation.
ATDC5 cells were submitted to chondrocytic using culture conditions as described in “Materials and Methods.” (A) Conventional RT-PCR was performed using total RNA from differentiated ATDC5 cells at the indicated time points of differentiation as described in “Materials and Methods.” Hpse mRNA expression is initially expressed at D5 and continues to be expressed throughout culture. Interestingly, HPSE mRNA expression peaks at D15–20 of culture. The Hpse peak expression was confirmed by QRTPCR. (B) QRTPCR reveals that Hpse mRNA significantly peaks in mid-stage culture (D15 and D20) and displays an increase in relative expression 73% at day 15 and 67.6% at day 20 after normalizing to ActB as described under “Materials and Methods.” C) Gene expression of early and late stage markers of chondrogenesis was investigated to determine the stage of ATDC5 differentiation at different time points. Collagen II/ColII mRNA was initially detected at D5. Its expression is maintained throughout differentiation. Collagen X/ColX transcript was initially detected at D10 but did not reach its peak until D30 of differentiation indicating late stage differentiation. Aggrecan/Agg mRNA, an early stage marker of chondrogenesis, was expressed and maintained throughout the duration of the culture. Perlecan/HSPG2 was detected at D0 and maintained expression throughout the entire differentiation process.
To determine the stage of differentiation mimicked by the cultures, early and late stage markers of chondrogenesis were examined using conventional PCR. Parallel studies established that collagen type II/COLII, an early stage marker of chondrogenesis, mRNA was detected at D5 of differentiation and increased until approximately D10 and then remained high through to D30. Collagen type X/COLX, a late stage marker of chondrogenesis, mRNA first was detected at D10 and gradually increased until D30. Aggrecan/Agg, an early stage marker of chondrogenesis, mRNA was expressed throughout the culture period, as was perlecan/HSPG2 (Figure 3C). Interestingly, up-regulation of Hpse mRNA occurred at the same time as did the initial up-regulation of collagen X, a marker normally expressed in the Hz at the time of terminal differentiation [56].
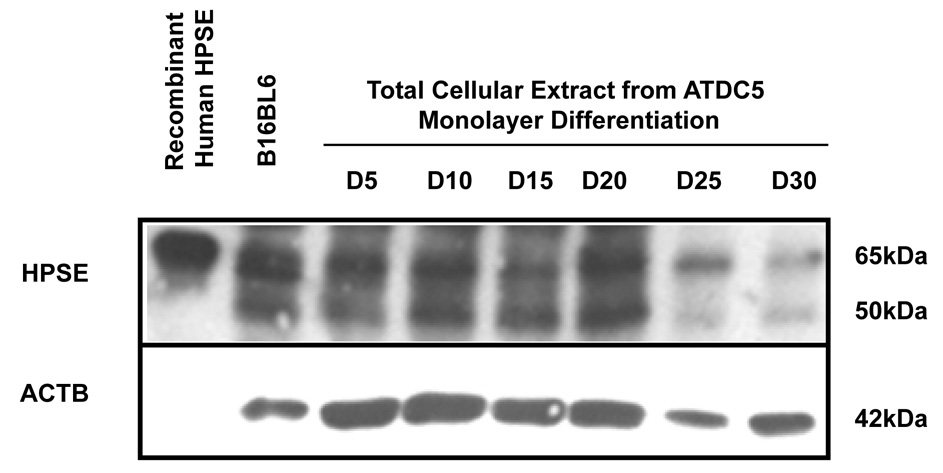
We extended our in vitro studies of HPSE expression by performing immunoblotting using an anti-HPSE polyclonal (1453) antibody raised against the entire 65kDa HPSE precursor. Recombinant human HPSE and total cellular extract from B16BL6 cells served as positive controls, and as additional molecular weight guides for the unprocessed (65kDa/pro-HPSE) and processed form (50kDa) of HPSE. Both the unprocessed and processed forms of HPSE were observed at all time points from differentiated ATDC5 cells. There was a modest increase in the processed form of HPSE at D10–20 when compared to D5. There was a significant decrease in both unprocessed and processed forms of HPSE at D25 and 30. (Figure 4) These trends at the protein level were similar to the observations when investigating Hpse expression at the mRNA level, although the degree of increase was not as dramatic as that observed for Hpse mRNA (compare Figure 3A to Figure 4).
Figure 4. HPSE expression peaks as ATDC5 cells approach terminal differentiation.
Total protein was extracted from differentiated ATDC5 cells at various time points and assayed for HPSE expression. Although, HPSE is not abundant in ATDC5 cells, there is an increase in HPSE expression as ATDC5 cells progress through chondrogenic differentiation. ATDC5 cells were submitted to chondrocytic using culture conditions as described in “Materials and Methods.” To determine protein expression, total cellular extract was electrophoresed, transferred, and immunoblotted using anti-heparanase polyclonal antibody 1453. The proform of HPSE is detected at 65kDa, and the enzymatically active form is detected at 50kDa. The proform was detected at high levels from D5 to D20 of chondrogenic differentiation, however, expression decreased at D25 and D30 of culture. The enzymatic form was initially detected at D5 and expression levels increased up until D20. At D25 and D30 there was little to no detection of the enzymatic form. Recombinant human HPSE and total cellular extract from B16BL6 cells, mouse melanoma cell line, was used as a positive control. Equivalent loading was confirmed by immunblotting with anti- ACTB. HPSE intensity was quantified as described in the “Materials and Methods.” Normalizing expression to ACTB indicates an increase in HPSE expression as the cells differentiate.
Because HPSE can be expressed as both an enzymatically unprocessed and a processed form [30], we performed an activity assay to determine if active HPSE was produced during ATDC5 cell differentiation. Consistent with the Western blotting studies, HPSE activity was initially low, and continued to increase as differentiation progressed. HPSE activity in the cell associated fraction was increased through day 21, and was detected through the end of the differentiation period (D33) (data not shown). Because it represented the time of peak increase of HPSE activity, D19 was used to assess the inhibitory effects of PI-88 on differentiating chondrocytic cultures (see below). To determine if the expression of HPSE was required for chondrogenic differentiation, we differentiated ATDC5 cells while perturbing HPSE activity using a competitive inhibitor, PI-88.
PI-88 decreases HPSE activity in ATDC5 cells in a dose-dependent manner
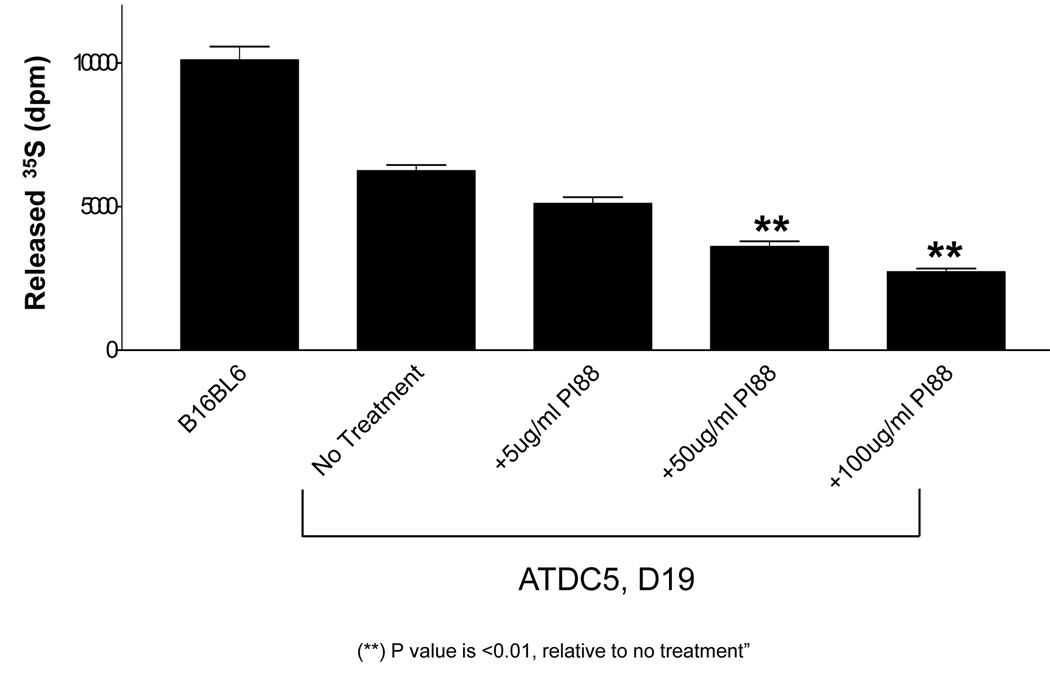
ECM pre-labeled metabolically with [35S]sulfate was treated with cellular extract from D19 differentiated ATDC5 cells in the absence and presence of 5, 50, and 100µg/ml of PI-88. The ability of endogenous HPSE to cleave radioactively-labeled HS chains was measured and quantified after chromatographic separation as described previously [31]. Total cellular extract from B16BL6 melanoma cells served as a positive control. Specificity for HS first was demonstrated by beta-elimination to confirm that the released fragments were not derived from the protein component of the proteoglycans. Second, nitrous acid degradation, which acts only on HS and not on other glycosaminoglycans, was used to verify that the released fragments were HS. A buffer control was used to determine background levels and subtracted from all experimental samples. There was a significant and dose-dependent decrease in HPSE activity in all cellular extracts containing PI-88. In the presence of 100µg/ml of PI-88, a 56% reduction in HPSE activity was observed when compared to no treatment control (Figure 5). For the remaining in vitro studies, 100µg/ml of PI-88 was used.
Figure 5. PI-88 inhibits HPSE activity in differentiated ATDC5 cells.
ATDC5 cells were differentiated for 35 days as described in “Materials and Methods.” Cellular extract from differentiated ATDC5 cells at D19 was treated with 5, 50, and 100µg/ml of PI-88. HPSE activity was inhibited by approximately 56% (p<0.01, 100µg/ml vs. control and 5µg/ml PI-88) using 100µg/ml of PI-88. To determine background, the amount of released 35S in the presence of buffer only was subtracted from all samples. There was a dose-dependent decrease in HPSE activity in the presence of increasing concentrations of PI-88.
PI-88 decreases the rate of GAG accumulation during ATDC5 differentiation
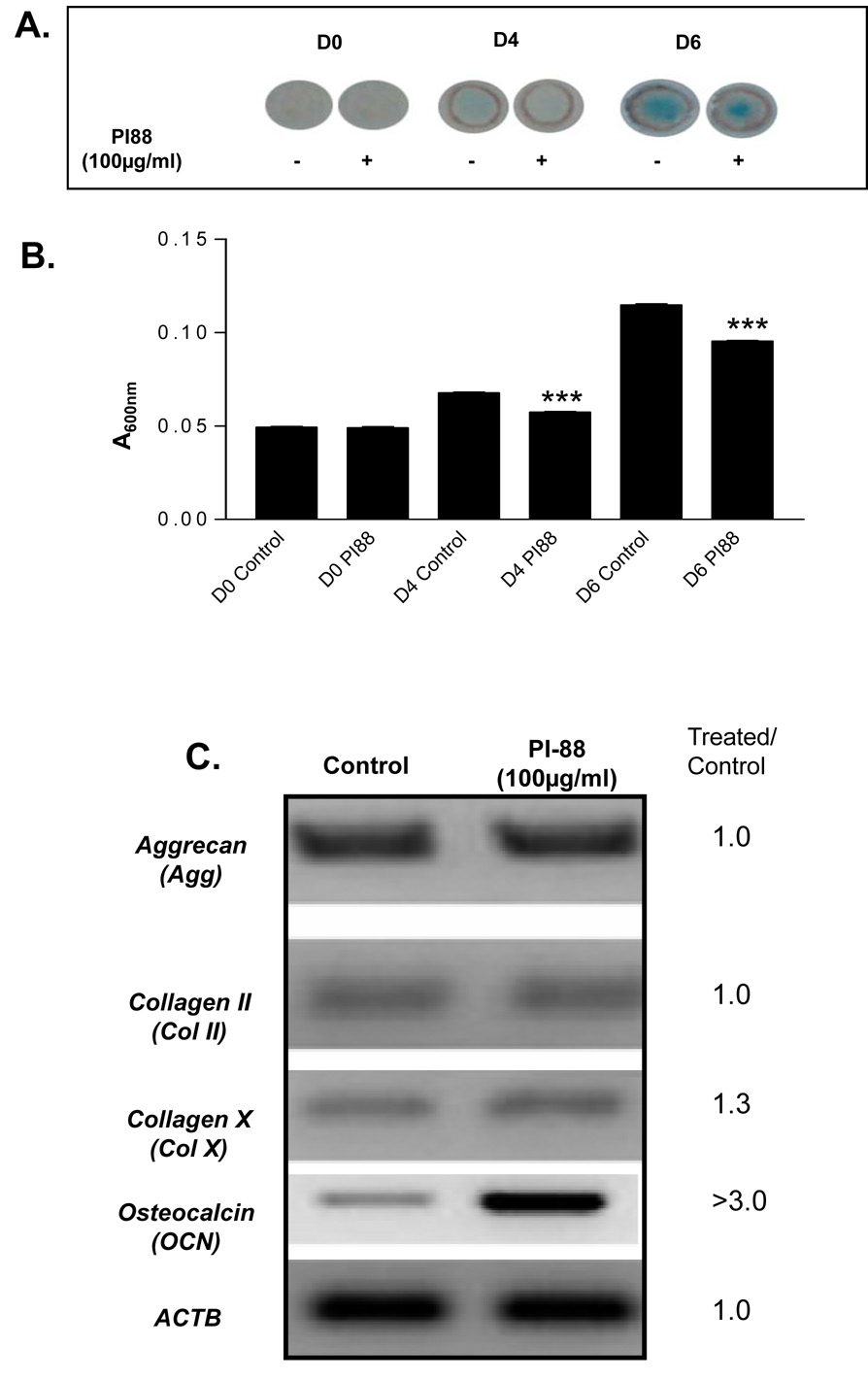
To determine the effects of PI-88 on chondrogenic differentiation, high density chondrogenic cultures of ATDC5 cells were treated with PI-88 (100µg/ml). GAG accumulation, which correlates with early chondrogenic differentiation, was assessed by staining micromasses with Alcian blue and quantitating dye accumulation spectrophotometrically. There was no indication of Alcian blue staining at D0 for either the control or experimental group (Figure 6A), normalized to 0.05 units in Figure 6B. At D4 and D6, both treated and control micromass cultures stained positively with Alcian blue for GAG accumulation as indicated by the presence of a blue-green circle in the center of the culture dish. The intensity of the staining, assessed by A600 nm, was significantly less in all PI-88 micromasses tested (Figure 6B). Thus, micromass cultures exposed to PI-88 accumulated a significantly smaller amount of GAG at both D4 and D6 of culture.
Figure 6. PI-88 decreases GAG accumulation and increases OCN transcript levels during high density chondrogenic cultures of ATDC5 cells.
ATDC5 cells were pretreated with 100µg/ml of PI-88, plated, and differentiated under micromass culture conditions in the presence of PI-88. (A) At D0, 4, and 6, micromasses were stained with Alcian blue, an indicator of GAG accumulation. Alcian blue staining was easily detected at D6 of high density chondrogenic differentiation. (B) After 6 days of culture, Alcian blue was extracted from all high density cultures and all time points to quantify GAG accumulation. D4 and D6 micromass cultures treated with 100µg/ml of PI-88 displayed significantly less Alcian blue staining compared to control micromass cultures at identical time points. At 100µg/ml, PI-88 decreases GAG accumulation in high density chondrogenic cultures. (C) Micromass cultures treated with PI-88 were analyzed for changes in chondrogenic markers, aggrecan/Agg and collagen II/CollII on D6 of culture. Osteocalcin (Ocn) mRNA, a marker of osteogenic differentiation, expression was assayed to evaluate the effects of PI-88 on osteogenesis and collagen X/CollX was assessed as an index to terminal chondrogenesis. Cultures treated with 100µg/ml of PI-88 displayed an increase in OCN mRNA indicating an increase in mineralization potential. A slight increase in CollX also was observed. Aggrecan/Agg and Collagen II/ColII mRNA levels remained unchanged on D6. ActB was used as a load control which remained constant.
To further investigate gene expression relevant to chondrogenic or osteogenic differentiation, RNA was extracted from both control and PI-88 treated micromass cultures on D6 and conventional RT-PCR was performed using specific primers for well accepted markers of chondrogenesis and osteogenesis. There was no change in AGG and COLII mRNA expression between ATDC5 cells treated with 100µg/ml of PI-88 and untreated controls. (Figure 6C). In contrast, examination of late stage markers of chondrogenesis, COLX and OCN, showed that while COLX was only slightly increased (1.3-fold), micromass cultures treated with 100µg/ml of PI-88 consistently displayed a significant increase in OCN expression (>3-fold) (Figure 6C). Because the OCN signal was saturated beyond the linear range, the 3-fold increase is likely underestimated.
PI-88 decreases cell migration in proliferative and terminally differentiated chondrocytes
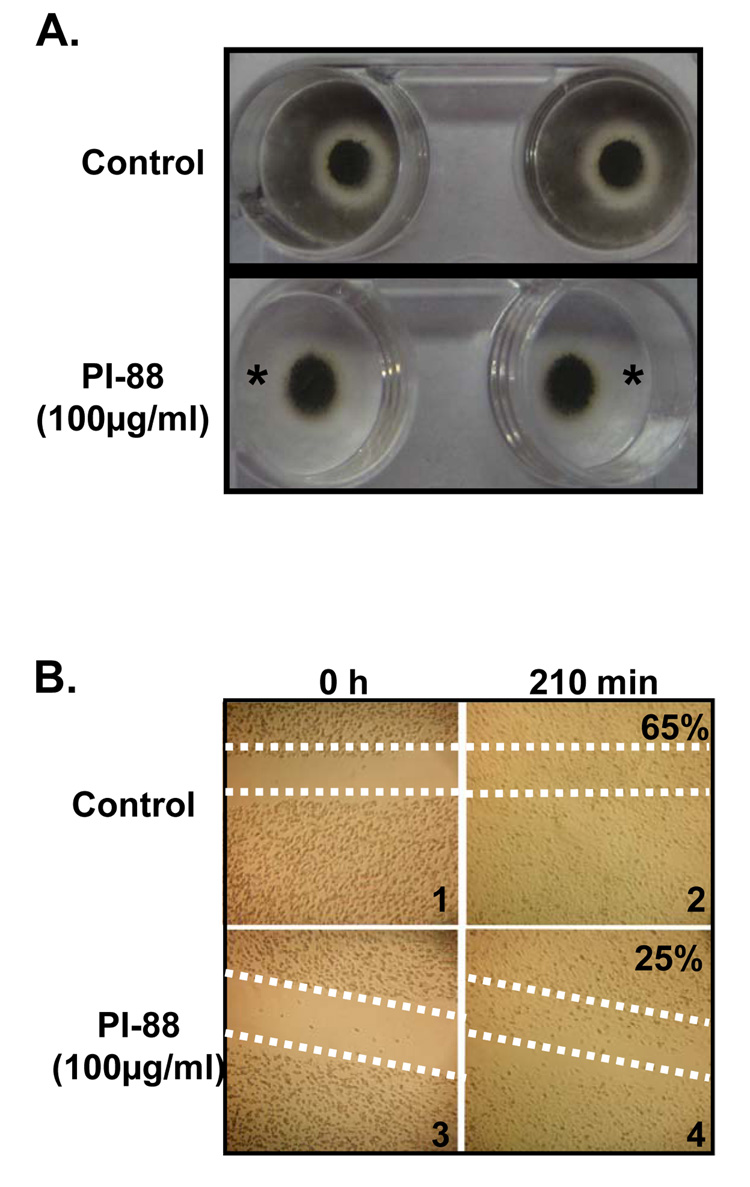
To further investigate the change in OCN, the marker of osteogenesis, high density cultures were grown under conditions conducive to mineralization in the presence of PI-88. When micromass cultures were stained with von Kossa, an indicator of mineralization, after 6 days in culture, an unexpected observation was made. Surprisingly, control cultures (Figure 7A) were found to stain intensely not only in the central high density portion of the micromass, but also in the periphery of the plate where it appeared that a significant number of the cells had migrated during the culture period. In contrast, in the presence of 100µg/ml of PI-88, while strong von Kossa staining, i.e. mineralization, was evident in the center of the dish, there was no staining in the periphery of the plates (Figure 7A). To determine if PI-88 had a direct inhibitory effect on migratory behavior of ATDC5 cells, we next tested the effects of PI-88 on cell migration. A standard wound healing scratch assay was performed using ATDC5 cells. The dotted lines in Figure 7B (panels 1 and 3) trace the initial area of cells removed following the scratch. Cells treated with PI-88 displayed a marked decrease in cell migration (Figure 7B, panel 4) illustrated by the persistence of a clear area into which the cells have not yet migrated. In contrast, control cells appeared to fill the scratch space after 210 min (Figure 7B panel 2). Quantitative densitometry using Image J indicatd that the coverage density in panel 2 was 65% versus 25% in PI-88 treated samples.
Figure 7. PI-88 decreases cellular migration of ATDC5 cells during proliferation and mineralization.
ATDC5 cells in both treated and untreated cultures were subjected to high density mineralization culture. To observe mineralization, micromass cultures were stained with von Kossa. (A) PI-88 caused a decrease in migratory cells during high density mineralization. This is indicated by an absence of a black halo surrounding the micromass. To confirm the effects of PI-88 on migration, the wound healing assay was employed . (B) Cells first were imaged immediately after the scratch was made (0h). At 210 min, cells were re-imaged to determine migration of ATDC5 cells in the wound. Cells treated with 100µg/ml of PI-88 migrated slower than untreated cells. After 210 min, untreated cells had occupied 65% of the original scratch area whereas treated cells had only occupied 25% of the space.
Discussion
This study identifies HPSE as a key regulator of the differentiation processes that take place during endochondral bone formation. In particular, expression is very high in a “ring” of Pc surrounding the region of growth plate that functions as a key signaling center regulating bone growth in both length and width dimensions [2]. This region also is the site where heparin binding growth factors, including FGFs [59], are expressed in developing cartilage. This finding supports a role for HPSE as a regulator between chondrogenic and osteogenic differentiation during endochondral bone formation, an axis in which the Pc is crucial for the proper invasion of blood vessels into the hypertrophic cartilage and for mineralization. [60]. During the chondrogenic process, cells in the upper growth plate proliferate and increase in cell number. In contrast, as chondrocytes differentiate, they exit the cell cycle and begin to express markers of terminal differentiation. A key regulator of this transition is the Ihh/PTHrP signaling pathway which modulates the balance of activity between proliferation and differentiation by controlling signaling between the PHz and the surrounding Pc [61–63]. Interestingly, perlecan/HSPG2 was recently shown by our laboratory to modulate hedgehog protein activity [64], indicating a role for HS in these processes, presumably through control of morphogen gradients as has been well documented in Drosophila [65]. It is thus of interest that the highest levels of HPSE/Hspe expression are in the Pc in the area bordering the PHz-Hz transition.
Further support for a function of HPSE in function of Pc comes from our recent studies in which we used a panel of antibodies specific for unique HS species present on HSPGs present in cartilage including glypican-3, perlecan/HSPG2, and syndecan 3, all of which are important in cartilage patterning and development [66–70]. These studies clearly showed that HS species are dynamic structures within developing growth plate cartilage and the Pc. In particular, GlcNS6S-IdoUA2S-GlcNS6S species were down regulated and localization of GlcNS6S-IdoUA-GlcNS6S species within the Hz of the growth plate was lost during normal development. Such regional differences in HS structures present within developing growth plates demonstrate that interactions with and responses to HS-binding proteins such as HPSE are temporospatially modulated during endochondral bone formation [12] The existence of an active tri-molecular complex involving HS, HBGFs, and HBGF receptors [71] suggests that HPSE, by virtue of its ability to degrade HS, provides an additional key molecule required for these receptor signaling systems to function.
The COJ is the site at which both perlecan/HSPG2 (Figure 1) and HS [12] abruptly disappear from bone, suggesting that HPSE activity plays a key role in perlecan/HSPG2 degradation and the onset of mineralization, an idea consistent with published findings that HPSE overexpression increases osteogenesis [43]. Interestingly, the COJ also is the site at which a number of important ECM degrading enzymes including MMPs have been found [72]. Highly basic, isoelectric point > 10, HS binding growth factor FGF18 that regulates skeletal vascularization and subsequent recruitment of osteoblasts/osteoclasts through regulation of early stages of chondrogenesis and VEGF expression, also is found in this region of the COJ [73]. Because heparin is a potent inhibitor of mineralization [74] and HPSE activity, it also is intriguing to speculate that HPSE driven catabolism of HS at the COJ not only supports HS dependent signaling and angiogenesis, but also removes a critical barrier to bone formation, namely perlecan/HSPG2 and HS.
Of interest, previously published studies showed that PI-88 accumulates in the Pc and Po in living rats given a single subcutaneous dose of [35S]PI-88 [75], presumably by binding to HS present there. To test the idea that active HPSE plays a role in bone growth, we cultured metatarsals in the presence of PI-88 which at high doses inhibited bone growth. We believe this reflects a direct effect of PI-88 on HPSE activity in the Pc, a site at which drug penetration was not expected to be a major problem even in the absence of a functional vasculature for drug delivery. Our observation of decreased growth plate expansion, ratiometric expansion of the bone marrow cavity, and decrease in total bone length in the presence of PI-88 is likely the result of premature hypertrophy and accelerated osteogenesis including Ocn expression, as a consequence of disruption of the Pz-Pc signaling axis owed to loss of HPSE digestion of HS. These observations are well in accord with the previous work identifying HPSE as an important molecule in bone formation. Zcharia et al. demonstrated that human HPSE could be overexpressed under the constitutive control of the ActB promoter [42]. Subsequent studies by the same group showed that while the HPSE transgene was weakly expressed throughout the bone marrow, there was a significant increase in expression in osteoblasts and osteocytes. Mice expressing this transgene displayed increased trabecular bone mass, cortical thickness, and bone formation rates [43]. Bone marrow stromal cells derived from transgenic mice expressing recombinant human HPSE and MC3T3 E1 osteoblastic cells receiving exogenous HPSE spontaneously underwent osteogenic differentiation. All of these findings, combined with our observations of HPSE/Hpse expression in the Pc and COJ reported in this work, are consistent with the idea that HSPGs suppress osteoblast function and that this inhibition is overcome by HS degradation with HPSE [43]. Furthermore, our data also suggest that HPSE present in the Pc directly contributes to bone lengthening by supporting HS-dependent growth factor signaling.
To mechanistically investigate the role of HPSE in endochondral bone formation, we used the well accepted ATDC5 cell in vitro model system, because it permits study of both chondrogenesis and osteogenesis within the same cell culture system [76]. ATDC5 also can mineralize spontaneously without the addition of an exogenous substrate such as β-glycerophosphate [77]. The mid-culture increases in HPSE expression are consistent with the elevation in Hpse expression seen in the PHz-Hz in the in situ hybridization experiments. One function of this short lived increase in expression at this critical time may be to liberate heparin binding growth factors sequestered in the PHz so that they diffuse to the Pc where they can interact with the growth signaling center located there.
To investigate further the importance of HPSE activity, we used PI-88 in the in vitro culture system. To reduce the time required for these experiments, we prepared high density cultures of ATDC5 cells which rapidly differentiate over a six day period. Biomarker analysis confirmed that by D6 terminal differentiation had occurred, and thus these cultures can be compared to the earlier studies using the longer monolayer culture period. In the presence of 100µg/ml of PI-88, an effective dose under these culture conditions, there was a 56% decrease in HPSE activity in D19 differentiated ATDC5 cells. We did not use the higher dose because of concerns about effects on cell attachment owing to the nonenzymatic activities of HPSE [32]. PI-88 treated cell cultures showed decreases in GAG accumulation at both days tested suggesting a role of HPSE activity in chondrocyte differentiation and ECM secretion. While there was only a modest effect of PI-88 on ColX expression under these conditions, there was a dramatic increase in Ocn expression, a late stage marker of osteogenesis. This is interesting in the context of the earlier observations of Kram et al [43] and suggests that HPSE activity must be carefully titrated at the transition from chondrogenesis to osteogenesis, as either its overexpression as seen with the transgene, or its inhibition, as seen here, tips the balance toward osteogenesis. One explanation for the high levels of Ocn seen in the presence of PI-88 may lie with the function of Runx2, the key transcription factor regulating Ocn expression. If inhibition of HPSE disrupts MAPK signaling, known to lie downstream of HPSE [78], then exit from the cell cycle and concomitant up-regulation of Runx2 may be sufficient to increase Ocn expression and support osteogenesis in the presence of PI-88 [79]. The increase in Ocn expression in the growth plate treated with PI-88 is indicative of an advanced stage of chondrocyte hypertrophy or a trans-differentiation of chondrocytes to an osteoblastic like cell, as described [80]. This is consistent with the histological findings shown in Figure 2B. The anti-proliferative and pro-differentiation effect of PI-88 is the one that forms the cornerstone of its testing in clinical trials as an anti-neoplastic agent [81].
When ATDC5 cells were cultured under conditions favoring osteogenesis, we were surprised to note the high number of cells migrating away from the central micromass and taking up residence in the perimeter of the cultures. This inhibition of migration was substantiated in the scratch assays. This novel finding is important because it reveals a new mechanism by which HPSE can modulate the balance between chondrogenic and osteogenic differentiation during endochondral bone formation. The ability of chondrocytes to migrate during transdifferentiation has only recently been addressed [82], but has important implications for cartilage and bone healing after injury and in conditions such as osteoarthritis.
In summary, the data presented here suggests that inhibition of HPSE activity by PI-88 in vitro and ex vivo leads to premature termination of chondrogenic growth and differentiation and favors premature osteogenic differentiation perhaps by promoting transdifferentiation of Hz cells. This effect appears to be due to actions at two distinct signaling centers at which HPSE is expressed: the PHz-Pc growth signaling center and the COJ where terminally differentiated cartilage gives way to trabecular bone and associated angiogenesis. Further understanding of the signal transduction pathways that are disrupted when HPSE activity is perturbed during endochondral ossification will provide valuable insight into skeletal abnormalities observed when HS biosynthetic enzymes and HSPGs are misexpressed during development [83].
Acknowledgments
We thank Dr. Israel Vlodavsky for providing the anti-HPSE polyclonal antibody 1453 and Progen Pharmaceuticals Ltd for providing us with PI-88. The authors wish to thank all the staff working in the Office of Lab Animal Medicine at the University of Delaware. The authors also thank Dr. Randall Duncan and his lab members for their input, as well as Drs. Melinda Duncan, George Dodge, Gary Laverty, and all members of the Carson and Farach-Carson laboratories for their discussions and insightful suggestions. We are grateful to Dr. Rosa Serra, University of Alabama at Birmingham, for her assistance in setting up the metatarsal culture assays. We especially wish to thank Ms. Sharron Kingston for her assistance in the preparation of this manuscript. The project described was partially supported by grants P20RR016458, DE13542 and P01 CA098912. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.DeLise AM, Fischer L, Tuan RS. Cellular interactions and signaling in cartilage development. Osteoarthritis Cartilage. 2000;8:309–334. doi: 10.1053/joca.1999.0306. [DOI] [PubMed] [Google Scholar]
- 2.Olsen BR, Reginato AM, Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 3.Ohta S, Muramatsu H, Senda T, Zou K, Iwata H, Muramatsu T. Midkine is expressed during repair of bone fracture and promotes chondrogenesis. J Bone Miner Res. 1999;14:1132–1144. doi: 10.1359/jbmr.1999.14.7.1132. [DOI] [PubMed] [Google Scholar]
- 4.Gritli-Linde A, Lewis P, McMahon AP, Linde A. The whereabouts of a morphogen: direct evidence for short- and graded long-range activity of hedgehog signaling peptides. Dev Biol. 2001;236:364–386. doi: 10.1006/dbio.2001.0336. [DOI] [PubMed] [Google Scholar]
- 5.Murakami S, Kan M, McKeehan WL, de Crombrugghe B. Up-regulation of the chondrogenic Sox9 gene by fibroblast growth factors is mediated by the mitogen-activated protein kinase pathway. Proc Natl Acad Sci U S A. 2000;97:1113–1118. doi: 10.1073/pnas.97.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pizette S, Niswander L. BMPs are required at two steps of limb chondrogenesis: formation of prechondrogenic condensations and their differentiation into chondrocytes. Dev Biol. 2000;219:237–249. doi: 10.1006/dbio.2000.9610. [DOI] [PubMed] [Google Scholar]
- 7.Tapp H, Hernandez DJ, Neame PJ, Koob TJ. Pleiotrophin inhibits chondrocyte proliferation and stimulates proteoglycan synthesis in mature bovine cartilage. Matrix Biol. 1999;18:543–556. doi: 10.1016/s0945-053x(99)00049-9. [DOI] [PubMed] [Google Scholar]
- 8.Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 9.Yano F, Kugimiya F, Ohba S, Ikeda T, Chikuda T, Ogasawara T, Ogata N, Takato T, Nakamura K, Kawaguchi H, Chung UI. The canonical Wnt signaling pathway promotes chondrocyte differentiation in a Sox9-dependent manner. Biochem Biophys Res Commun. 2005;333:1300–1308. doi: 10.1016/j.bbrc.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 10.Mukherjee A, Dong SS, Clemens T, Alvarez J, Serra R. Co-ordination of TGF-beta and FGF signaling pathways in bone organ cultures. Mech Dev. 2005;122:557–571. doi: 10.1016/j.mod.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Lee K, Lanske B, Karaplis AC, Deeds JD, Kohno H, Nissenson RA, Kronenberg HM, Segre GV. Parathyroid hormone-related peptide delays terminal differentiation of chondrocytes during endochondral bone development. Endocrinology. 1996;137:5109–5118. doi: 10.1210/endo.137.11.8895385. [DOI] [PubMed] [Google Scholar]
- 12.Gomes RR, Jr, Van Kuppevelt TH, Farach-Carson MC, Carson DD. Spatiotemporal distribution of heparan sulfate epitopes during murine cartilage growth plate development. Histochem Cell Biol. 2006;126:713–722. doi: 10.1007/s00418-006-0203-4. [DOI] [PubMed] [Google Scholar]
- 13.Atha DH, Stephens AW, Rimon A, Rosenberg RD. Sequence variation in heparin octasaccharides with high affinity for antithrombin III. Biochemistry. 1984;23:5801–5812. doi: 10.1021/bi00319a020. [DOI] [PubMed] [Google Scholar]
- 14.Chiao E, Fisher P, Crisponi L, Deiana M, Dragatsis I, Schlessinger D, Pilia G, Efstratiadis A. Overgrowth of a mouse model of the Simpson-Golabi-Behmel syndrome is independent of IGF signaling. Dev Biol. 2002;243:185–206. doi: 10.1006/dbio.2001.0554. [DOI] [PubMed] [Google Scholar]
- 15.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparin sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 16.Nicole S, Davoine CS, Topaloglu H, Cattolico L, Barral D, Beighton P, Hamida CB, Hammouda H, Cruaud C, White PS, Samson D, Urtizberea JA, Lehmann-Horn F, Weissenbach J, Hentati F, Fontaine B. Perlecan, the major proteoglycan of basement membranes, is altered in patients with Schwartz-Jampel syndrome (chondrodystrophic myotonia) Nat Genet. 2000;26:480–483. doi: 10.1038/82638. [DOI] [PubMed] [Google Scholar]
- 17.Ashikari S, Habuchi H, Kimata K. Characterization of heparan sulfate oligosaccharides that bind to hepatocyte growth factor. J Biol Chem. 1995;270:29586–29593. doi: 10.1074/jbc.270.49.29586. [DOI] [PubMed] [Google Scholar]
- 18.Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 19.Lindahl U, Thunberg L, Backstrom G, Riesenfeld J, Nordling K, Bjork I. Extension and structural variability of the antithrombin-binding sequence in heparin. J Biol Chem. 1984;259:12368–12376. [PubMed] [Google Scholar]
- 20.So CL, Kaluarachchi K, Tam PP, Cheah KS. Impact of mutations of cartilage matrix genes on matrix structure, gene activity and chondrogenesis. Osteoarthritis Cartilage. 2001;9 Suppl A:S160–S173. [PubMed] [Google Scholar]
- 21.Pacicca DM, Patel N, Lee C, Salisbury K, Lehmann W, Carvalho R, Gerstenfeld LC, Einhorn TA. Expression of angiogenic factors during distraction osteogenesis. Bone. 2003;33:889–898. doi: 10.1016/j.bone.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Gomes RR, Jr, Joshi SS, Farach-Carson MC, Carson DD. Ribozyme-mediated perlecan knockdown impairs chondrogenic differentiation of C3H10T1/2 fibroblasts. Differentiation. 2006;74:53–63. doi: 10.1111/j.1432-0436.2005.00055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomes RR, Jr, Farach Carson MC, Carson DD. Perlecan-stimulated nodules undergo chondrogenic maturation in response to rhBMP-2 treatment in vitro. Connect Tissue Res. 2003;44 Suppl 1:196–201. [PMC free article] [PubMed] [Google Scholar]
- 24.French MM, Gomes RR, Jr, Timpl R, Hook M, Czymmek K, Farach-Carson MC, Carson DD. Chondrogenic activity of the heparan sulfate proteoglycan perlecan maps to the N-terminal domain I. J Bone Miner Res. 2002;17:48–55. doi: 10.1359/jbmr.2002.17.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.French MM, Smith SE, Akanbi K, Sanford T, Hecht J, Farach-Carson MC, Carson DD. Expression of the heparan sulfate proteoglycan, perlecan, during mouse embryogenesis and perlecan chondrogenic activity in vitro. J Cell Biol. 1999;145:1103–1115. doi: 10.1083/jcb.145.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirn-Safran CB, Gomes RR, Brown AJ, Carson DD. Heparan sulfate proteoglycans: coordinators of multiple signaling pathways during chondrogenesis. Birth Defects Res C Embryo Today. 2004;72:69–88. doi: 10.1002/bdrc.20005. [DOI] [PubMed] [Google Scholar]
- 27.Zcharia E, Zilka R, Yaar A, Yacoby-Zeevi O, Zetser A, Metzger S, Sarid R, Naggi A, Casu B, Ilan N, Vlodavsky I, Abramovitch R. Heparanase accelerates wound angiogenesis and wound healing in mouse and rat models. Faseb J. 2005;19:211–221. doi: 10.1096/fj.04-1970com. [DOI] [PubMed] [Google Scholar]
- 28.Miao HQ, Navarro E, Patel S, Sargent D, Koo H, Wan H, Plata A, Zhou Q, Ludwig D, Bohlen P, Kussie P. Cloning, expression, and purification of mouse heparanase. Protein Expr Purif. 2002;26:425–431. doi: 10.1016/s1046-5928(02)00558-2. [DOI] [PubMed] [Google Scholar]
- 29.Podyma-Inoue KA, Yokote H, Sakaguchi K, Ikuta M, Yanagishita M. Characterization of heparanase from a rat parathyroid cell line. J Biol Chem. 2002;277:32459–32465. doi: 10.1074/jbc.M203282200. [DOI] [PubMed] [Google Scholar]
- 30.Levy-Adam F, Miao HQ, Heinrikson RL, Vlodavsky I, Ilan N. Heterodimer formation is essential for heparanase enzymatic activity. Biochem Biophys Res Commun. 2003;308:885–891. doi: 10.1016/s0006-291x(03)01478-5. [DOI] [PubMed] [Google Scholar]
- 31.D'Souza SS, Daikoku T, Farach-Carson MC, Carson DD. Heparanase Expression and Function During Early Pregnancy in Mice. Biol Reprod. 2007;77:433–441. doi: 10.1095/biolreprod.107.061317. [DOI] [PubMed] [Google Scholar]
- 32.Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr Pharm Des. 2007;13:2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- 33.Bame KJ. Heparanases: endoglycosidases that degrade heparan sulfate proteoglycans. Glycobiology. 2001;11:91R–98R. doi: 10.1093/glycob/11.6.91r. [DOI] [PubMed] [Google Scholar]
- 34.Freeman C, Browne AM, Parish CR. Evidence that platelet and tumour heparanases are similar enzymes. Biochem J. 1999;342(Pt 2):361–368. [PMC free article] [PubMed] [Google Scholar]
- 35.Sewell RF, Brenchley PE, Mallick NP. Human mononuclear cells contain an endoglycosidase specific for heparan sulphate glycosaminoglycan demonstrable with the use of a specific solid-phase metabolically radiolabelled substrate. Biochem J. 1989;264:777–783. doi: 10.1042/bj2640777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laskov R, Michaeli RI, Sharir H, Yefenof E, Vlodavsky I. Production of heparanase by normal and neoplastic murine B-lymphocytes. Int J Cancer. 1991;47:92–98. doi: 10.1002/ijc.2910470117. [DOI] [PubMed] [Google Scholar]
- 37.Matzner Y, Bar-Ner M, Yahalom J, Ishai-Michaeli R, Fuks Z, Vlodavsky I. Degradation of heparan sulfate in the subendothelial extracellular matrix by a readily released heparanase from human neutrophils. Possible role in invasion through basement membranes. J Clin Invest. 1985;76:1306–1313. doi: 10.1172/JCI112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mollinedo F, Nakajima M, Llorens A, Barbosa E, Callejo S, Gajate C, Fabra A. Major co-localization of the extracellular-matrix degradative enzymes heparanase and gelatinase in tertiary granules of human neutrophils. Biochem J. 1997;327(Pt 3):917–923. doi: 10.1042/bj3270917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlodavsky I, Goldshmidt O, Zcharia E, Metzger S, Chajek-Shaul T, Atzmon R, Guatta-Rangini Z, Friedmann Y. Molecular properties and involvement of heparanase in cancer progression and normal development. Biochimie. 2001;83:831–839. doi: 10.1016/s0300-9084(01)01318-9. [DOI] [PubMed] [Google Scholar]
- 40.Elkin M, Ilan N, Ishai-Michaeli R, Friedmann Y, Papo O, Pecker I, Vlodavsky I. Heparanase as mediator of angiogenesis: mode of action. Faseb J. 2001;15:1661–1663. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 41.Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–M108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 42.Zcharia E, Metzger S, Chajek-Shaul T, Aingorn H, Elkin M, Friedmann Y, Weinstein T, Li JP, Lindahl U, Vlodavsky I. Transgenic expression of mammalian heparanase uncovers physiological functions of heparan sulfate in tissue morphogenesis, vascularization, and feeding behavior. Faseb J. 2004;18:252–263. doi: 10.1096/fj.03-0572com. [DOI] [PubMed] [Google Scholar]
- 43.Kram V, Zcharia E, Yacoby-Zeevi O, Metzger S, Chajek-Shaul T, Gabet Y, Muller R, Vlodavsky I, Bab I. Heparanase is expressed in osteoblastic cells and stimulates bone formation and bone mass. J Cell Physiol. 2006;207:784–792. doi: 10.1002/jcp.20625. [DOI] [PubMed] [Google Scholar]
- 44.Shukunami C, Ishizeki K, Atsumi T, Ohta Y, Suzuki F, Hiraki Y. Cellular hypertrophy and calcification of embryonal carcinoma-derived chondrogenic cell line ATDC5 in vitro. J Bone Miner Res. 1997;12:1174–1188. doi: 10.1359/jbmr.1997.12.8.1174. [DOI] [PubMed] [Google Scholar]
- 45.Das SK, Wang XN, Paria BC, Damm D, Abraham JA, Klagsbrun M, Andrews GK, Dey SK. Heparin-binding EGF-like growth factor gene is induced in the mouse uterus temporally by the blastocyst solely at the site of its apposition: a possible ligand for interaction with blastocyst EGF-receptor in implantation. Development. 1994;120:1071–1083. doi: 10.1242/dev.120.5.1071. [DOI] [PubMed] [Google Scholar]
- 46.Addison WN, Azari F, Sorensen ES, Kaartinen MT, McKee MD. Pyrophosphate Inhibits Mineralization of Osteoblast Cultures by Binding to Mineral, Up-regulating Osteopontin, and Inhibiting Alkaline Phosphatase Activity. J Biol Chem. 2007;282:15872–15883. doi: 10.1074/jbc.M701116200. [DOI] [PubMed] [Google Scholar]
- 47.MacRae VE, Farquharson C, Ahmed SF. The restricted potential for recovery of growth plate chondrogenesis and longitudinal bone growth following exposure to pro-inflammatory cytokines. J Endocrinol. 2006;189:319–328. doi: 10.1677/joe.1.06609. [DOI] [PubMed] [Google Scholar]
- 48.McKee MD, Addison WN, Kaartinen MT. Hierarchies of extracellular matrix and mineral organization in bone of the craniofacial complex and skeleton. Cells Tissues Organs. 2005;181:176–188. doi: 10.1159/000091379. [DOI] [PubMed] [Google Scholar]
- 49.Ueda K, Yamanaka Y, Harada D, Yamagami E, Tanaka H, Seino Y. PTH has the potential to rescue disturbed bone growth in achondroplasia. Bone. 2007;41:13–18. doi: 10.1016/j.bone.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 50.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 51.Porzio MA, Pearson AM. Improved resolution of myofibrillar proteins with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochim Biophys Acta. 1977;490:27–34. doi: 10.1016/0005-2795(77)90102-7. [DOI] [PubMed] [Google Scholar]
- 52.Reiland J, Sanderson RD, Waguespack M, Barker SA, Long R, Carson DD, Marchetti D. Heparanase degrades syndecan-1 and perlecan heparan sulfate: functional implications for tumor cell invasion. J Biol Chem. 2004;279:8047–8055. doi: 10.1074/jbc.M304872200. [DOI] [PubMed] [Google Scholar]
- 53.Denker AE, Haas AR, Nicoll SB, Tuan RS. Chondrogenic differentiation of murine C3H10T1/2 multipotential mesenchymal cells: I. Stimulation by bone morphogenetic protein-2 in high-density micromass cultures. Differentiation. 1999;64:67–76. doi: 10.1046/j.1432-0436.1999.6420067.x. [DOI] [PubMed] [Google Scholar]
- 54.Atkinson BL, Fantle KS, Benedict JJ, Huffer WE, Gutierrez-Hartmann A. Combination of osteoinductive bone proteins differentiates mesenchymal C3H/10T1/2 cells specifically to the cartilage lineage. J Cell Biochem. 1997;65:325–339. [PubMed] [Google Scholar]
- 55.Izzo MW, Pucci B, Tuan RS, Hall DJ. Gene expression profiling following BMP-2 induction of mesenchymal chondrogenesis in vitro. Osteoarthritis Cartilage. 2002;10:23–33. doi: 10.1053/joca.2001.0478. [DOI] [PubMed] [Google Scholar]
- 56.Miller SA, Brown AJ, Farach-Carson MC, Kirn-Safran CB. HIP/RPL29 down-regulation accompanies terminal chondrocyte differentiation. Differentiation. 2003;71:322–336. doi: 10.1046/j.1432-0436.2003.7106002.x. [DOI] [PubMed] [Google Scholar]
- 57.Seghatoleslami MR, Tuan RS. Cell density dependent regulation of AP-1 activity is important for chondrogenic differentiation of C3H10T1/2 mesenchymal cells. J Cell Biochem. 2002;84:237–248. doi: 10.1002/jcb.10019. [DOI] [PubMed] [Google Scholar]
- 58.Naik MU, Naik UP. Calcium-and integrin-binding protein regulates focal adhesion kinase activity during platelet spreading on immobilized fibrinogen. Blood. 2003;102:3629–3636. doi: 10.1182/blood-2003-05-1703. [DOI] [PubMed] [Google Scholar]
- 59.Lazarus JE, Hegde A, Andrade AC, Nilsson O, Baron J. Fibroblast growth factor expression in the postnatal growth plate. Bone. 2007;40:577–586. doi: 10.1016/j.bone.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 60.Colnot C, Lu C, Hu D, Helms JA. Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev Biol. 2004;269:55–69. doi: 10.1016/j.ydbio.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 61.Akiyama H, Shigeno C, Iyama K, Ito H, Hiraki Y, Konishi J, Nakamura T. Indian hedgehog in the late-phase differentiation in mouse chondrogenic EC cells, ATDC5: upregulation of type X collagen and osteoprotegerin ligand mRNAs. Biochem Biophys Res Commun. 1999;257:814–820. doi: 10.1006/bbrc.1999.0494. [DOI] [PubMed] [Google Scholar]
- 62.Colnot C, de la Fuente L, Huang S, Hu D, Lu C, St-Jacques B, Helms JA. Indian hedgehog synchronizes skeletal angiogenesis and perichondrial maturation with cartilage development. Development. 2005;132:1057–1067. doi: 10.1242/dev.01649. [DOI] [PubMed] [Google Scholar]
- 63.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 64.Datta MW, Hernandez AM, Schlicht MJ, Kahler AJ, DeGueme AM, Dhir R, Shah RB, Farach-Carson C, Barrett A, Datta S. Perlecan, a candidate gene for the CAPB locus, regulates prostate cancer cell growth via the Sonic Hedgehog pathway. Mol Cancer. 2006;5:9. doi: 10.1186/1476-4598-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strigini M. Mechanisms of morphogen movement. J Neurobiol. 2005;64:324–333. doi: 10.1002/neu.20164. [DOI] [PubMed] [Google Scholar]
- 66.Arikawa-Hirasawa E, Watanabe H, Takami H, Hassell JR, Yamada Y. Perlecan is essential for cartilage and cephalic development. Nat Genet. 1999;23:354–358. doi: 10.1038/15537. [DOI] [PubMed] [Google Scholar]
- 67.Cano-Gauci DF, Song HH, Yang H, McKerlie C, Choo B, Shi W, Pullano R, Piscione TD, Grisaru S, Soon S, Sedlackova L, Tanswell AK, Mak TW, Yeger H, Lockwood GA, Rosenblum ND, Filmus J. Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J Cell Biol. 1999;146:255–264. doi: 10.1083/jcb.146.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costell M, Gustafsson E, Aszodi A, Morgelin M, Bloch W, Hunziker E, Addicks K, Timpl R, Fassler R. Perlecan maintains the integrity of cartilage and some basement membranes. J Cell Biol. 1999;147:1109–1122. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koyama E, Leatherman JL, Shimazu A, Nah HD, Pacifici M. Syndecan-3, tenascin-C, and the development of cartilaginous skeletal elements and joints in chick limbs. Dev Dyn. 1995;203:152–162. doi: 10.1002/aja.1002030204. [DOI] [PubMed] [Google Scholar]
- 70.Seghatoleslami MR, Kosher RA. Inhibition of in vitro limb cartilage differentiation by syndecan-3 antibodies. Dev Dyn. 1996;207:114–119. doi: 10.1002/(SICI)1097-0177(199609)207:1<114::AID-AJA11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 71.Wu ZL, Zhang L, Yabe T, Kuberan B, Beeler DL, Love A, Rosenberg RD. The involvement of heparan sulfate (HS) in FGF1/HS/FGFR1 signaling complex. J Biol Chem. 2003;278:17121–17129. doi: 10.1074/jbc.M212590200. [DOI] [PubMed] [Google Scholar]
- 72.Ortega N, Behonick D, Stickens D, Werb Z. How proteases regulate bone morphogenesis. Ann N Y Acad Sci. 2003;995:109–116. doi: 10.1111/j.1749-6632.2003.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 73.Liu Z, Lavine KJ, Hung IH, Ornitz DM. FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev Biol. 2007;302:80–91. doi: 10.1016/j.ydbio.2006.08.071. [DOI] [PubMed] [Google Scholar]
- 74.Hansen NM, Felix R, Bisaz S, Fleisch H. Aggregation of hydroxyapatite crystals. Biochim Biophys Acta. 1976;451:549–559. doi: 10.1016/0304-4165(76)90150-1. [DOI] [PubMed] [Google Scholar]
- 75.Levidiotis V, Freeman C, Punler M, Martinello P, Creese B, Ferro V, van der Vlag J, Berden JH, Parish CR, Power DA. A synthetic heparanase inhibitor reduces proteinuria in passive Heymann nephritis. J Am Soc Nephrol. 2004;15:2882–2892. doi: 10.1097/01.ASN.0000142426.55612.6D. [DOI] [PubMed] [Google Scholar]
- 76.Mushtaq T, Farquharson C, Seawright E, Ahmed SF. Glucocorticoid effects on chondrogenesis, differentiation and apoptosis in the murine ATDC5 chondrocyte cell line. J Endocrinol. 2002;175:705–713. doi: 10.1677/joe.0.1750705. [DOI] [PubMed] [Google Scholar]
- 77.Guicheux J, Palmer G, Shukunami C, Hiraki Y, Bonjour JP, Caverzasio J. A novel in vitro culture system for analysis of functional role of phosphate transport in endochondral ossification. Bone. 2000;27:69–74. doi: 10.1016/s8756-3282(00)00302-1. [DOI] [PubMed] [Google Scholar]
- 78.Patel VN, Knox SM, Likar KM, Lathrop CA, Hossain R, Eftekhari S, Whitelock JM, Elkin M, Vlodavsky I, Hoffman MP. Heparanase cleavage of perlecan heparan sulfate modulates FGF10 activity during ex vivo submandibular gland branching morphogenesis. Development. 2007;134:4177–4186. doi: 10.1242/dev.011171. [DOI] [PubMed] [Google Scholar]
- 79.Thomas DM, Johnson SA, Sims NA, Trivett MK, Slavin JL, Rubin BP, Waring P, McArthur GA, Walkley CR, Holloway AJ, Diyagama D, Grim JE, Clurman BE, Bowtell DD, Lee JS, Gutierrez GM, Piscopo DM, Carty SA, Hinds PW. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lian JB, McKee MD, Todd AM, Gerstenfeld LC. Induction of bone-related proteins, osteocalcin and osteopontin, and their matrix ultrastructural localization with development of chondrocyte hypertrophy in vitro. J Cell Biochem. 1993;52:206–219. doi: 10.1002/jcb.240520212. [DOI] [PubMed] [Google Scholar]
- 81.Miao HQ, Liu H, Navarro E, Kussie P, Zhu Z. Development of heparanase inhibitors for anti-cancer therapy. Curr Med Chem. 2006;13:2101–2111. doi: 10.2174/092986706777935230. [DOI] [PubMed] [Google Scholar]
- 82.Morales TI. Chondrocyte moves: clever strategies? Osteoarthritis Cartilage. 2007 doi: 10.1016/j.joca.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farach-Carson MC, Hecht JT, Carson DD. Heparan sulfate proteoglycans: key players in cartilage biology. Crit Rev Eukaryot Gene Expr. 2005;15:29–48. doi: 10.1615/critreveukaryotgeneexpr.v15.i1.30. [DOI] [PubMed] [Google Scholar]
- 84.Yang W, Gomes RR, Brown AJ, Burdett AR, Alicknavitch M, Farach-Carson MC, Carson DD. Chondrogenic differentiation on perlecan domain I, collagen II, and bone morphogenetic protein-2-based matrices. Tissue Eng. 2006;12:2009–2024. doi: 10.1089/ten.2006.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boudreaux JM, Towler DA. Synergistic induction of osteocalcin gene expression: identification of a bipartite element conferring fibroblast growth factor 2 and cyclic AMP responsiveness in the rat osteocalcin promoter. J Biol Chem. 1996;271:7508–7515. doi: 10.1074/jbc.271.13.7508. [DOI] [PubMed] [Google Scholar]