Summary
We describe the first NMR structure of a polytopic helical membrane protein. DsbB, a bacterial cytoplasmic membrane protein, plays a key role in disulfide bond formation. It re-oxidizes DsbA, the periplasmic protein disulfide oxidant, using the oxidizing power of membrane-embedded quinones. We determined the structure of an inter-loop disulfide bond form of DsbB, an intermediate in catalysis. Analysis of the structure and interactions with substrates DsbA and quinone reveals functionally relevant changes induced by these substrates. Analysis of the structure, dynamics measurements, and NMR chemical shifts around the inter-loop disulfide bond suggest how electron movement from DsbA to quinone through DsbB is regulated and facilitated. Our results demonstrate the extraordinary utility of NMR for functional characterization of polytopic integral membrane proteins and provide novel insights into the mechanism of DsbB catalysis.
Introduction
Disulfide bonds are essential for the folding, function, and stability of proteins exported from the cytoplasm. Although disulfide bond formation can occur spontaneously in the presence of oxygen, this process is inefficient for fast protein folding and often results in protein aggregation (Anfinsen et al., 1961). Both prokaryotes and eukaryotes have evolved elaborate enzymatic systems to ensure the fast and correct formation of disulfide bonds that occurs in vivo. In E. coli, the formation of disulfide bonds in the periplasm is catalyzed by the enzymes DsbA and DsbB and by their homologs in other gram-negative bacteria. DsbA has a Cys30-Pro31-His32-Cys33 active site embedded in its thioredoxin-like fold. This protein uses the disulfide bond formed in its active site to directly oxidize periplasmic substrates (Bardwell et al., 1991; Martin et al., 1993). DsbB spans the cytoplasmic membrane four times and has two periplasmic loops. This membrane protein re-oxidizes the reduced form of DsbA using the oxidizing power of membrane-embedded quinone (ubiquinone (UQ)-8 or menaquinone (MK)-8), linking disulfide bond formation to the electron transport chain (Bader et al., 1999). Since multiple bacterial virulence factors are secreted proteins and require disulfide bonds for proper folding and function, the DsbA/DsbB system is a potential antimicrobial drug target (Łasica and Jagusztyn-Krynicka, 2007).
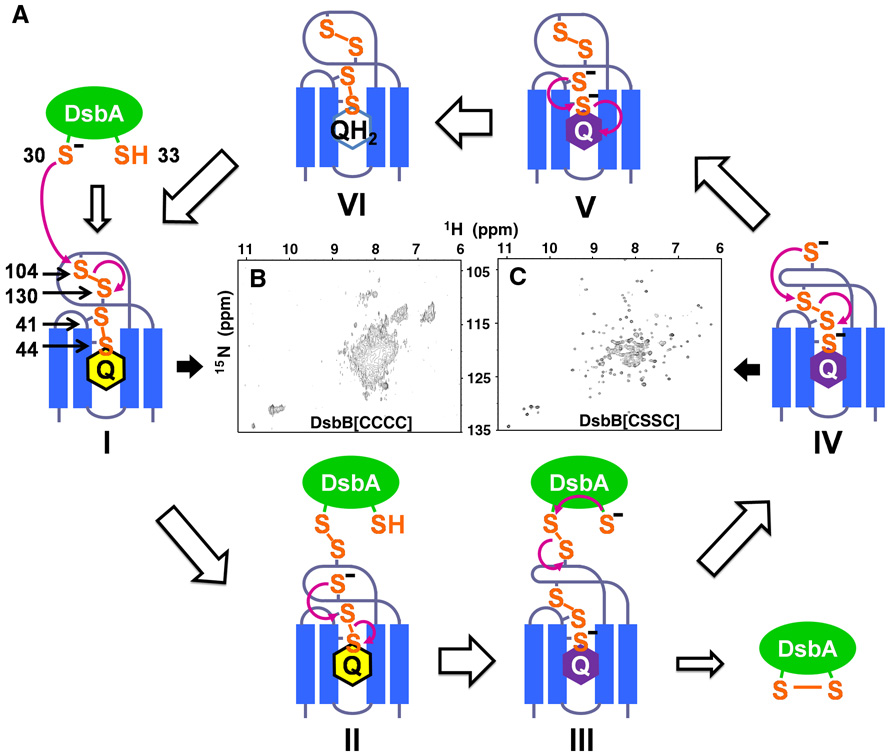
In the process of de novo disulfide bond formation, the spatially separated processes of DsbA oxidation and quinone reduction are linked by an electron transfer pathway consisting of two pairs of essential cysteines in DsbB, Cys41-Cys44 and Cys104-Cys130. The redox potential of ubiquinone (+113 mV), which is higher than that of DsbA (−122 mV), provides the thermodynamic driving force for the overall conversion of reduced DsbA to oxidized DsbA (Gennis and Stewart, 1996; Zapun et al., 1993). However, experimental measurements of the redox potential of the DsbB cysteine pairs indicate they are more reducing than the DsbA active site cysteine pair, suggesting that the transfer of reducing equivalent from DsbA to either one of the two cysteine pairs of DsbB would be energetically unfavorable (Inaba et al., 2005). Mutagenesis and kinetics studies suggest a disulfide rearrangement mechanism that can overcome this barrier (Kadokura and Beckwith, 2002; Tapley et al., 2007). Studies of disulfide bond intermediates in the mechanism of DsbB (Figure 1A) indicate that the reaction is initiated by the nucleophilic attack of Cys30 of DsbA on Cys104 of the Cys104-Cys130 disulfide in DsbB, resulting in a Cys30(DsbA)-Cys104(DsbB) intermolecular disulfide. The reduced Cys130, in turn, attacks the Cys41-Cys44 disulfide resulting in a Cys41-Cys130 inter-loop disulfide bond. The Cys44 thiolate and bound ubiquinone form a charge transfer complex that manifests as a characteristic purple color (Inaba et al., 2004).
Figure 1. Mechanism of DsbB catalyzed DsbA oxidation.
(A) Schematic representation of steps in DsbB catalyzed DsbA oxidation. (B) TROSY-based 15N-1H HSQC spectrum (500 MHz) of protonated wildtype DsbB and (C) DsbB[CSSC] in DPC micelles.
Typically, disulfide exchange reactions occur between two thiols and one disulfide bond. However, DsbB appears to utilize a unique mechanism, as described above, that involves the coordinated action of the four cysteines of DsbB before releasing oxidized DsbA from the DsbB-DsbA complex. We have proposed that such a concerted action of the DsbB cysteine residues is required for oxidation of the highly oxidizing DsbA (Kadokura and Beckwith, 2002). An alternative model has been proposed that suggests that electron transfer from DsbA to quinone proceeds through a sequential set of two dithiol/disulfide exchange reaction and then reduction of the quinone (Inaba et al., 2006; Inaba et al., 2005). However, there is growing evidence that supports the concerted mechanism of DsbB. In fact, resolution of the disulfide rearranged intermediate is the rate limiting step of the reaction which may well involve a concerted process of oxidized DsbA release, disulfide rearrangement, and quinone reduction (Tapley et al. 2007).
The inter-loop disulfide bond that forms between Cys41 and Cys130 is thought to play an important role in the reaction cycle of DsbB (Kadokura and Beckwith 2002; Tapley et al., 2007). In the previously-reported DsbB-DsbA crystal structure (corresponding to intermediate II in Figure 1A), information about this inter-loop disulfide bond was absent because it was necessary to mutate Cys130 to stabilize the DsbB-DsbA complex (Inaba et al., 2006a). Furthermore, several functional parts of the structure of DsbB were missing. To gain insights into the mechanism of DsbB catalysis, we solved the structure of the DsbB inter-loop disulfide bond intermediate. The detailed structures of DsbB we solved with NMR yield insights into how the cysteines of DsbB cooperate in a concerted reaction to move electrons from DsbA to quinones.
Results
Solution NMR approach to the structure of the polytopic membrane enzyme DsbB
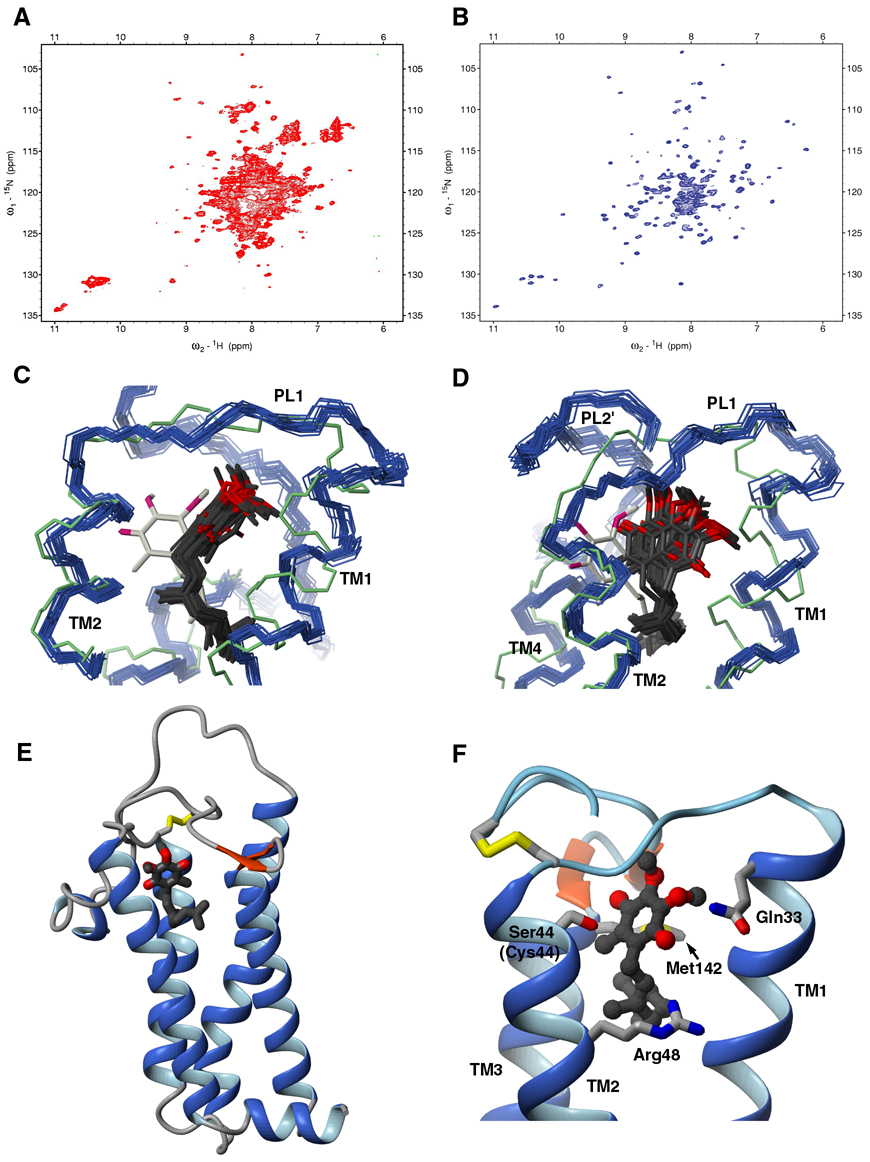
Oxidation of DsbA by DsbB is thought to proceed via multiple disulfide-bonded intermediates (Fig. 1A). Appropriate Ala or Ser mutations of Cys residues in DsbB can generate stable versions of these different intermediates which are amenable to structure determination. For example, mutation of Cys44 and Cys104 to Ser allows formation of a stable inter-loop disulfide bond corresponding to intermediate IV in Figure 1A. We will refer to this disulfide-bonded form as DsbB[CSSC]. This transient inter-loop disulfide bond couples DsbA oxidation and quinone reduction and promotes the forward reaction (Kadokura and Beckwith 2002). Structural information on this critical intermediate is likely to yield unique insights into the mechanism of electron transfer in DsbB. Interestingly, NMR spectra of wildtype DsbB are of low quality and indicative of conformational exchange, whereas the NMR spectrum of DsbB[CSSC] is of very high quality indicating that the conformational exchange has been quenched (Figures 1B and 1C).
NMR spectroscopy is a powerful tool for the study of the structure and function of enzymes. Not only can it be used for structure determination, but also for studies of the role of dynamics in function through relaxation measurements, identification of binding sites for substrates by chemical shift perturbation studies, and unique chemical information from the NMR chemical shifts. Polytopic α-helical membrane proteins like DsbB are notoriously difficult targets for structure determination by either X-ray crystallography or NMR spectroscopy. Using either approach, it is necessary to remove the protein from its native bilayer environment and stabilize it in some membrane mimetic environment, typically some type of detergent micelle. The resulting protein-detergent complexes are large by solution NMR standards and therefore require extensive deuteration of the protein to enhance the performance of the NMR experiments necessary for backbone resonance assignment. This results in a substantial loss of proton-proton NOEs which are typically utilized to determine structures from NMR data. This loss is particularly striking for α–helical membrane proteins as there are typically few if any long-range NH-NH NOEs to define the fold, unlike the β-barrel membrane proteins where the inter-strand NOEs readily provide adequate information to define the fold. As a result, successful application of NMR methods to α-helical membrane protein structure determination has thus far been largely limited to peptides where full deuteration is not required (Rastogi and Girvin, 1999) and homo-oligomers of single transmembrane peptides for which relatively few long-range NOEs are required to determine the global fold (Schnell and Chou, 2008). Thus, application of alternative methods are essential to determine the structure of DsbB[CSSC].
To address the need for technical improvement, we have used a combination of NOEs from a deuterated (2H,13C,15N-labeled) ILV methyl protonated sample, backbone dihedral angles from chemical shift analysis, paramagnetic relaxation enhancement (PRE) measurements from 9 nitroxide labeled samples, and extensive residual dipolar coupling (RDC) measurements (Table 1) to determine the structure of DsbB[CSSC]. DsbB[CSSC] samples were prepared by expression of the Cys41Ser/Cys104Ser mutant of DsbB, isolation of membranes, detergent extraction, and standard affinity purification. The protein was fully oxidized upon isolation. While we obtained a limited number of long-range NOEs involving the indole NH of Trp residues or the methyl groups from the ILV methyl protonated sample, these proved extremely useful as 50% of them were inter-helix NOEs and therefore played a key role in positioning the helices relative to one another. The residual dipolar couplings (RDCs) were measured using charged polyacrylamide gels, whose utility in aligning membrane proteins we have previously demonstrated (Cierpicki and Bushweller, 2004). The structure has been solved using data from DsbB samples in dodecyl phosphocholine (DPC), a detergent in which the wildtype protein retains full activity.
Table 1.
NMR and refinement statistics for DsbB protein structures.
| DsbB[CSSC] | DsbB[CSSC] – UQ2 | |
|---|---|---|
| NMR distance and dihedral constraints | ||
| Distance constraints | ||
| NOEs | ||
| Intra-residue | 41 | 44 |
| Inter-residue | ||
| Sequential (|i − j| = 1) | 191 | 191 |
| Medium-range (1 < |i − j| < 5) | 216 | 216 |
| Long-range (|i − j| > 4) | 39 | 39 |
| Intermolecular | NA | 10 |
| Ambiguous | 14 | 14 |
| Hydrogen bonds | 97 | 97 |
| Paramagnetic relaxation enhancements | ||
| Upper bound | 871 | 872 |
| Lower bound | 273 | 274 |
| Total dihedral angle restraints | ||
| ϕ | 144 | 144 |
| ψ | 151 | 151 |
| Residual dipolar couplings | ||
| 1DHN | 114 | 114 |
| 1DNC’ | 109 | 109 |
| 1DC’Cα | 114 | 114 |
| Structure statistics | 20 structures | 20 structures |
| Violations (mean and s.d.) | ||
| Distance constraints (Å) | 0.0099±0.0008 | 0.0098±0.0009 |
| Dihedral angle constraints (°) | 0.204±0.031 | 0.206±0.027 |
| Residual dipolar couplings (Hz) | ||
| HN | 2.64±0.08 | 2.63±0.06 |
| NC’ | 0.049±0.001 | 0.049±0.001 |
| C’Cα | 1.20±0.02 | 1.21±0.01 |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.0017±0.0000 | 0.0017±0.0000 |
| Bond angles (°) | 0.402±0.003 | 0.403±0.002 |
| Impropers (°) | 0.248±0.005 | 0.250±0.004 |
| Ramachandran plot statistics* (%) | ||
| Residues in most favored regions | 86.9 | 90.0 |
| Residues in additionally allowed regions | 10.0 | 9.2 |
| Residues in generously allowed regions | 3.1 | 0.8 |
| Residues in disallowed regions | 0 | 0 |
| Average pairwise r.m.s. deviation*(Å) | ||
| Heavy | 1.75 | 1.67 |
| Backbone | 0.80 | 0.70 |
Statistics applied in structure rigid regions (aa 4–97and 114–164, see Figure 3C).
Considering the available data and the structural characteristics of α-helical membrane proteins, a two step hybrid approach was used for structure calculations with DsbB. In the first step, the structure of each secondary structure element, i.e. the transmembrane helices and loops, was determined to high resolution by chemical shift derived backbone torsion angles, RDCs, and sequential/medium range NOEs. The structures of these elements were then fixed by experimentally verified hydrogen bond restraints and relatively restrictive backbone torsion angle restraints. In the second step, these segments were folded together using the RDC data, PRE data, and long-range methyl NOE restraints (details provided in Supplemental Data).
Structure of DsbB[CSSC]
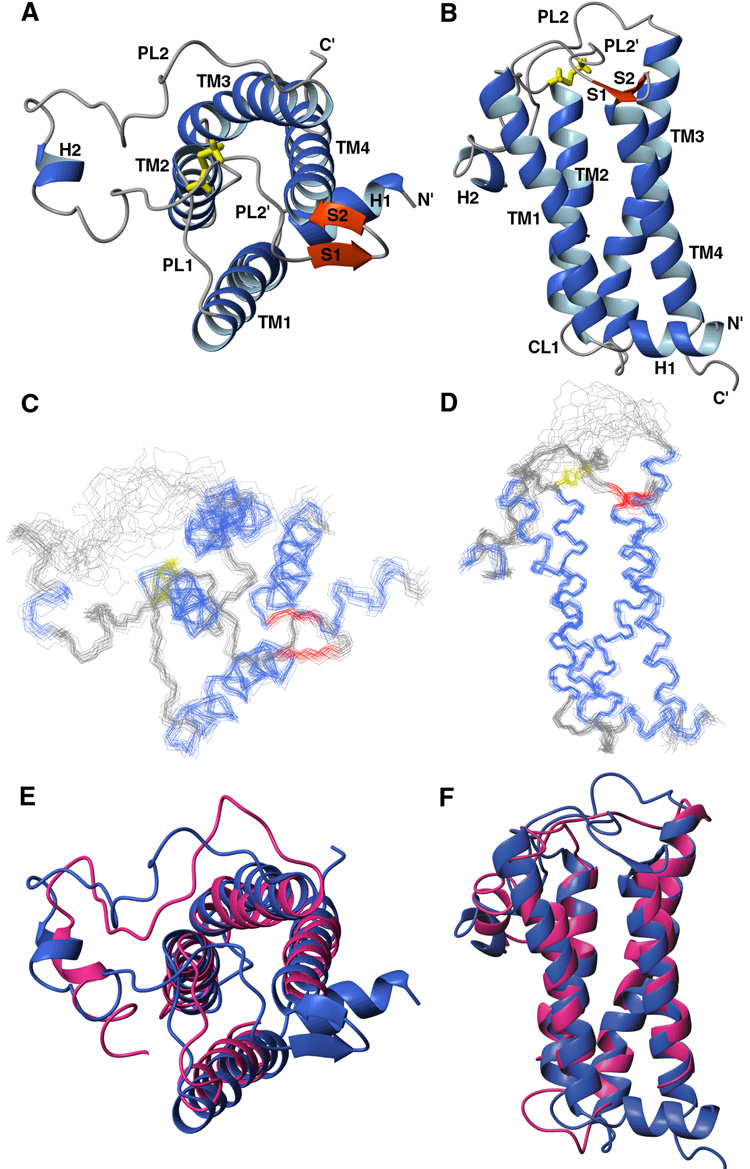
The structure of DsbB[CSSC] presented in Figure 2 shows that DsbB contains four transmembrane helices (TM1-TM4) arranged in a left-handed bundle. A previously unreported N-terminal amphipathic helix H1 (aa 2–9) precedes TM1 and is parallel to the membrane surface. TM1 is bent slightly outside the bundle creating a groove between TM1 and TM4 to accommodate the binding of UQ8, the quinone bound to DsbB in vivo under aerobic conditions. TM3 (aa 70–96) is the longest helix in DsbB, with its N-terminus protruding well above the lipid bilayer (see below). The functionally important second periplasmic loop connecting TM3 and TM4, containing two of the catalytically critical cysteine residues, can be divided into four segments based on secondary structure and dynamics. The N-terminal PL2 (aa 99–115) lacks any regular secondary structure. In addition, measurements of the 15N-{1H} heteronuclear NOE show that residues 99–110 have significantly increased mobility relative to the remainder of the protein (Figure 3C). PL2 is followed by a short amphipathic helical segment (H2, aa 116–119) which resides in the lipid bilayer (see below). PL2' (aa 120–135) is well-ordered and its C-terminal residues 125–135 adopt an extended conformation. PL2' crosses the top of TM2 where Cys41 meets Cys130 to form the inter-loop disulfide bond. At the C terminus of PL2', there is a short beta hairpin (aa 136–141) stabilized by the sidechain of the conserved Trp145 sitting on top of TM4.
Figure 2. Structure of DsbB[CSSC].
(A) Ribbon representation of the structure of DsbB[CSSC] viewed from the periplasmic end. The inter-loop disulfide bond (Cys41-Cys130) is in yellow. (B) Ribbon representation of the structure of DsbB viewed from the membrane plane. (C) Overlay of 20 conformers of DsbB[CSSC] viewed from the periplasmic end, with α-helices in blue, β-strands in red, disulfide bond in yellow. (D) Overlay of 20 conformers of DsbB[CSSC] viewed from the membrane plane. (E) Overlay of DsbB NMR structure (blue) and crystal structure (red) viewed from the periplasmic end. (F) Overlay of DsbB NMR and crystal structure viewed from the membrane plane.
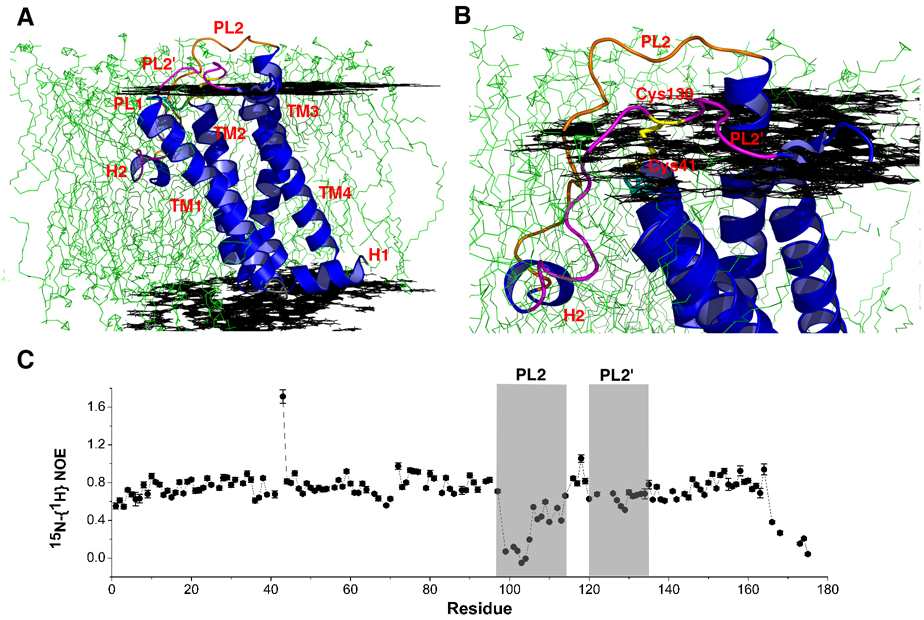
Figure 3. Docking of DsbB[CSSC] into a POPC lipid bilayer and periplasmic loop dynamics.
(A) Structure of DsbB[CSSC] docked into a POPC bilayer using EPR measurements. Black planes represent the level of the phosphate head group and green are individual phosholipid molecules from a molecular dynamics simulation. (B) Close-up view of DsbB[CSSC] docked into a POPC bilayer. (C) Plot of 15N-{1H} heteronuclear NOEs versus the primary sequence. PL2 (aa 97–115) and PL2’ (aa 120–135) are indicated with gray shadow. Depressed values in the N-terminus of PL2 (aa 99–105) indicate increased mobility in this region.
Comparison of the solution structure of DsbB[CSSC] with the 3.7 Å resolution crystal structure of DsbB in a DsbB-DsbA complex shows that the transmembrane portion of the two structures is similar (backbone r.m.s.d. of 1.6 Å for residues 15–33, 42–52, 71–96 and 143–159, see Figure 2E and 2F), suggesting that the transmembrane domain of DsbB does not undergo any significant conformational change upon going from intermediate II to intermediate IV (Figure 1A). The crystal structure lacks structural information for the N-terminal helix and the PL2’ loop. There is a significant difference between the solution structure and the crystal structure for helix H2. In the crystal structure, this was modeled as a continuous α helix from residue 112 to 126. In the solution structure, the well-defined portion of the H2 helix is much shorter (aa 116–119) and it is attached to the well-defined PL2’ loop.
EPR-based docking of DsbB[CSSC] into a POPC bilayer
EPR spectroscopy of nitroxide-labeled proteins provides a powerful approach to probe the depth of insertion, orientation, and dynamics of membrane proteins in their native bilayer environment in a site-specific manner. The necessary nitroxide labels are readily introduced by disulfide exchange with Cys residues introduced into the protein for this purpose. We have labeled 11 Cys mutants of DsbB using MTSSL. These nitroxide-labeled DsbB samples have been reconstituted into POPC vesicles and the depth of nitroxide insertion into the membrane bilayer measured using the differential effects of oxygen and nickel on the power saturation of the nitroxides (Altenbach et al., 2005). Using the depth of insertion data obtained for all nitroxides, we have docked the structure of DsbB into a bilayer (Figures 3A and 3B), showing the orientation of DsbB in its native bilayer environment. These results clearly show the hydrophobic portions of the TM helices to be embedded in the lipid portion of the bilayer, supporting the notion that the structure does indeed reflect that found in the bilayer. The C-terminus of TM3 extends above the bilayer, helping to position PL2 for interaction with DsbA. A portion of the second periplasmic loop including the short amphipathic helical segment (H2) is inserted quite deeply into the membrane, with several hydrophobic amino acid sidechains (Leu116, Trp119, and Val120) completely buried in the lipid portion of the bilayer. The sidechains of Asp117 and Lys118 are pointing towards the interfacial region but are also buried, presumably by means of a salt bridge interaction between the two which negates the charge. H2 is inserted deeper than a typical amphipathic helix. To verify its positon, we introduced nitroxide spin labels at Trp119 and Val120 in H2. The measured depths are 11.8 Å and 9.3 Å from the phosphate head group, respectively, consistent with the location shown in Figure 3. While it is possible that the orientation of this helix differs somewhat between the micelle and bilayer environments, the good agreement of all of the nitroxide depth data with the structure rules out any large-scale reorientation of this helix between the two environments. The importance of this helix was recently demonstrated in a study which showed that the amphipathic nature of this helix is essential for the ability of DsbB to selectively oxidize DsbA and not DsbC (Pan et al., 2008).
Mapping of the binding site for DsbA on DsbB identifies two sites for interaction
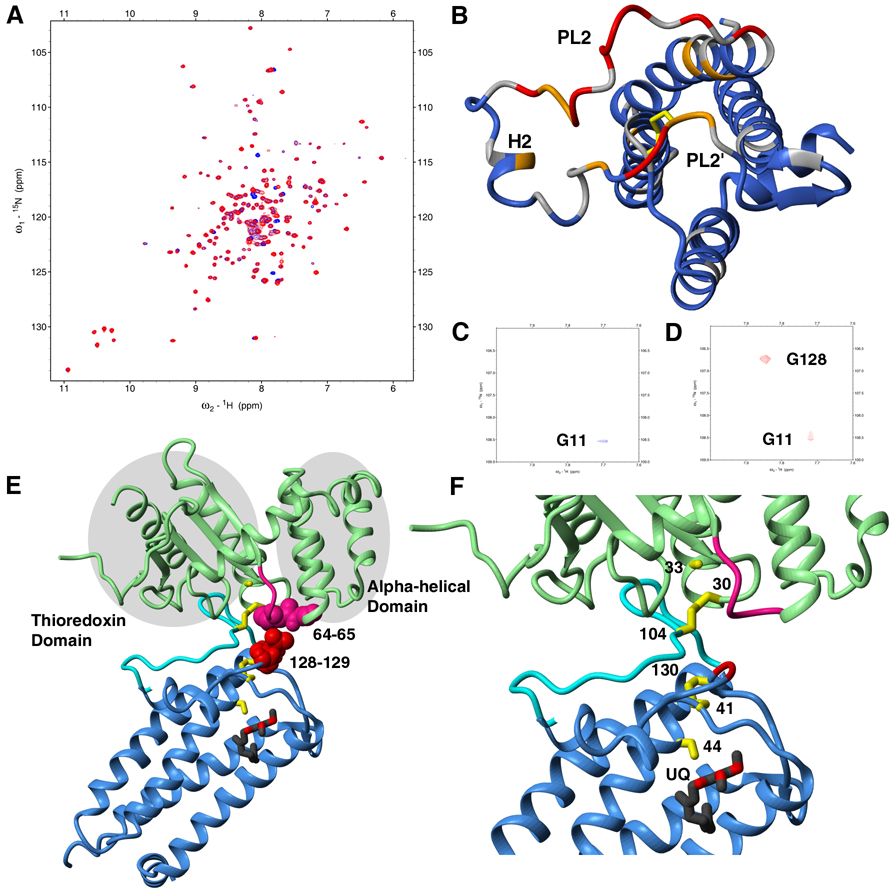
To identify the DsbA interaction surface on DsbB, we compared the transverse relaxation optimized spectroscopy (TROSY)-based 15N-1H heteronuclear single quantum coherence (HSQC) spectrum of DsbB[CSSC] alone and in the presence of the DsbA Cys30Ser/Cys33Ser mutant (DsbB[SS]), which mimics reduced DsbA (Figure 4A). Mapping the chemical shift differences onto the protein structure reveals that two regions of DsbB, residues 97–110 of PL2 and residues 126–131 of PL2', have significant changes upon DsbA binding (Figure 4B), suggesting contacts in both regions of DsbB. PL2 has been identified as a DsbA binding loop in the crystal structure of the DsbB-DsbA complex where electron density for PL2’ was missing (Inaba et al., 2006). To confirm that PL2’ also contacts DsbA, we performed a saturation transfer experiment (Takahashi et al., 2000) on a sample of 2H,13C,15N-labeled DsbB[CSSC] and unlabeled DsbA[SS]. This experiment provides an unequivocal approach to confirm spatial proximity by transfer of aliphatic proton saturation from a protonated protein to a deuterated protein. A total of five residues (Ser99, F106, M107, V108 in PL2 and Gly128 in PL2’) have been identified using this approach, proving that both PL2 and PL2’ interact with DsbA (Figure 4C, 4D and Figure S9). As further confirmation, we have introduced point mutations at Gly128 and Asp129 all of which adversely affect kcat and one of which (G128A) also resulted in an increase in Km (see Figure S10). As shown in Figure 3B, residues 97–110 of PL2 and residues 126–131 of PL2' are exposed above the bilayer where they can readily interact with DsbA. In the crystal structure of DsbB-DsbA, PL2 is intimately bound to DsbA and is structured (Inaba et al., 2006a). However, in the absence of DsbA, our heteronuclear NOE data shows that residues 99–105 constitute the most mobile region in DsbB (Figure 3C).
Fig. 4. DsbA binding and model of DsbB-DsbA complex.
(A) TROSY-HSQC spectra of DsbB[CSSC] in the presence (red) and in the absence (blue) of DsbA[SS]. (B) Mapping of NH chemical shift changes upon DsbA titration. Strong (>20 Hz), medium (8–20 Hz), and weak (<8 Hz) changes are colored red, orange, and blue, respectively. Residues with no information are colored gray. (C) Selected region of the saturation transfer TROSY-HSQC for saturation transfer from DPC to DsbB[CSSC]. (D) Selected region of the saturation transfer TROSY-HSQC for saturation transfer from DsbA[SS] and DPC to DsbB[CSSC] (see also Figure S9). (E) Model of DsbB-DsbA complex with rearranged intermolecular and inter-loop disulfide bonds. DsbA, the hinge loop between the thioredoxin and α-helical domains in DsbA, and PL2 of DsbB from the crystal structure are shown as green, pink, and cyan respectively. DsbB from the solution structure, with the exception of PL2, is shown as a blue ribbon. The six essential Cys residues are colored yellow. UQ2 is shown in a ball-and-stick representation. Gly128 and Glu129 (red space filling repesentation) of DsbB, which show significant chemical shift perturbations in DsbA titration experiment, are in close proximity to Met64 and Gly65 (pink space filling representation) in the hinge loop in DsbA. (F) Close-up view of the model of DsbB-DsbA complex. The six essential Cys residues are numbered.
We have overlayed the X-ray structure of the DsbB-DsbA complex (intermediate II in Figure 1A) with our solution structure of DsbB[CSSC] (intermediate IV in Figure 1A) to create a model of the inter-loop disulfide bond intermediate with DsbA bound (intermediate III in Figure 1A, see Figures 4E and 4F), for which there is currently no structural information. This model shows close proximity between Gly128 and Glu129 in DsbB PL2’ and the highly conserved loop in DsbA between the α-helical and thioredoxin domains (Martin et al., 1993), suggesting that this conserved loop of DsbA interacts with PL2’ in the vicinity of Cys130 thereby positioning Cys130 in spatial proximity to Cys41 and accelerating the rate of formation of the Cys130-Cys41 disulfide bond. Replacement of Met64 in the conserved loop of DsbA with shorter sidechain residues (Gly, Ala) was previously shown to result in an increased Km for DsbA, supporting the role of this loop in DsbB binding (Inaba et al., 2006a). As mentioned above, we have introduced mutations at DsbB Gly128 and Asp129, all of which result in decreased kcat values, consistent with a role for these residues in accelerating the rate of formation of the Cys130-Cys41 disulfide bond.
The Cys41-Cys130 inter-loop disulfide bond
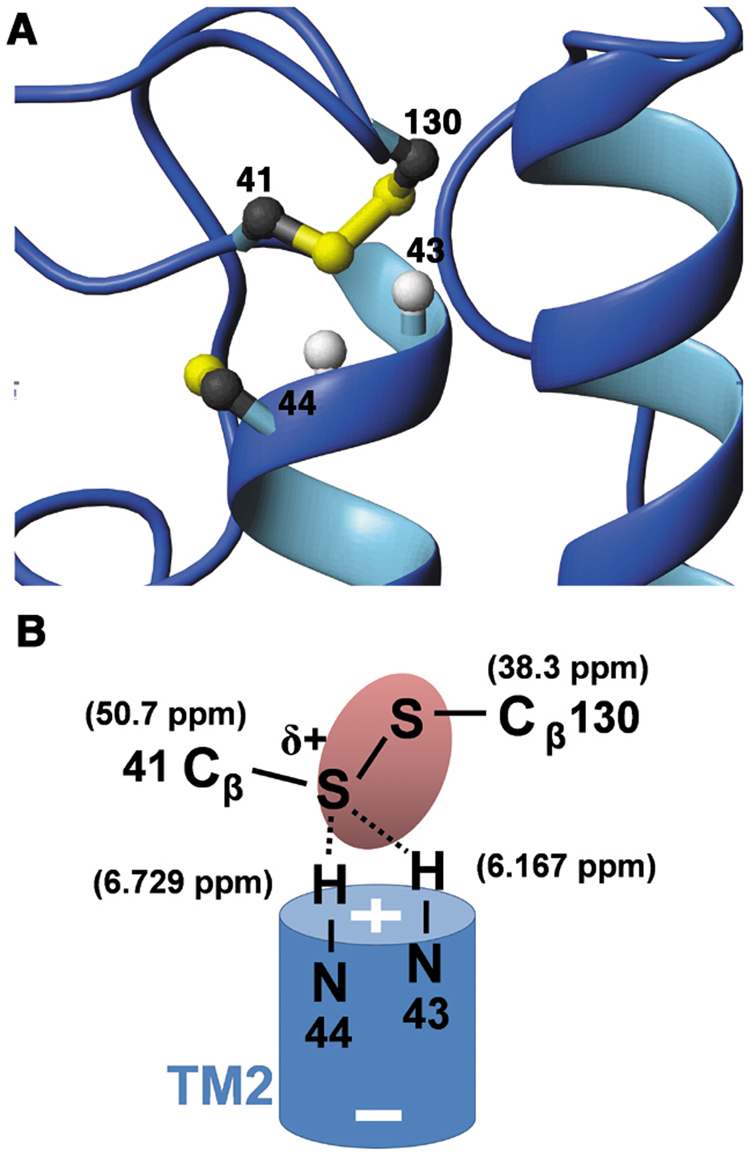
The Cys41-Cys130 inter-loop disulfide bond sits on top of the highly conserved N-terminus of TM2 (Figure 5A). The Cβ of Cys41 has a very unusual downfield chemical shift (50.7 ppm), one of the highest values in the BioMagResBank database and substantially higher than the average 41.6 ppm value found for disulfide-bonded cysteine residues (Zhang et al., 2003). In contrast, the Cys130 Cβ chemical shift (38.3 ppm) is close to the average value. Such a polarization of the Cβ chemical shifts across a disulfide bond is also observed in the active site of oxidized CXXC motifs of thioredoxin superfamily proteins where the first Cys, the one with the depressed pKa in the reduced form, has a Cβ chemical shift of ~44 ppm and the second Cys Cβ has a chemical shift of ~34 ppm (Sharma and Rajarathnam, 2000). The extremely high chemical shift indicates a withdrawal of electron density from the Cys41 Sγ, likely by means of hydrogen bonding and/or the effects of the TM2 helix dipole (Figure 5B). Leu43 and Ser44, located at the N-terminal end of TM2 right beneath the disulfide, have unusual upfield amide proton chemical shifts (6.167 ppm and 6.729 ppm, respectively), consistent with hydrogen bonding between these NH moieties and the Cys41 Sγ and resulting in a significant increase in electron density on the NH moieties. The partial positive charge at the Cys41 sulfur in DsbB makes it the preferred leaving group between the two sulfurs and will therefore promote the forward reaction of Cys104 attack on Cys130 with release of a Cys41 thiolate (Step (IV) -> (V) in Figure 1A), thereby lowering the activation energy for the overall reaction.
Fig. 5. The Cys41-Cys130 inter-loop disulfide bond.
(A) Closeup view of the Cys41-Cys130 disulfide bond. (B) Scheme of interactions around the inter-loop disulfide bond. The + and − signs refer to the TM2 helix dipole. The δ+ indicates the partial positive charge at the Cys41 sulfur. The oval is a schematic representation of the electron density around the disulfide bond.
Structure of the DsbB[CSSC] – UQ2 complex
Finally, our NMR analysis provides useful insights into how quinones (UQ8 and MK8) can promote the last step in the reoxidation of DsbB. An approximate location of the ubiquinone benzoquinone ring has been proposed based on the 3.7 Å resolution DsbB-DsbA crystal structure (Inaba et al., 2006a). To obtain NMR data on a DsbB[CSSC] – UQ2 complex, we prepared a 2H,13C,15N-labeled quinone-free DsbB[CSSC] sample using an E. coli strain which is unable to synthesize quinones (Inaba et al., 2004). Subsequently, we added UQ2 (non-deuterated). UQ2 is an established substrate of DsbB with higher detergent solubility, making it feasible to diffuse into the protein-detergent complex. Interestingly, NMR spectra of the ubiquinone-free DsbB show clear evidence for a loss of structural integrity (compare Figures 6A and 6B), indicating that the quinone cofactor contributes to the structural integrity of DsbB. This result strongly implies that previous studies which used UQ-free DsbB to draw conclusions about the disulfide exchange mechanism of DsbB should be viewed with caution. The DsbB[CSSC] – UQ2 sample made it possible to identify 10 intermolecular (DsbB-quinone) NOEs and 3 intramolecular (quinone-quinone) NOEs as well as PRE data for one MTSSL-labeled sample (Leu30Cys) to define the ubiquinone binding site and conformation (Figure S8B).
Fig. 6. Characterization of the UQ binding site.
(A) TROSY-based 15N-1H HSQC spectrum of UQ-free DsbB[CSSC]. (B) TROSY-based 15N-1H HSQC spectrum of DsbB[CSSC]-UQ2. (C, D) Overlay of 20 conformers of the solution structure of the DsbB[CSSC] – UQ2 complex (DsbB in blue, carbon and oxygen atoms of UQ2 in black and red). The DsbB crystal structure and proposed quinone orientation is also shown (DsbB in green, carbon and oxygen atoms of UQ in ivory and red). For clarity, both are shown from two different angles. (E) Ribbon representation of the DsbB[CSSC] – UQ2 complex. UQ2 is shown in a ball-and-stick representation. Carbon and oxygen atoms of UQ2 are colored black and red, respectively. (F) Close-up view of the UQ binding site on DsbB. Sidechains of quinone interacting residues are shown and labeled (Gln33, Cys44, Arg48, Met142).
UQ2 binds at the periplasmic end of DsbB among TM1, TM2 and TM4, a site close to, but different from, the proposed ubiquinone binding site in the crystal structure of the DsbB-DsbA complex (Figures 6C and 6D). The benzoquinone ring of UQ2 inserts into a site consisting of highly conserved residues from the C-terminus of TM1, PL1, and the N-terminus of TM2 (residue 29–48) (Figure 6E). The isoprenoid tail of UQ2 runs between TM1 and TM4. Cys44, which is mutated to serine in DsbB[CSSC], is the essential cysteine that directly interacts with UQ (Inaba et al., 2004). Arg48, Gln33, and Met142 are all found in proximity to the quinone (Figure 6F). Previous mutagenesis studies showed the importance of Arg48 and Met142 for quinone binding (Inaba et al., 2006a; Kadokura et al., 2000). We have mutated Gln33 to Ala and showed that this results in a loss of the characteristic chromophore seen for the CCSC mutant of DsbB (Figure S1), suggesting that Gln33 also plays an important role in the interaction of DsbB with quinone. In comparison to UQ2, the endogenous UQ8 found bound to DsbB has 6 more isoprenyl units. Comparison of 15N-1H TROSY-HSQC spectra of DsbB[CSSC] in complex with UQ2 and UQ8 shows that there are significant chemical shift changes between the two in TM1 and TM4 (Figure S2), suggesting that the remaining isoprenoid tail of UQ8 packs between TM1 and TM4.
Discussion
DsbB uses two pairs of redox-active cysteines to re-oxidize DsbA utilizing the oxidizing power of quinones. An inter-loop disulfide bond that forms between Cys41 and Cys130 has been shown to play a vital role in the reaction cycle of this enzyme. Here we solved the structure of a DsbB intermediate with this inter-loop disulfide bond (DsbB[CSSC]) using NMR spectroscopy. Our analysis illuminates several features of the reaction mechanism of this protein. In particular, our data provide a structural basis for how a highly concerted process of electron transfer from DsbA to quinones through DsbB can be performed. In part this is made possible because the NMR structure presents features and information that were missing or not obtainable from the X-ray crystallographic analysis of the DsbB-DsbA complex.
Binding of DsbA to DsbB – Functional Effects
Upon titration with DsbA, DsbB shows chemical shift changes in PL2 and for residues in the vicinity of Cys130 in PL2’. The changes seen in PL2 agree with the crystal structure of the DsbB-DsbA complex which showed that PL2 binds to DsbA (Inaba et al., 2006). The chemical shift changes, saturation transfer effects, and mutagenesis for PL2’ indicate that there are contacts between DsbA and this loop in the DsbB-DsbA complex. This region of DsbB was missing in the DsbB-DsbA crystal structure. In the NMR solution structure of the inter-loop disulfide form of DsbB, PL2’ adopts a well-defined extended conformation. The interaction between DsbA and the Cys130-containing PL2’ is likely functionally significant in two ways. First, after formation of the initial DsbB-DsbA intermediate (II in Figure 1A), it is necessary for Cys130 to migrate to the location of the Cys41-Cys44 disulfide bond for nucleophilic attack on Cys41. The interaction of the Cys130-containing loop with DsbA, as observed here, will help to guide Cys130 to the appropriate location for attack on Cys41. In so doing, the rate of the forward reaction will be accelerated, i.e. the activation energy will be lowered. Consistent with this, methionine marking in the DsbB-DsbA crystal structure showed that Cys130 in PL2’ moves away from Cys104 and is on top of TM2 in the DsbB-DsbA complex (which lacks the inter-loop disulfide bond), a position ready to attack the Cys41-Cys44 disulfide bond (Inaba et al., 2006). Second, the interaction between PL2’ and DsbA will also help to prevent the unproductive back reaction in which Cys130 attacks the DsbA Cys30 – DsbB Cys104 intermolecular disulfide bond, i.e. this interaction will help to sequester Cys130 from the intermolecular disulfide bond.
We have used our structure and the crystal structure of the DsbB-DsbA complex (II in Figure 1A) to create a model (Figures 4E and 4F) of the DsbB-DsbA inter-loop disulfide intermediate (III in Figure 1A). This model shows the highly conserved loop in DsbA between the thioredoxin and α–helical domains is in close proximity to the region of PL2’ (Figure 4E). Therefore, it is plausible that this loop in DsbA mediates the interaction of DsbA with PL2’ in DsbB. In addition, this model shows that all six essential Cys residues in this complex are aligned in a straight line. Such an arrangement makes it possible for a highly concerted series of disulfide exchange reactions to occur, as has been suggested previously (Kadokura and Beckwith 2002; Tapley et al., 2007). Indeed, stopped flow kinetics measurements clearly show that DsbA binding is followed by rapid disulfide exchange resulting in intermediate III in Figure 1A, identified by its characteristic purple color associated with the Cys44 thiolate – quinone charge transfer complex (Tapley et al., 2007). This rapid exchange is followed by the slower quinone reduction step and release of DsbA.
Role of periplasmic loop mobility in electron transfer
Heteronuclear NOE measurements show that PL2 is highly mobile in our structure (Figure 3C). We propose that the increased mobility of this loop is functionally important in two ways. First, DsbA substrates are proteins undergoing oxidative folding and are therefore likely to be unstructured. Thus, the mobile PL2’ may mimic the in vivo substrates of DsbA and thereby enhance the recognition of DsbB by DsbA. Secondly, the mobility of this loop likely plays a critical role in the final steps of the mechanism. Reoxidation of DsbA will release an intermediate with a reduced Cys104 (IV in Figure 1A). While this loop is highly structured when bound to DsbA, upon release from DsbA this loop apparently becomes highly mobile. Cys104 must traverse a significant distance to reach the Cys130-Cys41 inter-loop disulfide bond and attack Cys130 to reform the Cys130-Cys104 disulfide bond. Subsequently, Cys41 will attack Cys44 resulting in quinone reduction and reformation of the Cys41-Cys44 disulfide bond. Such a large movement of Cys104 to this site would be facilitated by the mobility of PL2. Intriguingly, the increased dynamics of this loop appears to be conserved among functional homologs as the eukaryotic functional homologs of DsbB, Ero1p and Erv2p, also use mobile regions to interact with protein disulfide isomerase (PDI), the eukaryotic functional homolog of DsbA (Sevier et al., 2005).
Basis for the disulfide exchange pathway within DsbB
Our data also provides an explanation for the selective attack of Cys104 on Cys130 of the Cys41-Cys130 disulfide bond that results in the re-formation of the Cys104-Cys130 disulfide bond. First, as described above, the increased mobility of the PL2 loop ensures that upon departure of DsbA, Cys104 can readily achieve nucleophilic attack on Cys130 in the inter-loop disulfide bond. Second, the inter-loop Cys41-Cys130 disulfide bond is positioned on top of the highly conserved N-terminus of TM2 (Figure 5A). Whereas the sulfur of Cys130 is in the interfacial region of the bilayer and accessible for attack by Cys104, the sulfur of Cys41 is located deeper in the interfacial region (Figure 3B) and is not accessible for Cys104 attack due to a steric block by residues of TM2, providing a rationale for selective attack of Cys104 at Cys130. Finally, chemical shift analysis shows that the Cys41-Cys130 inter-loop disulfide bond in DsbB is highly polarized by hydrogen bonding and/or the effects of the TM2 helix dipole. If this step proceeds via an SN2 mechanism, the partial positive charge at the Cys41 sulfur makes it the preferred leaving group and will therefore promote the forward reaction of Cys104 attack on Cys130, thereby lowering the activation energy for the overall reaction.
Our results provide a rationale for how the unidirectional electron transfer between the two periplasmic loops of DsbB occurs: the direction of electron flow is regulated both sterically (positioning of the Cys41-Cys130 disulfide) as well as electrostatically (partial positive charge at Cys41), and the reaction is facilitated by the mobility of the PL2’ loop. Since both Ero1p and Erv2p also have a CXXC active site at the N-terminal end of an α-helix that functions in an analogous manner to the Cys41-Cys44 pair of DsbB (Sevier et al., 2005), a similar mechanism may operate for the unidirectional interdomain transfer of electrons within Ero1p and Erv2p.
Role of structural homology to Trx, Grx, and DsbA
The sulfur of Cys41 is located at the top of the TM2 helix in DsbB, the same structural location seen for the N-terminal cysteine of the Cys-Xaa-Xaa-Cys active site motif in thioredoxin superfamily members such as thioredoxins, glutaredoxins, and DsbA. In these proteins, this is the cysteine that shows a substantially depressed pKa (Kallis and Holmgren, 1980; Yang and Wells, 1991; Nelson and Creighton, 1994). Structural and molecular dynamics simulation studies of Grx3 indicate that a substantial portion of the stabilization of the thiolate anion in Grx3 results from hydrogen bonding to the backbone amides of the residues two and three amino acids C-terminal to the cysteine (Foloppe et al., 2001). This is the identical situation as we see for DsbB where hydrogen bonding can occur to the Cys41 sulfur from the backbone amides of Leu43 and Ser44, the exact sites where we see unusual upfield chemical shifts. This would suggest that the structural arrangement around Cys41 in DsbB will facilitate the transient formation of a thiolate anion at Cys41 after formation of the Cys104-Cys130 disulfide bond (intermediate V in Figure 1A), helping to promote electron transfer to the quinone by means of charge repulsion with the Cys44 thiolate anion.
Structure of DsbB-UQ2 complex is consistent with a charge transfer mechanism
Mutagenesis studies and quantum chemistry simulation of the quinone spectral transition have suggested a charge transfer mechanism between UQ and DsbB (Inaba et al., 2006b). It has been suggested that a Cys44 thiolate anion transfers partial charge to the benzoquinone ring of ubiquinone and could also form a covalent adduct. The positively charged guanidinium group of the conserved Arg48 likely stabilizes this charge transfer complex or adduct by interactions with the Cys44 thiolate anion or the benzoquinone ring. A similar cysteine-flavin charge transfer interaction has been observed in flavin dependent disulfide oxidoreductases (Miller et al., 1990). The structure of the DsbB-UQ2 complex is consistent with the proposed mechanism and provides further structural insights (Figure 6F). Based on the structures and rotations about the Cα-Cβ bond, the Cys44 Sγ can come as close as 1.9 Å away from the benzoquinone ring C5/C6 (sites of methyl and isoprenyl substitution on the benzoquinone ring, respectively), a distance close enough for charge transfer from the Cys44 thiolate to the benzoquinone ring or for covalent bond formation to the quinone, as has been suggested previously (Inaba et al., 2006b). While we cannot rule out the possibility that the C44S mutation we introduced has altered the binding of UQ, the close approach we observe between this residue and the quinone is consistent with previous mutagenesis and modeling studies of the enzyme. The sidechains of Arg48 and Cys44 are positioned on the same side of TM2 and perpendicular to the benzoquinone ring of UQ2. Energetically reasonable rotations of the Arg48 sidechain allows its guanidinium group to be within hydrogen bonding distance to O1 of the benzoquinone ring or interact with the Cys44 thiolate anion, in agreement with its role in stabilizing the charge transfer/adduct complex. In addition, Gln33, which we have identified here to be important for the interaction of DsbB with ubiquinone (Figure S1), may help stabilize the charge transfer complex by hydrogen bonding to the O1/O2 of the benzoquinone ring.
Use of NMR to study the structure, dynamics, and function of integral membrane proteins
Integral membrane proteins constitute approximately one-third of any genome and are the targets of more than half of all currently used drugs, making them important targets for structure determination. While there are more than 50,000 structures of soluble proteins deposited in the PDB, there are only ~150 structures of integral membrane proteins, thus there is a profound need for further efforts in this important area. This structure of DsbB represents the first structure of a polytopic helical membrane protein to be solved using NMR methods. The use of a combination of approaches (methyl NOEs, PREs, chemical shift analysis, and RDCs) to determine the backbone structure to high-resolution establishes a viable approach for approaching future membrane protein structure determinations using NMR. The methods employed here should be applicable to much larger systems and therefore make a large body of membrane proteins accessible for structure determination by NMR. More importantly, the functional insights gained into DsbB function using NMR spectroscopy likely could not be obtained from other methods. Our results clearly establish NMR spectroscopy as a potent tool in our arsenal for the characterization of the structure, dynamics, and function of integral membrane proteins.
Experimental Procedures
Protein expression and purification
Details of the expression and purification of labeled DsbB and DsbA are provided in the Supplemental Data.
Optimization of sample for NMR
DPC (FOS-CHOLINE-12, Anatrace) was chosen to carry out all NMR experiments because DsbB retains activity and long-term stability in this detergent. The oligomerization status of DsbB[CSSC] in DPC micelles was characterized by size exclusion chromatography using a Bio-Silect SEC 250-5 Column (Bio-Rad Laboratories) (Figure S3). The apparent size of DsbB[CSSC] in DPC micelles is 40 kDa, consistent with a DsbB[CSSC] monomer.
NMR backbone resonance assignments
NMR experiments were carried out at 40 °C on a Varian Inova 600 spectrometer equipped with a cryogenic probe. Spectra were processed using NMRPipe and analyzed using Sparky (Goddard, T. D., and Kneller, D. G. SPARKY 3, University of California, San Francisco). A total of 98% of the backbone resonance assignments were obtained using TROSY-based HNCO, HN(CA)CO, HNCA, HN(CO)CA and HN(CA)CB experiments on a 1.2 mM 2H, 13C, 15N-labeled DsbB[CSSC] sample (Figure S4).
Paramagnetic relaxation enhancement (PRE) measurements
Nitroxide spin labels were introduced via single cysteine mutants into DsbB[CSSC]. Nine different sites (Ala14, Leu30, Val72, Leu87, Tyr89, Gln122, Phe137, Gly139, and Val161) were mutated. 2H, 15N-labeled DsbB[CSSC] single cysteine mutants were labeled with (1-oxyl-2,2,5,5-tetramethyl-η3-pyrroline-3-methyl) methanethiosulfonate or (1-acetyl-2,2,5,5-tetramethyl-η3-pyrroline-3-methyl) methanethiosulfonate (MTSSL and dMTSSL, Toronto Research Chemicals Inc.) according to established protocols (Liang et al., 2006). NMR spectroscopy and mass spec analysis were used to confirm the labeling is complete and that there is no labeling on Cys41 or Cys130. TROSY-HSQC spectra were recorded and assignments of the DsbB[CSSC] mutants were carried out by comparison to DsbB[CSSC] spectra.
PRE distance restraints were derived from peak intensity ratios of paramagnetic and diamagnetic labeled DsbB[CSSC] mutants (Ipara/Idia) according to previously described procedures (Liang et al., 2006). The global rotational correlation time of DsbB[CSSC] (16 ns) was determined using the [15N,1H]-TRACT experiment and was used to approximate the correlation time of the electron-nuclear interaction. The HN transverse relaxation rates (R2) were determined by HSQC based T2 measurements on a 2H,15N-labeled DsbB[CSSC] sample. Intensity data was scaled by peaks unperturbed by the nitroxide label (>26 Å distance) to compensate for the concentration differences of the MTSSL and dMTSSL labeled samples. PREs were classified into 3 categories based on Ipara/Idia ratios for distance calculation: (a) for Ipara/Idia smaller than 0.2, an upper distance limit of 16 Å was assigned, (b) for Ipara/Idia bigger than 0.8, a lower distance limit of 21 Å was assigned, (c) for Ipara/Idia between 0.2 and 0.8, distances were calculated as described previously (Liang et al., 2006) and a ±2 Å error range was employed. In total, nine nitroxide spin labels made it possible to delineate 871 lower-limit constraints and 272 upper-limit constraints (Figure S5).
Residual dipolar coupling (RDC) measurements
Alignment of the protein for RDC measurements was achieved using charged polyacrylamide gels as described previously (Cierpicki and Bushweller, 2004). A 1.2 mM 2H,13C,15N-labeled DsbB[CSSC] sample in deuterated DPC micelles was soaked into ~4% (final concentration) positively charged polyacrylamide gel containing 25% (3-acrylamidopropyl)-trimethylammonium chloride (APTMAC, Sigma-Aldrich, Inc.). The gel was vertically compressed from 21 mm to 16 mm in a 4.2 mm inner diameter Shigemi NMR tube (Shigemi). TROSY-HNCO based experiments were used to record one bond HN, NC', and C'Cα RDCs (Figure S6). Subsequently, the gel was released to 17 mm and a second dataset was recorded. The accuracy of the measurements was estimated by comparing the 16 and 17 mm sets of data. The amplitude (11.2 Hz) and rhombicity (0.63) of RDCs was derived from powder pattern analysis.
NOEs and PREs between DsbB and UQ2
Assignment of the backbone amide resonances of DsbB and proton resonances of UQ2 in the DsbB[CSSC]-UQ2 complex was achieved on a 0.4 mM 2H,13C,15N-labeled UQ-free DsbB[CSSC] sample supplemented with 2.0 mM UQ2 in deuterated DPC micelles (details provided in Supplemental Data).
Intermolecular NOEs between UQ2 and DsbB[CSSC] were recorded using a 3D 15N-edited NOESY spectrum (200 ms mixing time) on the DsbB[CSSC]-UQ2 sample. Distances between backbone amide protons in α helices of DsbB[CSSC] are known, therefore they were used to calibrate the distance constraints. Intermolecular NOEs between UQ2 and DsbB[CSSC] were classified into 2 distance ranges: 1.8–3.3, 1.8–4.7 Å, corresponding to distances of (i,i+1), (i,i+2) backbone amide protons plus an extra 0.5 Å allowance for UQ2 methyl groups. Intramolecular NOEs of UQ2 were analyzed in a 2D 1H-1H NOESY experiment used for assignment.
Using the above NOE distance restraints, the position and conformation of the UQ2 isoprenoid tail has been determined. However, the benzoquinone ring of UQ2 in the calculated structures assumes two conformations differing by a 180 degree flip along the C3–C6 axis. This results from there being only two unique restraints between the benzoquinone ring and DsbB[CSSC]. To uniquely define the conformation of the benzoquinone ring, we used PRE effects on the UQ2 5' methyl group. Two 10 µM samples were prepared of Leu30->Cys 2H,15N-labeled DsbB[CSSC] with 600 µM UQ2 in D2O, one MTSSL labeled and one dMTSSL labeled. The R2 values for the UQ2 methyl groups were measured using a standard CPMG experiment. The R2 enhancements of 5'-CH3, 10-CH3, and 15-CH3 of UQ2 are 3.84, 4.29 and 3.44 Hz, respectively. This data indicates that in terms of distances to the Leu30Cys-MTSSL paramagnetic center, the 5'-CH3 is intermediate in distance between the 10 and 15 methyls that were calculated to be 14.4–16.5 and 17.4–19.6 Å away, respectively. A distance range of 14.4–19.6 Å was therefore assigned between the Leu30Cys-MTSSL paramagnetic center and the 5'-CH3, which uniquely defined the conformation of the UQ2 benzoquinone ring.
Structure calculations
Structure calculations were carried out by using the simulated annealing protocol in XPLOR-NIH (Details provided in Supplemental Data).
EPR measurements and docking of DsbB[CSSC] into POPC bilayer
Eleven MTSSL spin-labeled DsbB mutants (A14R1, L23R1, L30R1, G56R1, V72R1, L87R1, Q122R1, L137R1, A152R1, V161R1, and A163R1) were reconstituted in POPC vesicles by dialysis and analyzed by EPR measurements. Details of the measurements and docking procedure are provided in the Supplemental Data.
Supplementary Material
Acknowledgments
This work was supported by grants R01 GM078296 to J.H.B. and R01 GM41883 to J.B. from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Coordinates have been deposited in the PDB (2K73 and 2K74).
References
- Altenbach C, Froncisz W, Hemker R, McHaourab H, Hubbell WL. Accessibility of nitroxide side chains: absolute Heisenberg exchange rates from power saturation EPR. Biophys. J. 2005;89:2103–2112. doi: 10.1529/biophysj.105.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB, Haber E, Sela M, White FH., Jr. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc. Natl. Acad. Sci. U.S.A. 1961;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- Cierpicki T, Bushweller JH. Charged gels as orienting media for measurement of residual dipolar couplings in soluble and integral membrane proteins. J. Am. Chem. Soc. 2004;126:16259–16266. doi: 10.1021/ja046054g. [DOI] [PubMed] [Google Scholar]
- Foloppe N, Sagemark J, Nordstrand K, Berndt KD, Nilsson L. Structure, dynamics and electrostatics of the active site of glutaredoxin 3 from Escherichia coli: comparison with functionally related proteins. J. Mol. Biol. 2001;310:449–470. doi: 10.1006/jmbi.2001.4767. [DOI] [PubMed] [Google Scholar]
- Gennis RB, Stewart V. In: Escherichia coli and Salmonella, Cellular and Molecular Biology. Neidhardt FC, editor. Washington, D. C.: American Society for Microbiology; 1996. p. 222. [Google Scholar]
- Inaba K, Murakami S, Suzuki M, Nakagawa A, Yamashita E, Okada K, Ito K. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006a;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- Inaba K, Takahashi YH, Fujieda N, Kano K, Miyoshi H, Ito K. DsbB elicits a red-shift of bound ubiquinone during the catalysis of DsbA oxidation. J. Biol. Chem. 2004;279:6761–6768. doi: 10.1074/jbc.M310765200. [DOI] [PubMed] [Google Scholar]
- Inaba K, Takahashi YH, Ito K. Reactivities of quinone-free DsbB from Escherichia coli. J. Biol. Chem. 2005;280:33035–33044. doi: 10.1074/jbc.M506189200. [DOI] [PubMed] [Google Scholar]
- Inaba K, Takahashi YH, Ito K, Hayashi S. Critical role of a thiolate-quinone charge transfer complex and its adduct form in de novo disulfide bond generation by DsbB. Proc. Natl. Acad. Sci. U.S.A. 2006b;103:287–292. doi: 10.1073/pnas.0507570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Bader M, Tian H, Bardwell JC, Beckwith J. Roles of a conserved arginine residue of DsbB in linking protein disulfide-bond-formation pathway to the respiratory chain of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10884–10889. doi: 10.1073/pnas.97.20.10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Beckwith J. Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J. 2002;21:2354–2363. doi: 10.1093/emboj/21.10.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallis GB, Holmgren A. Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J. Biol. Chem. 1980;255:10261–10265. [PubMed] [Google Scholar]
- Łasica AM, Jagusztyn-Krynicka EK. The role of Dsb proteins of Gram-negative bacteria in the process of pathogenesis. FEMS Microbiol. Rev. 2007;31:626–636. doi: 10.1111/j.1574-6976.2007.00081.x. [DOI] [PubMed] [Google Scholar]
- Liang B, Bushweller JH, Tamm LK. Site-directed parallel spin-labeling and paramagnetic relaxation enhancement in structure determination of membrane proteins by solution NMR spectroscopy. J. Am. Chem. Soc. 2006;128:4389–4397. doi: 10.1021/ja0574825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JL, Bardwell JC, Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature. 1993;365:464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- Miller SM, Massey V, Ballou D, Williams CH, Jr., Distefano MD, Moore MJ, Walsh CT. Use of a site-directed triple mutant to trap intermediates: demonstration that the flavin C(4a)-thiol adduct and reduced flavin are kinetically competent intermediates in mercuric ion reductase. Biochemistry. 1990;29:2831–2841. doi: 10.1021/bi00463a028. [DOI] [PubMed] [Google Scholar]
- Nelson JW, Creighton TE. Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry. 1994;33:5974–5983. doi: 10.1021/bi00185a039. [DOI] [PubMed] [Google Scholar]
- Pan JL, Sliskovic I, Bardwell JC. Mutants in DsbB that appear to redirect oxidation through the disulfide isomerization pathway. J. Mol. Biol. 2008;377:1433–1442. doi: 10.1016/j.jmb.2008.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi VK, Girvin ME. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature. 1999;402:263–268. doi: 10.1038/46224. [DOI] [PubMed] [Google Scholar]
- Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier CS, Kadokura H, Tam VC, Beckwith J, Fass D, Kaiser CA. The prokaryotic enzyme DsbB may share key structural features with eukaryotic disulfide bond forming oxidoreductases. Protein Sci. 2005;14:1630–1642. doi: 10.1110/ps.051355705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Rajarathnam K. 13C NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR. 2000;18:165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Nakanishi T, Kami K, Arata Y, Shimada I. A novel NMR method for determining the interfaces of large protein-protein complexes. Nat. Struct. Biol. 2000;7:220–223. doi: 10.1038/73331. [DOI] [PubMed] [Google Scholar]
- Tapley TL, Eichner T, Gleiter S, Ballou DP, Bardwell JC. Kinetic characterization of the disulfide bond-forming enzyme DsbB. J. Biol. Chem. 2007;282:10263–10271. doi: 10.1074/jbc.M611541200. [DOI] [PubMed] [Google Scholar]
- Yang YF, Wells WW. Identification and characterization of the functional amino acids at the active center of pig liver thioltransferase by site-directed mutagenesis. J. Biol. Chem. 1991;266:12759–12765. [PubMed] [Google Scholar]
- Zapun A, Bardwell JC, Creighton TE. The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry. 1993;32:5083–5092. doi: 10.1021/bi00070a016. [DOI] [PubMed] [Google Scholar]
- Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J. Biomol. NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.