Abstract
Multilocus sequence typing (MLST) has been applied to 266 Campylobacter jejuni isolates, mainly from veterinary sources, including cattle, sheep, poultry, pigs, pets, and the environment, as well as isolates from human cases of campylobacteriosis. The populations of veterinary and human isolates overlap, suggesting that most veterinary sources should be considered reservoirs of pathogenic campylobacters. There were some associations between source and sequence type complex, indicating that host or source adaptation may exist. The pig isolates formed a distinct group by MLST and may well represent a potential pig-adapted clone of C. jejuni. A subset (n = 82) of isolates was reanalyzed with a second MLST scheme which provided a unique set of isolates that had been analyzed at a total of 12 loci. The distribution of isolates among the complexes in each of the two schemes was similar but not identical. In addition to isolates from human outbreaks, one group of isolates that were not epidemiologically linked was also identical at all 12 loci. This group of isolates is believed to represent another stable strain of C. jejuni.
Campylobacter jejuni and C. coli are major causes of acute bacterial enteritis in humans worldwide. In 2001, there were 56,420 reported cases in England and Wales (Communicable Disease Surveillance Centre), which, according to a recent intestinal infectious diseases survey, is an underestimate of about eightfold (29). Campylobacters colonize many animals but appear to have evolved for optimal growth in the avian gut as a commensal. Many poultry flocks worldwide are colonized with these organisms (9). A major human risk factor for the acquisition of campylobacter infection is thought to be the handling or consumption of contaminated poultry meat. Despite this, previous studies using typing methods have suggested that the ranges of campylobacter types found in humans and chickens do not totally overlap. These studies concluded that some isolates infecting humans do not colonize chickens, and conversely, some isolates colonizing chickens do not infect humans (4, 15, 16). The implications from these findings are that other sources of campylobacter infection may be important in human disease and possibly that not all campylobacters are pathogenic to humans. Other food-producing animals such as cattle, sheep, and pigs carry this organism in their guts; however, the relative risk of human infection associated with these potential sources is unclear.
C. jejuni is known to be a highly diverse species. This is exemplified by the wide range of phenotypes and genotypes detectable by a number of techniques, such as serotyping, pulsed-field gel electrophoresis (PFGE), and amplified fragment length polymorphism (AFLP) (32). The recent application of multilocus sequence typing (MLST) to C. jejuni, in agreement with previous studies, has shown that the organism is genetically diverse, yet it has a weakly clonal population structure. This means that there is evidence of frequent recombination within a clonal framework (5, 6, 20, 28). MLST is similar to multilocus enzyme electrophoresis (MLEE) in that it measures variation in housekeeping genes located around the genome. The advantage of MLST is that this variation is determined at the level of DNA sequence, thus making the technique both highly reproducible and portable (17). These previous MLST studies on C. jejuni concentrated mainly on isolates of human origin, and there is a general paucity of information regarding where strains isolated from veterinary sources fit into the overall population structure of this organism.
For this study, MLST was applied to study the genetic relationships of 266 isolates of veterinary and human origin. Veterinary isolates were obtained from poultry, cattle, sheep, pigs, and pets as well as from the environment in and around broiler houses.
MATERIALS AND METHODS
Campylobacter isolates.
C. jejuni isolates (n = 266) were selected to represent a wide range of veterinary sources, some of which may act as reservoirs of potentially pathogenic organisms. The majority of isolates were from the United Kingdom (n = 231); however, isolates were also included that were from Denmark (n = 13), Czech Republic (n = 9), The Netherlands (n = 8), South Africa (n = 3), France (n = 1), and Sweden (n = 1). Isolates were selected from poultry (n = 70), cattle (n = 63), sheep (n = 40), pigs (n = 22), and pets (n = 8) as well as from the environment in and around broiler houses (n = 9). Human isolates were included for comparison (n = 51). The three remaining isolates were from diverse origins, including an ostrich (n = 1), a giraffe (n = 1), and a water source (n = 1). The poultry isolates were obtained from cloacal swabs of live broiler chickens, whereas the cattle, sheep, and pig isolates were obtained from fecal samples taken from animals at slaughter during a national abattoir survey in the United Kingdom. The pet isolates were obtained from fecal samples taken at a pet boarding facility in the United Kingdom, and the human isolates were mainly from fecal samples, predominantly from sporadic cases, although some human outbreak isolates were also included.
Bacterial growth and preparation of genomic DNA.
All C. jejuni isolates were grown on 10% (vol/vol) sheep blood agar plates with actidione (250 μg/ml) and Skirrow's supplement (10 μg of vancomycin per ml, 2.5 IU of polymyxin B per ml, 5 μg of trimethoprim per ml) at 42°C in a microaerobic environment (7.5% [vol/vol] CO2, 7.5% [vol/vol] O2, 85% [vol/vol] N2) for 24 to 48 h. Genomic DNA was extracted by the cetyltrimethylammonium bromide-NaCl method (2). DNA pellets were resuspended in 100 μl of distilled water and stored at 4°C.
Loci and primers used.
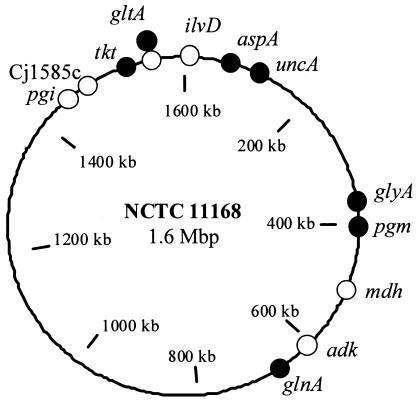
All PCR products were amplified by use of previously described primers (6), referred to as scheme A. A subset of isolates (n = 82) were also analyzed by a second MLST scheme, scheme B. The loci for scheme B were selected prior to completion of the genome sequence of C. jejuni NCTC11168 and before the scheme A loci were made accessible. Figure 1 shows the positions of these loci compared to scheme A in the now completed genome of NCTC11168 (24). The loci were as follows: dihydroxy acid dehydratase (ilvD), adenylate kinase (adk), citrate synthase (gltA), d-lactate dehydrogenase (Cj1585c; reannotated as putative oxidoreductase), glucose-6-phosphate isomerase (pgi), and malate dehydrogenase (mdh). Initially, the MLST scheme consisted of seven loci, as that was deemed to be the minimum number of loci required to provide sufficient discrimination for an MLST scheme (8). The katA locus was removed at an early stage, as it was found to be very variable, apparently with evidence of evolution being driven by a strong selective pressure (R. J. Meinersmann, personal communication). The region of the gltA locus sequenced was the same for both schemes. The primer sequences for scheme B are given in Table 1. Each primer was used for both amplification and sequencing reactions. Alternative primer sequences are given and were used when necessary.
FIG. 1.
Positions of the loci for both scheme A (filled circles) and scheme B (open circles) in the genome of C. jejuni strain NCTC11168.
TABLE 1.
Primer sequences used for PCR amplification and sequence determination of the loci for MLST scheme B
| Locus | Amplicon size (bp) | Primer (direction [sequence]) |
|---|---|---|
| ilvD | 619 | Forward (5′-GAT GGT ATA GCT ATG GGA CA-3′) |
| Reverse (5′-CCT GCT TCA CGC GAA ATG GCA A-3′) | ||
| adk | 528 | Forward (5′-ATC ATA GGT GCA CCA GGT AGT GGA-3′) |
| Reverse (5′-TCA TGT CTG CAA CGA TAG GTT CGA-3′) | ||
| gltA | 617 | Forward (5′-TTA ATG CAC CGT GGC TAT CCT A-3′) |
| Reverse (5′-AAC ACC TTC ATT AGC TCC ACC A-3′) | ||
| Cj1585ca | 604 | Forward 1 (5′-GCA GCA GGT ACA AGT TTA AGT GGA-3′) |
| Reverse 1 (5′-GCA CAG GCC TTA AAT TCC AA-3′) | ||
| 452 | Forward 2 (5′-GAT GGA GTA CTT GTA GTG AT-3′) | |
| Reverse 2 (5′-ATC AAC AAA GGC ATT AAG GC-3′) | ||
| pgi | 622 | Forward (5′-TTG TGG TGT AAA AGC CTT GCG TGA-3′) |
| Reverse (5′-TGA GTG CAA TAG GAG TTA AAC CTA-3′) | ||
| mdh | 617 | Forward (5′-TGC TTT TTA GTG CAG GTT TTG CTA-3′) |
| Reverse (5′-CCA TTT TCA TGA TTT CAA TCA CA-3′) |
The open reading frame originally annotated as d-lactate dehydrogenase in the genome sequence was described as a putative oxidoreductase once annotation was completed.
Amplification and nucleotide sequence determination.
PCRs were carried out in 25-μl reaction volumes, with a typical reaction comprising the following: ∼10 ng of C. jejuni chromosomal DNA, 175 ng of each PCR primer, 1× PCR buffer (Invitrogen), 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, and 1.25 U of Taq DNA polymerase (Invitrogen). The reaction conditions were an initial denaturation at 95°C for 5 min, followed by 25 cycles of the following: 94°C for 45 s, primer annealing at 55 or 60°C for 45 s, and extension at 72°C for 1 min 30 s. The PCR products were purified either by precipitation with 20% (wt/vol) polyethylene glycol-2.5 M NaCl (2) or by use of the Qiaquick multiwell PCR purification kit (Qiagen), and the concentration was estimated by agarose gel electrophoresis. Sequence reactions were carried out in BigDye Ready reaction mix (Applied Biosystems) used in accordance with the manufacturer's instructions or by use of the CEQ DTCS sequencing system (Beckman Coulter). Unincorporated dye terminators were removed by precipitation with 95% ethanol, and the sequenced products were separated and detected with an ABI Prism 3700, ABI 377 automated DNA sequencer (Applied Biosystems) or a CEQ 2000 DNA sequencer (Beckman Coulter). Sequences were assembled and edited by use of Seqman (DNASTAR; Lasergene).
Allele and ST assignment.
For scheme A, alleles and sequence types (STs) were assigned by submitting the DNA sequence to the Campylobacter MLST database (http://campylobacter.mlst.net). For scheme B, the loci and STs were assigned arbitrary numbers in the order of identification. The organization of isolates into clonal complexes was carried out by use of the program BURST (based upon related STs), which is part of the START (Sequence Type Assignment Recombination Tests) group of programs (E. J. Feil and M. C. Chan; http://campylobacter.mlst.net) (14). Isolates were grouped together if they shared five, six, or seven of the seven total scheme A loci or four, five, or six of the six total scheme B loci. Only one member of each outbreak was included in these analyses. To avoid artifacts in complex assignment due to chaining effects, the clonal complexes were verified by cross-checking with the Campylobacter MLST database, in the case of scheme A, and by UPGMA(unweighted pair group method with arithmetic means) analysis, in the case of scheme B.
Linkage analysis.
Linkage analysis was carried out by using the index of association (IA), as defined previously (14, 27). We examined whether alleles were randomly associated, that is, at linkage equilibrium, indicating a freely recombining population, or nonrandomly associated, that is, at linkage disequilibrium, implying a clonal population structure. If there is linkage equilibrium, i.e., a random association between alleles of different loci, IA = 0. If IA is significantly different from 0, it indicates that recombination has been rare or absent and that the population has a clonal structure (19).
Statistical analysis.
The tests for association were carried out by use of Pearson's chi-square test. A two-way frequency table of sample source by ST complex was created, and the observed number of isolates from each source within the ST complex was compared with that expected on the assumption of independence of the row and column categories. The expected value was obtained by assuming that the isolates from each source are distributed among the clonal complexes according to the proportion of the total number of isolates occupied by each complex, e.g., since 30.8% of the data set constitutes the ST21 complex, the assumption is that 30.8% of the isolates from each source would be part of the ST21 complex. Only one representative of each of the outbreaks was included in this analysis. StatXact software was used to calculate the exact significance probabilities.
Invasion assay.
The invasion assay used was a gentamicin protection assay based on the method of Elsinghorst (7), with some modifications (10). Briefly, a monolayer of INT407 cells (ca. 5 × 105 cells per ml) was inoculated with broth-grown bacteria at a ratio of 50 to 200 bacteria per INT407 cell. To avoid variation in invasiveness due to motility, the bacteria were centrifuged onto the monolayer at room temperature for 15 min at 800 × g. Invasion was allowed to occur for 3 h at 37°C in 5% CO2, after which nonassociated bacteria were removed by washing three times with Hanks balanced salt solution (Sigma). Associated bacteria were removed during a 2-h incubation at 37°C in 5% CO2 with 2 ml of a 250-μg/ml solution of gentamicin (Sigma). Finally, the monolayer was washed three more times, and the internalized bacteria were then released by lysis of the INT407 cells with 1% Triton X-100 (Sigma) and were enumerated by plate count. Due to variations between assays, all isolates to be compared were included in the same assay, within which each isolate was assayed in triplicate. Each assay was repeated at least three times to verify the results. For each assay, a one-way analysis of variance followed by the Newman-Keuls multiple comparison test was carried out by use of GraphPad Prism software.
RESULTS
Genetic relatedness of isolates.
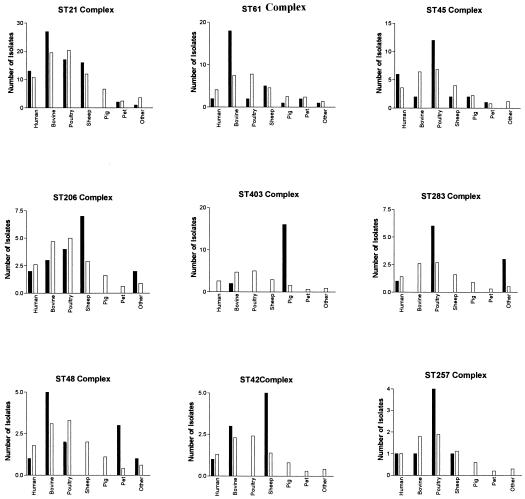
Overall, the 266 isolates were grouped into 19 clonal complexes, based on the fact that the isolates within one complex are identical at five, six, or all of the seven MLST loci (Table 2). There are highly significant associations among STs of bovine (P < 0.001), pig (P < 0.001), sheep (P < 0.001), and poultry (P < 0.001) samples, and by comparing the observed and expected sample frequencies, we can see which complexes are associated with particular sources. The ST21 complex contains isolates from most sources, whereas some of the other main complexes have over- or under-representations of isolates from particular sources. The most obvious example of this is the ST403 complex, which consists of a total of 18 isolates, 16 of which are from pigs. There were also more than expected bovine isolates in the ST61 complex, sheep isolates in the ST42 and ST206 complexes, and poultry isolates in the ST45 complex.
TABLE 2.
Distribution of all 266 isolates among ST complexes when MLST scheme A was used
| ST complex | ST | Isolate | Source | Countrya | Serotype
by:
|
ST complex | ST | Isolate | Source | Countrya | Serotype
by:
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LEP | Penner method | LEP | Penner method | |||||||||||
| 21 | 8 | C26 | Cow | UK | ||||||||||
| 19 | S11 | Sheep | UK | |||||||||||
| S51 | Sheep | UK | ||||||||||||
| Ch59 | Poultryb | UK | ||||||||||||
| C132(1) | Poultry | UK | ||||||||||||
| C132(4) | Poultry | UK | ||||||||||||
| 21 | 99/118 | Cow | NL | 1 | ||||||||||
| 99/188 | Human O/B1 | UK | 8 | 2 | ||||||||||
| 99/197 | Human O/B1 | UK | 8 | 2 | ||||||||||
| 99/208 | Human O/B1 | UK | 8 | 2 | ||||||||||
| 99/236 | Human O/B1 | UK | 4 | 2 | ||||||||||
| 99/206 | Human | DK | 50 | 2 | ||||||||||
| 99/217 | Human | DK | 50 | 2 | ||||||||||
| 88 126 | Cow | UK | ||||||||||||
| 88 231 | Cow | UK | ||||||||||||
| 88 238 | Cow | UK | NTc | |||||||||||
| 88 77 | Cow | UK | ||||||||||||
| 90 134 | Cow | UK | ||||||||||||
| 93 372 | Pet | UK | ||||||||||||
| 88 219 | Cow | UK | ||||||||||||
| 88 224 | Cow | UK | ||||||||||||
| 88 106 | Cow | UK | ||||||||||||
| 00 043 | Human | UK | 1 | |||||||||||
| S12 | Sheep | UK | ||||||||||||
| S14 | Sheep | UK | ||||||||||||
| S80 | Sheep | UK | ||||||||||||
| C2 | Cow | UK | ||||||||||||
| C17 | Cow | UK | ||||||||||||
| C20 | Cow | UK | ||||||||||||
| C44 | Cow | UK | ||||||||||||
| C50 | Cow | UK | ||||||||||||
| C262(2) | Poultry | UK | ||||||||||||
| C262(3) | Poultry | UK | ||||||||||||
| C262(4) | Poultry | UK | ||||||||||||
| H01 37 | Human | UK | ||||||||||||
| gm1 | Human | UK | ||||||||||||
| 43 | 00 87 | Human | UK | |||||||||||
| 47 | C254(1) | Poultry | UK | |||||||||||
| HF 7 | Pet | UK | ||||||||||||
| 50 | H01 35 | Human | UK | |||||||||||
| 94/194 | Poultry | UK | ||||||||||||
| 53 | 99/198 | Human O/B2 | DK | NT | 2 | |||||||||
| 99/199 | Human O/B2 | DK | 50 | 2 | ||||||||||
| 99/200 | Human O/B2 | DK | NT | 2 | ||||||||||
| 99/203 | Human O/B2 | DK | NT | 2 | ||||||||||
| 99/211 | Poultry | NL | NT | 2 | ||||||||||
| 91 28 | Cow | UK | ||||||||||||
| 104 | C204(10) | Poultry | UK | |||||||||||
| C204(16) | Poultry | UK | ||||||||||||
| H01 10 | Human | UK | ||||||||||||
| 262 | EX1182 | Environment | UK | |||||||||||
| EX1286 | Poultry | UK | ||||||||||||
| S46 | Sheep | UK | ||||||||||||
| S52 | Sheep | UK | ||||||||||||
| S83 | Sheep | UK | ||||||||||||
| C32 | Cow | UK | ||||||||||||
| C41 | Cow | UK | ||||||||||||
| 266 | C3 | Cow | UK | |||||||||||
| 300 | S2 | Sheep | UK | |||||||||||
| S20 | Sheep | UK | ||||||||||||
| 351 | 99/111 | Poultry | NL | |||||||||||
| 373 | 99/121 | Cow | NL | |||||||||||
| 376 | 89 80 | Cow | UK | |||||||||||
| 482 | 88 312 | Cow | UK | |||||||||||
| 486 | 00 037 | Poultry | UK | |||||||||||
| 487 | 00 014 | Poultry | UK | |||||||||||
| 489 | 94 006 | COW | UK | |||||||||||
| 490 | 00 038 | Human | UK | |||||||||||
| 623 | Ch87 | Poultry | UK | |||||||||||
| 630 | PS673.1 | Pig | UK | |||||||||||
| 399 | PS418 | Pig | UK | |||||||||||
| 483 | 99 373 | Human | UK | |||||||||||
| 546 | 99 339 | Human | UK | |||||||||||
| 502 | 88 26 | Cow | UK | |||||||||||
| 540 | 00 016 | Poultry | UK | |||||||||||
| 544 | 00 062 | Human | UK | |||||||||||
| 559 | C1 | Cow | UK | |||||||||||
| 561 | C13 | Cow | UK | |||||||||||
| 615 | S13 | Sheep | UK | |||||||||||
| 616 | S15 | Sheep | UK | |||||||||||
| 621 | C36 | Cow | UK | |||||||||||
| 626 | H01 51 | Human | UK | |||||||||||
| 635 | S87 | Sheep | UK | |||||||||||
| 640 | S24 | Sheep | UK | |||||||||||
| 641 | S41 | Sheep | UK | |||||||||||
| 642 | S48 | Sheep | UK | |||||||||||
| 668 | 94/174 | Poultry | UK | |||||||||||
| 61 | 60 | 99/209 | Human | UK | NT | 16, 50 | ||||||||
| 61 | 99/215 | Cow | NL | 50 | 16 | |||||||||
| 91 29 | Cow | UK | ||||||||||||
| 93 562 | Cow | UK | ||||||||||||
| 93 563 | Giraffe | UK | ||||||||||||
| S63 | Sheep | UK | ||||||||||||
| S82 | Sheep | UK | ||||||||||||
| S85 | Sheep | UK | ||||||||||||
| S95 | Sheep | UK | ||||||||||||
| C15 | Cow | UK | ||||||||||||
| C19 | Cow | UK | ||||||||||||
| C21 | Cow | UK | ||||||||||||
| C23 | Cow | UK | ||||||||||||
| C47 | Cow | UK | ||||||||||||
| C49 | Cow | UK | ||||||||||||
| 219 | 00 015 | Poultry | UK | |||||||||||
| 00 022 | Poultry | UK | ||||||||||||
| C37 | Cow | UK | ||||||||||||
| 477 | 88/34 | Cow | UK | |||||||||||
| 478 | 88 3 | Cow | UK | |||||||||||
| 479 | 88 5 | Cow | UK | |||||||||||
| 480 | 88 6 | Cow | UK | |||||||||||
| 500 | 89 116 | Cow | UK | |||||||||||
| 554 | PS567 | Pig | UK | NT | ||||||||||
| 618 | C16 | Cow | UK | |||||||||||
| 620 | C35 | Cow | UK | |||||||||||
| 622 | C42 | Cow | UK | |||||||||||
| 628 | H01 28 | Human | UK | |||||||||||
| 636 | S22 | Sheep | UK | |||||||||||
| 45 | 45 | EX145 | Poultry | UK | ||||||||||
| 99/97 | Human | NL | ||||||||||||
| EX146 | Poultry | UK | ||||||||||||
| 99/189 | Human O/B3 | SC | 50 | 55 | ||||||||||
| 99/192 | Human O/B3 | SC | 50 | 55 | ||||||||||
| 99/212 | Human O/B3 | SC | 50 | 55 | ||||||||||
| 99/218 | Human O/B3 | SC | 50 | 55 | ||||||||||
| 99/194 | Cow | UK | 50 | 55 | ||||||||||
| 99/202 | Cow | DK | NT | |||||||||||
| 99/216 | Human | FR | NT | 58 | ||||||||||
| S58 | Sheep | UK | ||||||||||||
| Ch84 | Poultry | UK | ||||||||||||
| Ch112 | Poultry | UK | ||||||||||||
| 94/229 | Poultry | UK | NT | |||||||||||
| 94/242 | Poultry | UK | NT | |||||||||||
| 137 | 99/389 | Human | UK | |||||||||||
| CH115 | Poultry | UK | ||||||||||||
| 241 | Ch140 | Poultry | UK | |||||||||||
| 334 | 99/13 | Poultry | CZ | |||||||||||
| 633 | S9 | Sheep | UK | |||||||||||
| 665 | 94/438 | Poultry | UK | 38 | ||||||||||
| 672 | 93/415 | Poultry | UK | 38 | ||||||||||
| 675 | HF 8 | Pet | UK | |||||||||||
| 675 | HF 8 | Pet | UK | |||||||||||
| 206 | 46 | 99/66 | Poultry | DK | ||||||||||
| 99/210 | Poultry | DK | 13 | 9 | ||||||||||
| 99/220 | Poultry | DK | NT | 1, 9 | ||||||||||
| 206 | EX303 | Environment | UK | |||||||||||
| S21 | Sheep | UK | ||||||||||||
| S79 | Sheep | UK | ||||||||||||
| S89 | Sheep | UK | ||||||||||||
| C31 | Cow | UK | ||||||||||||
| 221 | 99/18 | Human | CZ | |||||||||||
| 227 | C228(5) | Poultry | UK | |||||||||||
| 271 | S74 | Sheep | UK | |||||||||||
| 273 | S81 | Sheep | UK | |||||||||||
| S86 | Sheep | UK | ||||||||||||
| 471 | EX1692 | Environment | UK | |||||||||||
| S18 | Sheep | UK | ||||||||||||
| 543 | 00 060 | Human | UK | |||||||||||
| 562 | C28 | Cow | UK | |||||||||||
| 645 | C228(3) | Cow | UK | |||||||||||
| 403 | 55 | 99/238 | Cow | UK | ||||||||||
| C25 | Cow | UK | ||||||||||||
| 270 | PS762 | Pig | UK | 22 | ||||||||||
| PS852 | Pig | UK | 29 | |||||||||||
| PS857 | Pig | UK | NT | |||||||||||
| 403 | PS549.1 | Pig | UK | NT | ||||||||||
| PS830 | Pig | UK | NT | |||||||||||
| PS838 | Pig | UK | NT | |||||||||||
| PS843 | Pig | UK | NT | |||||||||||
| PS849 | Pig | UK | NT | |||||||||||
| 435 | PS484 | Pig | UK | 23 | ||||||||||
| 550 | PS220 | Pig | UK | 23 | ||||||||||
| 551 | PS304 | Pig | UK | NT | ||||||||||
| 552 | PS355 | Pig | UK | 23 | ||||||||||
| PS623 | Pig | UK | NT | |||||||||||
| 553 | PS444 | Pig | UK | 23 | ||||||||||
| 556 | PS706 | Pig | UK | 35 | ||||||||||
| 557 | PS799 | Pig | UK | NT | ||||||||||
| 283 | 267 | 81116d | Human O/B4a | UK | 6 | 6 | ||||||||
| 8280 | Human O/B4a | UK | 6 | |||||||||||
| 8269 | Human O/B4a | UK | 6 | |||||||||||
| 8279 | Human O/B4a | UK | 6 | |||||||||||
| 8272 | Environment | UK | 6 | 6 | ||||||||||
| Ch114 | Poultry | UK | ||||||||||||
| 383 | C130(2) | Poultry | UK | |||||||||||
| C130(4) | Poultry | UK | ||||||||||||
| 564 | EX524 | Environment | UK | |||||||||||
| EX497 | Poultry | UK | ||||||||||||
| EX543 | Environment | UK | ||||||||||||
| EX496 | Poultry | UK | ||||||||||||
| 625 | Ch146 | Poultry | UK | |||||||||||
| 48 | 48 | 99/201 | Cow | NL | 50 | 50 | ||||||||
| C6 | Cow | UK | ||||||||||||
| HF 9 | Pet | UK | ||||||||||||
| 473 | EX2200 | Environment | UK | |||||||||||
| 474 | 99/27 | Poultry | CZ | |||||||||||
| 475 | 99/96 | Poultry | DK | |||||||||||
| C4(1) | Cow | UK | ||||||||||||
| 476 | 88 139 | Cow | UK | |||||||||||
| 541 | 00 048 | Human | UK | |||||||||||
| C22 | Cow | UK | ||||||||||||
| 674 | HF 1 | Pet | UK | |||||||||||
| 676 | HF 10 | Pet | UK | |||||||||||
| 42 | 42 | 99/219 | Human | NL | ||||||||||
| S4 | Sheep | UK | ||||||||||||
| 501 | 88 138 | Cow | UK | |||||||||||
| 575 | C7 | Cow | UK | |||||||||||
| 629 | C34 | Cow | UK | |||||||||||
| 632 | S3 | Sheep | UK | |||||||||||
| 634 | S31 | Sheep | UK | |||||||||||
| 638 | S6 | Sheep | UK | |||||||||||
| 643 | S84 | Sheep | UK | |||||||||||
| 257 | 17 | Ch61 | Poultry | UK | ||||||||||
| 257 | S75 | Sheep | UK | |||||||||||
| C120(2) | Poultry | UK | ||||||||||||
| H01 43 | Human | UK | ||||||||||||
| 496 | 99/15 | Poultry | CZ | |||||||||||
| 560 | C9 | Cow | UK | |||||||||||
| 565 | C120(4) | Poultry | UK | |||||||||||
| 354 | 354 | S45 | Sheep | UK | ||||||||||
| S50 | Sheep | UK | ||||||||||||
| 472 | EX2072 | Environment | UK | |||||||||||
| 491 | 00/052 | Human | UK | |||||||||||
| 542 | 00/054 | Human | UK | |||||||||||
| 627 | H01 38 | Human | UK | |||||||||||
| 177 | 563 | 8287 | Human O/B4b | UK | 58 | |||||||||
| 8286 | Human O/B4b | UK | 58 | |||||||||||
| 8274 | Human O/B4b | UK | 58 | |||||||||||
| 8277 | Human O/B4b | UK | 58 | |||||||||||
| 673 | HF 2 | Pet | UK | |||||||||||
| 22 | 497 | 99/258 | Human | SA | ||||||||||
| 499 | 99/69 | Poultry | DK | |||||||||||
| 545 | 00 064 | Human | UK | |||||||||||
| 617 | C5 | Cow | UK | |||||||||||
| 52 | 52 | 99/204 | Sheep | UK | ||||||||||
| 539 | 00/032 | Poultry | UK | |||||||||||
| 614 | 99/30 | Poultry | CZ | |||||||||||
| 669 | 94/266 | Poultry | UK | 5 | ||||||||||
| 49 | 49 | 99/191 | Poultry | UK | ||||||||||
| C24 | Cow | UK | ||||||||||||
| 156 | 99/16 | Poultry | CZ | |||||||||||
| 353 | 133 | 99/14 | Poultry | CZ | ||||||||||
| 353 | OF25 | Poultry | UK | |||||||||||
| OF26 | Poultry | UK | ||||||||||||
| 443 | 443 | 99/23 | Poultry | CZ | ||||||||||
| 547 | 99 422 | Environment | UK | |||||||||||
| 631 | 99/265 | Human | SA | |||||||||||
| 433 | 498 | EX2289 | Environment | UK | ||||||||||
| 566 | C29(5) | Cow | UK | |||||||||||
| 460 | 670 | 94/184 | Poultry | UK | ||||||||||
| 573 | 644 | Ch3 | Poultry | UK | ||||||||||
| UAe | 59 | 99/214 | Human | UK | ||||||||||
| 442 | 99/260 | Human | SA | |||||||||||
| 481 | 93/564 | Ostrich | UK | |||||||||||
| 484 | 00/026 | Poultry | UK | |||||||||||
| 485 | 00/036 | Poultry | UK | |||||||||||
| 488 | 00/017 | Poultry | UK | |||||||||||
| 495 | 99/12 | Poultry | CZ | |||||||||||
| 548 | 99/369 | Poultry | UK | |||||||||||
| 549 | PS162 | Pig | UK | 35 | ||||||||||
| 558 | PS831 | Pig | UK | 35 | ||||||||||
| 619 | C18 | Cow | UK | |||||||||||
| 624 | Ch88 | Poultry | UK | |||||||||||
| 637 | 99/68 | Poultry | DK | |||||||||||
| 639 | S7 | Sheep | UK | |||||||||||
| 664 | PS835 | Pig | UK | NT | ||||||||||
| 666 | 94/300 | Poultry | UK | |||||||||||
| 667 | 94/318 | Poultry | UK | |||||||||||
| 671 | HF17 | Pet | UK | |||||||||||
UK, United Kingdom; DK, Denmark; CZ, Czech Republic; NL, The Netherlands; SA, South Africa; SC, Scotland; FR, France; SW, Sweden.
Poultry isolates were chicken cecal samples.
NT, not typeable.
81116 was originally isolated from outbreak 4a; however, the isolate included in this study was a laboratory-passaged version of the same strain.
UA, isolates that were unassigned to any clonal complex defined so far (last search, June 2003).
Confirmation of genetic stability by using MLST.
An investigation of epidemiologically linked isolates provides an opportunity to investigate the robustness of the MLST technique in an organism known to be prone to genetic instability (31). As expected, the human outbreak isolates within this study had identical MLST types within each outbreak (Table 2). These isolates were also identical by a range of other genotyping schemes, such as PFGE, AFLP, phage typing, and ribotyping (data not shown). One set of outbreak isolates, originally isolated in 1981 (human O/B4) (23), was found to be divided into two groups according to Penner serotype (25). This observation was confirmed more recently by other genotyping methods (18). In this study, this group of isolates was also divided into two groups by MLST: human O/B4a and human O/B4b (ST267 and ST563, respectively; the former is part of the ST283 complex and the latter is part of the ST177 complex). The outbreak 4a group of isolates was previously found to be highly similar (with a level of homology of 95% by AFLP) to a group of isolates from a broiler house environment almost 20 years later and was thought to represent a stable strain of C. jejuni (18). The group of isolates from the broiler house environment, comprising isolates EX524, EX497, EX543, and EX496, are all in the group ST564 (Table 2). Interestingly, ST267 and ST564, both within the ST283 complex, differ at just one locus, glyA, by MLST, which in fact is represented by a single nucleotide change, probably having arisen by point mutation. The second outbreak group (O/B4b), as expected, was very different by MLST (ST563). ST563 is part of the ST177 complex, as defined previously (6), which contains isolates from wild birds and the sand of bathing beaches.
Reanalysis of a subset of isolates by use of a second MLST scheme.
A subset of the isolates (n = 82) included in this study was reanalyzed with a second MLST scheme, utilizing five more unique loci, as gltA was shared by both schemes. Comparison of the complex assignment for each scheme, using just these 82 isolates, revealed that the isolates were grouped into seven complexes with scheme A and five complexes with scheme B (Table 3). The IA increased to 2.50 when 68 unique STs using 12 loci from both schemes were analyzed. The human outbreak isolates remained identical at all 12 loci, as expected (Table 4). Interestingly, another group of isolates were also identical at all 12 loci (Table 5). This group comprised six isolates of human, bovine, and poultry origin, from the United Kingdom, Denmark, and The Netherlands, from three different years within a 12-year period. Since this group appeared to be so highly related, the phenotypes of the isolates were analyzed in more detail. The serotype of each isolate was determined by use of the Laboratory of Enteric Pathogens (LEP; Health Protection Agency, Colindale, United Kingdom) method (11), and their invasion potentials were determined by use of an in vitro gentamicin protection assay (7). The results are given in Table 5. Two of the isolates were nontypeable by the LEP method, but of the remaining four, two different serotypes, serotypes 1 and 50, were obtained. Three of the five isolates tested in the invasion assay had similar levels of invasion, whereas isolates 88/238 and 99/118 had significantly greater invasion potentials than the other isolates within this group (P < 0.001).
TABLE 3.
Comparison of the main ST complexes for scheme A and scheme B
| Scheme | Lineage | ST | Isolate(s) | Scheme | Lineage | ST | Isolate(s) | |
|---|---|---|---|---|---|---|---|---|
| A | 21 | 21 | 00/043,a 88/106, 88/224, 88/219, 93/372, 90/134, 88/77, 88/238,a 88/231,a 88/126, 99/118, 99/236, 99/208, 99/197, 99/188, 99/217,a 99/206a | |||||
| 53 | 99/211, 99/203, 99/200, 99/199, 99/198, 91/28 | |||||||
| 262 | EX1286, EX1182 | |||||||
| 373 | 99/121 | |||||||
| 376 | 89/80 | |||||||
| 482 | 88/312 | |||||||
| 486 | 00/037 | |||||||
| 487 | 00/014 | |||||||
| 489 | 94/006 | |||||||
| 490 | 00/038 | |||||||
| 502 | 88/26 | |||||||
| 540 | 00/016 | |||||||
| 45 | 45 | 99/202, 99/194, 99/192, 99/97, EX145 | ||||||
| 483 | 99/373 | |||||||
| 48 | 48 | 99/201 | ||||||
| 473 | EX2200 | |||||||
| 474 | 99/27 | |||||||
| 475 | 99/96 | |||||||
| 476 | 88/139 | |||||||
| 541 | 00/048 | |||||||
| 49 | 49 | 99/191 | ||||||
| 156 | 99/16 | |||||||
| 61 | 60 | 99/209 | ||||||
| 61 | 93/563, 93/562, 91/29, 99/215 | |||||||
| 219 | 00/022, 00/015 | |||||||
| 478 | 88/3 | |||||||
| 479 | 88/5 | |||||||
| 480 | 88/6 | |||||||
| 206 | 46 | 99/220, 99/210, 99/66 | ||||||
| 206 | EX303 | |||||||
| 221 | 99/18 | |||||||
| 471 | EX1692 | |||||||
| 443 | 443 | 99/23 | ||||||
| 631 | 99/265 | |||||||
| B | 1 | 1 | 00/043,a 00/037, 88/238,a 88/231,a 99/217,a 99/206,a, 99/118 | |||||
| 2 | 99/121 | |||||||
| 3 | EX1182 | |||||||
| 4 | EX1286 | |||||||
| 5 | 94/006, 99/211, 99/203, 99/200, 99/199, 99/198 | |||||||
| 7 | 90/134 | |||||||
| 8 | 91/28 | |||||||
| 9 | 99/27 | |||||||
| 10 | 99/96 | |||||||
| 14 | 99/236, 99/208, 99/197, 99/188 | |||||||
| 15 | 00/015, 99/215, 99/209 | |||||||
| 16 | 88/312 | |||||||
| 18 | 00/022 | |||||||
| 20 | 93/563 | |||||||
| 21 | EX2200, 99/201 | |||||||
| 22 | 00/016 | |||||||
| 24 | 93/562 | |||||||
| 25 | 88/224 | |||||||
| 26 | 93/372 | |||||||
| 27 | 88/139 | |||||||
| 28 | 88/34 | |||||||
| 29 | 88/3, 88/5 | |||||||
| 30 | 88/6 | |||||||
| 31 | 88/77 | |||||||
| 32 | 00/014 | |||||||
| 52 | 00/038 | |||||||
| 53 | 91/29 | |||||||
| 56 | 89/80 | |||||||
| 58 | 00/048 | |||||||
| 59 | 88/219 | |||||||
| 60 | 88/106, 88/26, 88/126 | |||||||
| (13)b | 13 | EX303, EX1692 | ||||||
| 42 | 99/66 | |||||||
| 43 | 99/220, 99/210 | |||||||
| (33) | 33 | EX145 | ||||||
| 34 | 99/192 | |||||||
| 36 | 00/036 | |||||||
| 37 | 99/194 | |||||||
| 55 | 99/373 | |||||||
| (40) | 40 | 99/30 | ||||||
| 45 | 99/204 | |||||||
| 46 | 00/032 | |||||||
| 47 | EX2072 | |||||||
| (38) | 38 | 99/16 | ||||||
| 54 | 99/191 |
Highly related group of isolates that were identical at all 12 loci.
No founder member was assigned for ST complexes in parentheses, so the first ST was chosen as the group name for identification purposes.
TABLE 4.
ST complexes for the isolates analyzed at all 12 loci
| Complex | ST | Isolate(s) |
|---|---|---|
| 1 | 1 | 00/043, 88/238, 88/231, 99/217, 99/206, 99/118 |
| 2 | 90/134 | |
| 3 | 99/236, 99/208, 99/197, 99/188 | |
| 4 | 88/224 | |
| 5 | 93/372 | |
| 6 | 88/77 | |
| 7 | 88/219 | |
| 8 | 88/106, 88/126 | |
| 19 | 99/211, 99/203, 99/200, 99/199, 99/198 | |
| 20 | 91/28 | |
| 33 | EX1182 | |
| 34 | EX1286 | |
| 37 | 89/80 | |
| 43 | 99/27 | |
| 51 | 88/312 | |
| 55 | 00/037 | |
| 58 | 94/006 | |
| 62 | 88/26 | |
| (10)a | 10 | EX145 |
| 11 | 99/192 | |
| 13 | 99/194 | |
| (14) | 14 | 99/66 |
| 15 | 99/220, 99/210 | |
| (16) | 16 | 99/201 |
| 42 | EX2200 | |
| 65 | 00/048 | |
| (23) | 22 | 99/209 |
| 23 | 99/215 | |
| 24 | 93/563 | |
| 25 | 93/562 | |
| 26 | 91/29 | |
| 30 | 00/015 | |
| 31 | 00/022 | |
| 48 | 88/5 | |
| 49 | 88/6 | |
| (29) | 29 | EX303 |
| 40 | EX1692 | |
| (39) | 39 | 99/23 |
| 67 | 99/265 |
No founder member was assigned for ST complexes in parentheses, so the first ST was chosen as the group name for identification purposes.
TABLE 5.
Highly related group of isolates
| Isolate | Source | Year | Countrya | ST (scheme A) | ST (scheme B) | ST (schemes A and B) | LEP serotype | Invasion potentialb |
|---|---|---|---|---|---|---|---|---|
| 00/043 | Human | 2000 | UK | 21 | 1 | 1 | 1 | 0.019 (0.0077) |
| 88/238 | Cow | 1988 | UK | 21 | 1 | 1 | NTc | 0.090 (0.0070) |
| 99/217 | Human | 1999 | DK | 21 | 1 | 1 | 50 | 0.020 (0.0062) |
| 99/206 | Poultry | 1999 | NL | 21 | 1 | 1 | 50 | 0.032 (0.0064) |
| 99/118 | Cow | 1999 | NL | 21 | 1 | 1 | 1 | 0.174 (0.020) |
| 88/231 | Cow | 1988 | UK | 21 | 1 | 1 | NT | NDd |
UK, United Kingdom; DK, Denmark; NL, The Netherlands.
Invasion potential was determined as the percentage of the inoculum applied to the INT407 cell monolayer that survived gentamicin treatment. Each isolate was tested in at least three independent assays. These data are from a single assay in which each isolate was tested in triplicate. Numbers in parentheses are standard deviations.
NT, nontypeable by the LEP method.
ND, not done.
DISCUSSION
Previous investigations of the population structure of C. jejuni have indicated that the population is weakly clonal, consisting of large clonal complexes, some of which may be associated with certain sources, such as the sand of bathing beaches (6). These studies used a database comprising largely human isolates (61%) (5), with some isolates from food sources. For this study, the MLST scheme developed by Dingle et al. (6) was applied to 215 isolates from veterinary and veterinary-related sources. These were then compared with 51 human isolates in an attempt to identify potential overlaps between the veterinary and human populations and to investigate the presence of host specificity among C. jejuni strains. A subset of the isolates was also analyzed by a second MLST scheme which contributed 5 additional loci, providing a unique set of isolates that were analyzed at a total of 12 loci.
The population analyzed, as described previously (5, 6, 20), appears to have a weakly clonal structure, that is, there is clonal structure as well as some evidence of recombination. The data from this study also clearly show that campylobacters from veterinary and human populations do overlap. Each complex detected contained isolates of human and veterinary origin, apart from the ST403 complex, which contained only pig and bovine isolates. The distribution of STs among isolates from cattle, sheep, poultry, pets, and humans has been reported previously (5, 6) and indicates that these isolates have perhaps adapted to infect a number of hosts, supporting the hypothesis that each animal isolate is a potential human pathogen. This is also in agreement with a previous study in which MLEE was used to investigate the relationship between animal and human isolates (1). In that study, both human and animal isolates shared electrophoretic types, with very little distinction between the two populations.
The association of source with ST complex may indicate that isolates within ST complexes have adapted to a particular niche or type of niche. The presence of clonal complexes within an otherwise recombining population suggests that there must be some selective pressure to maintain this structure, which may be provided by niche adaptation (5). Other mechanisms that may have influenced this structure include clonal expansion, geographical or ecological isolation, host immune selection, or barriers to genetic exchange (19).
The associations with source observed in this study are similar to those reported previously (5). The ST45 and ST257 complexes were found to predominantly contain isolates of human and poultry origin, with significantly more than the expected number of poultry isolates in the ST45 complex in this study. There also appears to be a strong association between the ST61 and ST42 complexes and bovine and sheep isolates, respectively, which may be a reflection of the number of isolates from these sources included in the study. The presence of host, source, or niche adaptation contradicts the hypothesis that all campylobacters are potential human pathogens and instead indicates that nonpathogenic campylobacters may exist (4, 15, 16).
There was a very strong association between the ST403 complex and isolates from pigs. When compared to the Campylobacter MLST database (http://campylobacter.mlst.net), the ST403 complex was found to contain isolates from other food products as well as from infected humans, indicating that isolates within this complex do have the capacity to cause disease in humans. Interestingly, a large number of these human isolates are from the Dutch Caribbean island of Curaçao (B. Duim, personal communication). The reason that a large number of human isolates from a distinct geographical location are clonally related to C. jejuni isolates from pigs in the United Kingdom is unclear and warrants further investigation. In spite of this, a number of the STs were unique to pigs and may themselves represent pig-adapted isolates. In a recent report, subtypes of both C. jejuni and C. coli isolated from pigs were found to be clustered into one of only four genotypes by ribotyping and flaA restriction fragment length polymorphism, demonstrating the persistence of clonal types within this host (22). Moreover, subspecies of both C. jejuni and C. coli isolated from pigs were identified by MLEE (21). Further identification to the species level of the pig isolates in our study revealed that they were hippurate-negative C. jejuni isolates (Stephen On, personal communication) and may represent an unusual pig-associated clone of C. jejuni.
Interestingly, the pig isolates had been serotyped by the LEP method (11) prior to inclusion in this study. The majority were nontypeable (9 of 16; 56%); however, four different serotypes (HS23, -35, -29, and -22) were present among the remaining isolates, confirming that isolates with identical or very similar genotypes can express different antigens, in this case detected by LEP serotyping. Moreover, it is unlikely that this clonal group of pig isolates would have been detected if serotyping alone had been used. Serotype does not appear to be a good indicator of clonal complex (5). Most clonal complexes were found to contain multiple serotypes; however, there were exceptions, as in the case of the ST22 complex, which contained isolates of HS serotype 19 only (5).
One group of human outbreak isolates within this study was found to be part of the ST177 complex when compared to the available database. This complex was previously thought to be associated with isolates from the sand of bathing beaches and from wild bird feces and thus was presumed to represent a potentially nonpathogenic group of isolates (6). Despite the presence of human isolates within this ST complex, the association with wild bird feces might still be plausible, given that the source of the human outbreak was a water source which was contaminated with fecal matter from wild birds or bats (23). It is possible that these isolates are more adapted to colonize the intestines of wild birds and to survive in a particular environment but that when the opportunity arises, they are capable of causing human disease.
A number of the isolates from this study were reanalyzed by use of a second MLST scheme. The largest clonal complex in each scheme shared similar isolates; however, there were some differences in the exact clustering which were probably due to the different levels of discrimination between the two schemes. Most of the variation in scheme B seemed to be accounted for by two loci, d-lactate dehydrogenase and adenylate kinase. Adenylate kinase was the most variable of all the scheme B loci, with 8.9% variable sites across the 401-bp length. The putative oxidoreductase was the least variable, but difficulties in amplification were faced for a number of isolates and alternative primers had to be used.
Isolates which were epidemiologically linked, such as those from outbreaks, were clustered together by both schemes, confirming their identities as clonally related isolates. In contrast, apparently unrelated isolates differed in their organization within each scheme, which is supported by the observation that C. jejuni subtyping results generally only correlate if isolates are clonally related (T. M. Wassenaar and D. G. Newell, Abstr. 11th Int. Workshop Campylobacter Helicobacter Related Organisms, abstr. H39, 2001). Nevertheless, one other interesting group of isolates that shared all 12 loci was also identified. This group of apparently epidemiologically unlinked isolates were from different geographical locations, sources, and times, yet they were genotypically highly related. It is hypothesized, therefore, that this group of isolates represents another clonal strain of C. jejuni that is similar to the one previously reported (18).
Despite their genetic relatedness, these isolates were found to vary in serotype as well as virulence potential, as determined by in vitro assays of invasion. This is not too surprising, given that it is well established for other bacteria that identity or similarity of serotype, biotype, or other phenotypic character does not indicate genetic identity (26). The genotypic and phenotypic diversity that exists within C. jejuni is not due only to point mutations, horizontal DNA transfer, and genomic rearrangements detected by MLST (5, 6) and other genotyping methods such as fla typing (12, 13, 30) and PFGE (31). It may also be due to polymorphisms within homonucleotide stretches throughout the genome (24) that may rapidly alter the phenotype of the organism (33) through variation in gene expression or posttranslational modification.
Of those isolates whose STs were not yet assigned to a complex, the majority (9 of 18; 50%) were of poultry origin. Poultry isolates were previously found to have a broader distribution among the clonal complexes than human isolates (6), and it is possible that these isolates are more genetically diverse than those from other sources. This has also been observed in a previous study (3) in which many more fla types were present among poultry isolates than among human isolates. This genetic diversity among poultry isolates fits with the theory of bottleneck selection, in which it is proposed that C. jejuni undergoes variation only during growth inside the host (Wassenaar and Newell, Abstr. 11th Int. Workshop Campylobacter Helicobacter Related Organisms). Such diversity is believed to increase the chance of a subset of bacteria to survive the environmental stresses to which they are exposed outside the host. Since C. jejuni has fastidious growth requirements and appears to be adapted for growth in the avian gut, it is likely that most of the variation occurs within this environment, resulting in a greater diversity of types isolated from poultry. With the addition of more poultry isolates into this MLST study, most of these so far unassigned isolates would most likely become part of larger complexes.
This study set out to investigate whether the populations of veterinary and human isolates overlap and whether other potential sources of C. jejuni infections in humans could be identified. It appears that the populations do overlap, with STs shared between human isolates and those from various other sources. There are, however, STs that appear to be associated with a single source only and some complexes that are more associated with isolates of a particular source. It will only be possible to determine whether some of these STs are host specific by conducting a similar study on a much larger scale. In conclusion, it appears that isolates from cattle, sheep, poultry, pets, the environment, and even some from pigs may all have the potential to cause disease in humans, given the opportunity, and so should all be considered potential sources of human infection.
Association between clonal complex and source of isolation.
The distribution of isolates from the various sources in the nine largest clonal complexes is given in Fig. 2. The largest complex for scheme A was the ST21 complex, which contained 34 of 82 (42%) of the isolates, with ST21 as the founder member. For scheme B, the ST1 complex was the largest, containing 52 of 82 (63%) of the isolates, with ST1 as the founder member. In total, 61 isolates were present in both the ST21 and ST1 complexes. Seventeen isolates were present in the ST1 complex (scheme B) but absent from the ST21 complex (scheme A) (isolates 99/27, 99/96, 00/015, 99/215, 99/209, 00/022, 93/563, EX2200, 99/201, 93/562, 88/139, 88/34, 88/3, 88/5, 88/6, 91/29, and 00/048). In contrast, all of the isolates that form the ST21 complex (scheme A) were present in the ST1 complex of scheme B. An IA of 0.51 was obtained when 59 unique STs identified for the panel of isolates using the six loci from scheme B were analyzed. This compares to an IA of 0.93 when any six loci selected from scheme A (the example given shows data excluding uncA) were analyzed. Both values again indicate that the population has some degree of clonality and that individual clonal complexes are sufficiently stable to be clearly identified by use of sequence data from six or seven loci.
FIG. 2.
Observed (black bars) and expected (white bars) numbers of isolates from each source within nine of the largest scheme A ST complexes. “Other” includes environmental and other diverse isolates. Only one representative from each outbreak was included in this analysis.
Reanalysis of the subset of isolates at 12 unique loci.
When analyzed at all 12 loci (10, 11, or 12 loci in common), the isolates were grouped into seven complexes (Table 4).
The most common complex was the ST21 complex, which comprised 82 isolates (30.8% of the data set) divided among 33 STs, followed by the ST61 complex, which comprised 29 isolates (10.9% of the data set) divided among 14 STs, and the ST45 complex, with 28 isolates (10.5% of the data set) divided among 13 STs. The ST21 and ST45 complexes were reported previously (5, 6) as the largest among the population of C. jejuni analyzed, which comprised mainly human isolates.
Six of the 19 complexes contained three or fewer members. These STs were assigned to a complex based on the data in the Campylobacter MLST database (http://campylobacter.mlst.net). In some cases, no founder was identified within this set of isolates, even when the complex consisted of more than three members (ST433, ST443, ST460, ST573, ST22, ST283, and ST177 complexes). Again, the founders of these complexes were identified by comparison with the larger database of isolates. Eighteen STs remain unassigned to a complex (last database query, June 2003).
The IA for 149 unique STs within the whole data set using the seven loci from scheme A was 1.29, indicating significant linkage disequilibrium or that there is a certain degree of clonality within the population. This compares to an IA of −0.37 for unique STs within the dominant clonal complex typified by ST21, which suggests that recombination within the clonal complex is high. This indicates, as previously proposed (6, 20), that the C. jejuni population is weakly clonal.
Acknowledgments
We thank Lise Petersen, Mogens Madsen, Birgitta Duim, Iva Steinhauserova, Al Lastovica, Exeter Public Health Laboratory, and CAMPYNET for providing some of the isolates that were used in this study. We also thank Kate Dingle, Frances Colles, Martin Maiden, and Lynne Richardson for helpful advice and DNA sequencing. Thanks also go to Robin Sayers for help with statistical analysis, Adrian Whatmore for useful discussions, Stephen On for further characterization of the pig isolates, and Jenny Frost (HPA, Colindale, United Kingdom) for LEP serotyping.
This work was funded by Department of Environment, Food and Rural Affairs, United Kingdom, project number OZO602.
REFERENCES
- 1.Aeschbacher, M., and J.-C. Piffaretti. 1989. Population genetics of human and animal enteric Campylobacter strains. Infect. Immun. 57:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, S. F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, K. Struhl, L. M. Albright, D. M. Coen, and A. Varki (ed.). 1994. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 3.Clow, K. 2000. The genotypic and phenotypic comparison of Campylobacter jejuni isolates from humans and poultry. Ph.D. thesis. University of Reading, Reading, United Kingdom.
- 4.Clow, K., S. Park, P. Hawtin, and D. Newell. 1998. The genotypic comparison of Campylobacter jejuni strains from humans and poultry, p. 25-32. In A. Aspan and R. Mulder (ed.), COST Action 97. Development of monitoring procedures, rapid detection methods and techniques. Molecular epidemiology of Campylobacter and Salmonella, vol. 4. Office for Official Publications of the European Communities, Gare, Luxembourg.
- 5.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. A. Wareing, and M. C. J. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elsinghorst, E. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236:405-420. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 9.Evans, S. 1997. Epidemiological studies of Salmonella and Campylobacter in poultry. Ph.D. thesis. University of London, London, United Kingdom.
- 10.Fearnley, C. 2002. The variations in virulence of Campylobacter jejuni strains associated with poultry. PhD thesis. University of Birmingham, Birmingham, United Kingdom.
- 11.Frost, J. A., A. N. Oza, R. T. Thwaites, and B. Rowe. 1998. Serotyping scheme for Campylobacter jejuni and Campylobacter coli based on direct agglutination of heat-stable antigens. J. Clin. Microbiol. 36:335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanninen, M. L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrington, C. S., F. M. Thomson-Carter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 15.Koenraad, P., R. Ayling, W. Hazeleger, and D. Newell. 1995. The speciation and subtyping of Campylobacter isolates from sewage plants and waste water from a connected poultry abattoir using molecular techniques. Epidemiol. Infect. 115:485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korolik, V., L. Moorthy, and P. J. Coloe. 1995. Differentiation of Campylobacter jejuni and Campylobacter coli strains by using restriction endonuclease DNA profiles and DNA fragment polymorphisms. J. Clin. Microbiol. 3:1136-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manning, G., B. Duim, T. Wassenaar, J. A. Wagenaar, A. Ridley, and D. G. Newell. 2001. Evidence for a genetically stable strain of Campylobacter jejuni. Appl. Environ. Microbiol. 67:1185-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maynard-Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meinersmann, R. J., C. M. Patton, G. M. Evins, I. K. Wachsmuth, and P. I. Fields. 2002. Genetic diversity and relationships of Campylobacter species and subspecies. Int. J. Syst. E vol. Microbiol. 52:1789-1797. [DOI] [PubMed] [Google Scholar]
- 21.Moore, J. E., M. M. Garcia, and R. H. Madden. 2002. Subspecies characterisation of porcine Campylobacter coli and Campylobacter jejuni by multilocus enzyme electrophoresis typing. Vet. Res. Commun. 26:1-9. [DOI] [PubMed] [Google Scholar]
- 22.Moore, J. E., J. Lanser, M. Heuzenroeder, R. M. Ratcliff, B. C. Millar, and R. H. Madden. 2002. Molecular diversity of Campylobacter coli and C. jejuni isolated from pigs at slaughter by flaA-RFLP analysis and ribotyping. J. Vet. Med. B 49:388-393. [DOI] [PubMed] [Google Scholar]
- 23.Palmer, S. R., P. R. Gully, J. M. White, A. D. Pearson, W. G. Suckling, D. M. Jones, J. C. Rawes, and J. L. Penner. 1983. Water-borne outbreak of campylobacter gastroenteritis. Lancet i:287-290. [DOI] [PubMed]
- 24.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 25.Penner, J. L., A. D. Pearson, and J. N. Hennessey. 1983. Investigation of a waterborne outbreak of. Campylobacter jejuni enteritis with a serotyping scheme based on thermostable antigens. J. Clin. Microbiol. 18:1362-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selander, R. K., and J. M. Musser. 1990. Population genetics of bacterial pathogens, p. 11-36. In B. H. Iglewski and V. L. Clark (ed.), Molecular basis of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 27.Souza, V., T. T. Nguyen, R. R. Hudson, D. Pinero, and R. E. Lenski. 1992. Hierarchical analysis of linkage disequilibrium in Rhizobium populations: evidence for sex? Proc. Natl. Acad. Sci. USA 89:8389-8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tompkins, D. S., M. J. Hudson, H. R. Smith, R. P. Eglin, J. G. Wheeler, M. M. Brett, R. J. Owen, J. S. Brazier, P. Cumberland, V. King, and P. E. Cook. 1999. A study of infectious intestinal disease in England: microbiological findings in cases and controls. Commun. Dis. Public Health 2:77-152. [PubMed] [Google Scholar]
- 30.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1995. Variation of the flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology 141:95-101. [DOI] [PubMed] [Google Scholar]
- 31.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wassenaar, T. M., J. A. Wagenaar, A. Rigter, C. Fearnley, D. G. Newell, and B. Duim. 2002. Homonucleotide stretches in chromosomal DNA of Campylobacter jejuni display high frequency polymorphism as detected by direct PCR analysis. FEMS Microbiol. Lett. 212:77-85. [DOI] [PubMed] [Google Scholar]