Abstract
As part of an effort to develop detectors for selected species of bacterial spores, we screened phage display peptide libraries for 7- and 12-mer peptides that bind tightly to spores of Bacillus subtilis. All of the peptides isolated contained the sequence Asn-His-Phe-Leu at the amino terminus and exhibited clear preferences for other amino acids, especially Pro, at positions 5 to 7. We demonstrated that the sequence Asn-His-Phe-Leu-Pro (but not Asn-His-Phe-Leu) was sufficient for tight spore binding. We observed equal 7-mer peptide binding to spores of B. subtilis and its most closely related species, Bacillus amyloliquefaciens, and slightly weaker binding to spores of the closely related species Bacillus globigii. These three species comprise one branch on the Bacillus phylogenetic tree. We did not detect peptide binding to spores of several Bacillus species located on adjacent and nearby branches of the phylogenetic tree nor to vegetative cells of B. subtilis. The sequence Asn-His-Phe-Leu-Pro was used to identify B. subtilis proteins that may employ this peptide for docking to the outer surface of the forespore during spore coat assembly and/or maturation. One such protein, SpsC, appears to be involved in the synthesis of polysaccharide on the spore coat. SpsC contains the Asn-His-Phe-Leu-Pro sequence at positions 6 to 10, and the first five residues of SpsC apparently must be removed to allow spore binding. Finally, we discuss the use of peptide ligands for bacterial detection and the use of short peptide sequences for targeting proteins during spore formation.
A limited number of bacteria produce endospores (or spores) upon nutrient deprivation of vegetatively growing cells. The spore is dormant and highly resistant to extreme temperatures, radiation, desiccation, harsh chemicals, and physical damage. These properties allow spores to survive adverse environments—for many years in some cases—until encountering conditions that trigger germination and vegetative cell outgrowth (17). The genus Bacillus includes a diverse collection of gram-positive, rod-shaped, aerobic, spore-forming soil bacteria (20). In these bacteria, the sporulation process begins with a final round of DNA replication followed by an asymmetric septation that produces large and small genome-containing compartments: the mother cell and prespore, respectively (7). The mother cell septal membrane migrates around the prespore, surrounding it with two opposing membranes. Between these membranes, the prespore, now called the forespore, is encircled first by a thin germ cell wall and then by a thick layer of modified peptidoglycan, the cortex (8). Multiple layers of a proteinaceous spore coat are then deposited onto the cortex. In the well-studied case of Bacillus subtilis, the coat layers (i.e., under, inner, and outer coats) are composed of over two dozen different proteins (6, 12). For B. subtilis, the outer coat may be tightly covered by a thin and presently poorly characterized surface-exposed layer (25, 26). For other Bacillus species (e.g., B. anthracis), an additional prominent and loose-fitting layer called the exosporium forms the outermost surface of the spore (9). The assembly of the cortex, coat, and (when present) exosporium occurs by using mother cell components that are synthesized at appropriate times. The assembly process depends on specific but poorly understood interactions among these components and on secondary enzymatic (e.g., proteolytic) and nonenzymatic modifications (5, 12). The assembly stage is followed by a period of maturation in which covalent changes (e.g., glycosylation and cross-linking) occur in the outer layers (12, 21), and the spore assumes its properties of resistance and dormancy. Approximately 8 h after the initiation of sporulation, the mother cell lyses to release the mature spore.
Recently, our laboratory has been involved in the discovery and development of small-molecule ligands that can be used in a variety of detector platforms for species-specific capture and/or tagging of spores of B. anthracis, the causative agent of anthrax. Because it is impractical to use pathogenic spores (or in some cases even nonpathogenic variants of these spores) in the early stages of detector development, we constructed a model detection system involving a safe prototypical spore and a standard small-molecule ligand capable of binding this spore. In this paper, we describe this system: peptide ligands that bind tightly to spores of B. subtilis. The peptide ligands were identified by biopanning a library of phage-displayed peptides against B. subtilis spores. The selected peptide ligands contain a common amino-terminal tetrapeptide sequence and have other similarities. These ligands were found to bind tightly only to B. subtilis and closely related spores. A consensus sequence derived from the ligands was used to identify B. subtilis proteins that may employ the conserved peptide ligand sequence for docking to the outer surface of the forespore during spore coat assembly and/or maturation.
MATERIALS AND METHODS
Bacterial strains and bacteriophage.
The Bacillus strains used in this study and their sources are as follows: B. subtilis (trpC2) 1A700 (originally designated 168), B. amyloliquefaciens 10A1 (originally H), B. licheniformis 5A36 (originally ATCC 14580), and B. pumilus 8A3 (originally ATCC 7061) from the Bacillus Genetic Stock Center, Ohio State University, Columbus; B. globigii (also called B. atrophaeus and B. subtilis variety “niger”), B. thuringiensis subsp. kurstaki, B. cereus T, and B. anthracis Sterne (nonpathogenic) from the U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, Frederick, Md.; and B. mycoides ATCC 10206 and B. megaterium ATCC 14581 from the American Type Culture Collection, Manassas, Va. Escherichia coli strain ER2537 [F′ lacIq Δ(lacZ)M15 proA+B+/fhuA2 supE thi Δ(lac-proAB) Δ(hsdMS-mcrB)5 (rk− mk− McrBC−)], a host for M13 phage, and M13 phage display (Ph.D.) peptide libraries were obtained from New England Biolabs. The Ph.D.-7 and -12 libraries contained random 7- and 12-mer peptides, respectively, fused to the amino terminus of the minor phage coat protein pIII. This protein is present in five copies at one end of the filamentous phage particle. Thus, each phage displayed five copies of a particular peptide, the sequence of which was encoded in a single recombinant pIII gene.
General M13 methods and DNA sequence analysis.
M13 phage were propagated, titers were determined, and relevant regions of genomic DNAs of selected phage were sequenced according to the procedures in the Phage Display Peptide Library kit (New England Biolabs), except that M13 genomic DNA was prepared according to the QIAprep Spin M13 protocol (Qiagen).
Preparation of spores.
Except for B. pumilus, spores were prepared by growing cells at 37°C in either liquid Difco sporulation medium (DSM) (18) with shaking (150 rpm) or on solid DSM (1.5% agar) until sporulation was essentially complete—typically 48 to 72 h. Spores prepared on liquid or solid media behaved identically in our studies. B. pumilus spores were prepared by growing cells on solid DSM at 30°C. Spores were collected by centrifugation, washed extensively with cold (4°C) sterile distilled water, sedimented through a two-step gradient of 20 and 50% Renografin (Bracco Diagnostics), and extensively washed again with cold water (12, 18). Spores were stored protected from light in sterile distilled water at 4°C and washed every 2 weeks to prevent germination. Spores were quantitated microscopically with a Petroff-Hausser counting chamber, and spore preparations were checked before use for the absence of germinating spores. Only freshly prepared spores of B. globigii were used in the studies shown, because these spores gradually lose their capacity for peptide binding over several months.
Biopanning phage-displayed peptides.
The phage display libraries were screened (or biopanned) for peptides that bind B. subtilis spores by gently mixing approximately 109 spores and 1011 phage in 1 ml of sterile TBST (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% Tween 20) for 10 min at room temperature. The spore-phage complexes were collected by centrifugation (12,000 × g) at 4°C for 10 min, and the supernatant was removed. The complexes were washed 10 times with 1 ml (each) of ice-cold TBST, with centrifugation (12,000 × g) at 4°C for 5 min after each wash. After the final wash, spore-phage complexes were quickly resuspended with a vortex mixer in 1 ml of elution buffer (0.2 mM glycine-HCl [pH 2.2], 1 mg of bovine serum albumin per ml) and then gently mixed for 5 min at room temperature. This sample was centrifuged as described above for 5 min. The supernatant, which contained eluted phage, was quickly removed and neutralized by the addition of 150 μl of 1 M Tris-HCl (pH 9.1) to prevent phage killing. During these procedures, a small sample of input phage, supernatants from the initial collection of spore-phage complexes and selected washes, and eluted phage were saved for titering.
The eluted phage (except for the small sample for titering) were amplified by infecting E. coli strain ER2537. The resulting phage stock was used for a second round of biopanning, which was performed exactly as described above. A total of four rounds of biopanning were performed, after which the final eluted phage were plated to obtain single plaques. These plaques (up to 30) were used to prepare phage stocks, from which genomic DNAs were extracted. The sequence of the region of each genomic DNA encoding a putative spore-binding peptide was determined.
Fluorescent labeling of M13 phage.
Samples of M13 phage displaying a particular peptide were fluorescently labeled by using the Alexa Fluor 488 protein labeling kit (Molecular Probes). The labeling conditions were essentially those provided by Molecular Probes, except that 4 × 1012 phage particles were labeled instead of a 1-mg protein sample. The Alexa Fluor 488 dye contains a succinimidyl ester moiety that reacts with a limited number of exposed primary amines of phage coat proteins (primarily pVIII). Labeled phage were precipitated from the reaction mixture by adding 1/6 volume of polyethylene glycol (PEG)-NaCl (autoclaved 20% [wt/vol] PEG 8000, 2.5 M NaCl), mixing thoroughly, and allowing the sample to stand for 1 h at room temperature. The labeled phage were collected by centrifugation (10,600 × g) at 4°C for 15 min, and the supernatant was removed. The phage pellet was suspended in 1 ml of phosphate-buffered saline (PBS) (22), and the phage were precipitated again with PEG-NaCl for 20 min and collected as described above. The phage were suspended in 0.4 ml of PBS and stored in the dark at 4°C.
The phage concentration and the degree of labeling were determined by measuring the absorbance of a diluted labeled-phage sample at 494 nm (dye absorption maximum) and 280 nm and employing conversion factors equivalent to those provided by Molecular Probes: phage/ml = [A280 − (A494 × 0.11)] × dilution factor × (5 × 1012 phage/ml); and dye molecules/phage = A494 × dilution factor × (8.5 × 1015 molecules/ml)/(phage/ml).
Preparation of peptide-phycoerythrin conjugates.
Peptides were attached to R-phycoerythrin (Prozyme) by using the heterobifunctional cross-linker sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (Pierce) as previously described (13).
Standard assay for peptide binding to spores.
Spores (107) and either fluorescently labeled M13 phage (1011) or phycoerythrin (0.135-mg/ml final concentration) displaying a selected peptide were mixed in 20 μl of PBS and incubated with gentle shaking for 10 to 60 min at room temperature. The sample was then diluted with 180 μl of PBST (PBS-0.5% Tween 20) and centrifuged at 800 × g for 5 min at 4°C. The supernatant was removed, the pellet was suspended in 200 μl of PBST, and the sample was centrifuged again. The latter step was repeated twice. The supernatant was removed, and the pellet was suspended in 200 μl of PBST. This suspension was analyzed by fluorescence-activated cell sorting (FACS) using a Becton Dickinson FACSCalibur instrument. Spores were identified by their light-scattering properties, and 20,000 spores were analyzed for associated fluorescence.
Construction of recombinant M13 phage.
To construct recombinant M13 phage displaying a specified peptide, we prepared double-stranded replicative form (RF) genomic DNA from a random library phage as described in the Qiagen Plasmid Purification Handbook. The peptide-encoding KpnI-EagI fragment in the RF DNA of the library phage was excised and replaced with a chemically synthesized KpnI-EagI fragment encoding the specified peptide. The construct was verified by DNA sequence analysis. The recombinant M13 RF DNA was transformed into E. coli strain ER2537 to produce phage. As with the Ph.D.-7 phage, the recombinant phage contain the specified peptide fused to the amino terminus of coat protein pIII.
RESULTS
Identification of phage-displayed peptides that bind B. subtilis spores.
To identify 7-mer peptides that bind spores of B. subtilis, we biopanned a Ph.D.-7 phage display library (New England Biolabs, 2 × 109 independent clones) as described in Materials and Methods. Briefly, approximately 109 spores were mixed with 1011 Ph.D.-7 phage, spore-phage complexes were harvested by centrifugation, the complexes were washed 10 times, and phage were eluted from the complexes. The eluted phage pool (5 × 104 PFU) was amplified and used as input phage in the next round of biopanning exactly as described above. A total of four rounds of biopanning were performed. Titers of samples of input phage, initial supernatants, supernatants of selected washes, and eluted phage were determined. The results showed that after the first round of biopanning, 102 to 104 more phage were present in the washes and eluted phage samples, indicating that a large enrichment for spore-binding phage had occurred (data not shown).
We used a sample of the eluted phage from the fourth round of biopanning (6 × 108 PFU) to purify plaques of individual phage, 30 of which were used to prepare genomic DNA for sequence analysis of the peptide-encoding regions. The results showed that these phage genomes encoded 13 unique peptide sequences (Table 1). Phage displaying the same peptide also contained the same peptide-encoding genomic DNA sequence, indicating that they were probably siblings. All of the peptides contained the sequence Asn-His-Phe-Leu at the amino terminus. Although the sequences at positions 5 to 7 were not identical, there were clear preferences for amino acids. For example, 6 of 13 (46%) position 5 residues and 12 of 39 (31%) position 5-to-7 residues were Pro. In addition, 10 of 39 (26%) position 5-to-7 residues were basic amino acids, while no acidic amino acids were found at these positions.
TABLE 1.
Sequences of 7-mer peptides that bind B. subtilis spores
| Sequence (no. of phage)a | Residue at position in peptide:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| 1 (6) | Asn | His | Phe | Leu | Ile | Lys | Pro |
| 2 (3) | Asn | His | Phe | Leu | Arg | Ser | Pro |
| 3 (3) | Asn | His | Phe | Leu | Pro | Arg | Trp |
| 4 (8) | Asn | His | Phe | Leu | Pro | Lys | Val |
| 5 | Asn | His | Phe | Leu | Leu | Pro | Pro |
| 6 | Asn | His | Phe | Leu | Pro | Pro | Arg |
| 7 | Asn | His | Phe | Leu | Pro | Thr | Gly |
| 8 | Asn | His | Phe | Leu | Met | Pro | Lys |
| 9 | Asn | His | Phe | Leu | Lys | Gly | Thr |
| 10 | Asn | His | Phe | Leu | Pro | Gln | Asn |
| 11 | Asn | His | Phe | Leu | Leu | Trp | Arg |
| 12 (2) | Asn | His | Phe | Leu | Ile | Arg | Lys |
| 13 | Asn | His | Phe | Leu | Pro | Thr | Ala |
Numbers in parentheses indicate the number of phage with this sequence.
To confirm that this family of peptides bound to B. subtilis spores and did not arise because of preferential amplification of phage displaying these peptides, we examined phage enrichment without amplification. A phage mixture was prepared containing 99.9% phage from the Ph.D.-7 library and 0.1% phage displaying peptide 4 (Asn-His-Phe-Leu-Pro-Lys-Val; Table 1). A sample of this mixture containing 1010 total phage was mixed with 109 spores, and a single round of biopanning was performed. The eluted phage were plaque purified, and 10 plaques were used to determine the sequences of peptide-encoding genomic DNA. Seven of the 10 phage examined contained the sequence for peptide 4, indicating a 700-fold enrichment of the peptide 4 phage. This large enrichment was almost certainly due to binding of peptide 4 phage to spores.
We also biopanned a Ph.D.-12 phage display library (New England Biolabs, 2 × 109 independent clones) for 12-mer peptides that bind spores of B. subtilis as described for the Ph.D.-7 library. Ten plaques of round 4 eluted phage were used to determine peptide-encoding genomic sequences. Eight unique peptide sequences were found, and again, they contained the sequence Asn-His-Phe-Leu at the amino terminus (Table 2). Also, the amino acids found at positions 5 to 7 were similar to those of the 7-mer peptides, with at least one Pro residue found in this region in all sequences except sequence 1. The latter sequence had a Pro residue at position 8. One 12-mer sequence (peptide 4) included Glu at position 6 (between two Pro residues), the only acidic residue found in positions 5 to 7 of a spore-binding peptide. The sequences in positions 8 to 12 were not highly restricted, except for the prevalence of Pro residues (i.e., 11 of 40 residues). The failure to find the Asn-His-Phe-Leu sequence internally in the 12-mer (or 7-mer) peptides strongly indicated that this sequence must be present at the amino terminus of the peptide to permit spore binding.
TABLE 2.
Sequences of 12-mer peptides that bind B. subtilis spores
| Sequence (no. of phage)a | Residue at position in peptide:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| 1 (2) | Asn | His | Phe | Leu | Lys | Ser | Gln | Pro | Gly | Val | Val | Thr |
| 2 | Asn | His | Phe | Leu | Asn | Arg | Pro | Ala | Gln | Ser | Gln | Val |
| 3 | Asn | His | Phe | Leu | Pro | Pro | Lys | Met | Gly | Pro | Thr | Asp |
| 4 | Asn | His | Phe | Leu | Pro | Glu | Pro | Arg | Leu | Val | Met | Pro |
| 5 (2) | Asn | His | Phe | Leu | Ala | Pro | Gln | Pro | Pro | Val | Lys | Pro |
| 6 | Asn | His | Phe | Leu | Met | Pro | Asn | Pro | Leu | Leu | Ala | Met |
| 7 | Asn | His | Phe | Leu | Ile | Pro | Pro | Glu | Pro | Leu | Arg | Glu |
| 8 | Asn | His | Phe | Leu | Pro | Leu | Asn | Pro | Pro | Ala | Pro | Ser |
Numbers in parentheses indicate the number of phage with this sequence.
Peptide sequence requirements for spore binding.
To identify the tightest-binding 7-mer peptide or peptides listed in Table 1, we performed a competitive biopanning experiment. A phage pool containing equal amounts of each of the 13 phage displaying a unique 7-mer peptide sequence was prepared, and its approximate composition was confirmed by DNA sequence analysis of 33 plaques from the pool. A sample of the phage pool containing a total of 1.3 × 1012 phage was mixed with 108 B. subtilis spores, and this mixture was used in a first round of biopanning following the standard protocol in Materials and Methods. The eluted phage were amplified, and 1.3 × 1012 of these phage were mixed with 108 spores for a second round of biopanning. Two more rounds (3 and 4) of biopanning were performed following this procedure. Eluted phage from round 4 were plaque purified, and 31 plaques were used to analyze peptide-encoding genomic sequences for phage identification. Comparison of the frequencies of phage appearance in the original phage pool with those in the round 4 eluted phage showed no statistically significant differences for 10 of the unique phage (Table 3). Phage displaying peptides 5, 7, and 9 were not detected in the final eluted phage, although two or three copies of each were present in the 33 phage identified in the original pool. Statistically, this result may not be meaningful, and inspection of the missing peptide sequences did not reveal a common feature. Taken together, these results indicate that the spore binding properties of most (and perhaps all) of the 13 unique peptides are similar under the conditions examined here. In addition, repeated washing of complexes of spores and phage displaying the spore-binding peptides were highly resistant to repeated washing, indicating tight binding.
TABLE 3.
Composition of input and 4th round eluted phage pools for competitive biopanning with phage displaying 7-mer peptide ligands
| Sequencea | Position 5-to-7 sequence | No. of phage with indicated peptide sequence detected in:
|
|
|---|---|---|---|
| Input | 4th round eluate | ||
| 1 | Ile-Lys-Pro | 2 | 2 |
| 2 | Arg-Ser-Pro | 3 | 3 |
| 3 | Pro-Arg-Trp | 2 | 6 |
| 4 | Pro-Lys-Val | 5 | 8 |
| 5 | Leu-Pro-Pro | 2 | 0 |
| 6 | Pro-Pro-Arg | 1 | 2 |
| 7 | Pro-Thr-Gly | 3 | 0 |
| 8 | Met-Pro-Lys | 1 | 3 |
| 9 | Lys-Gly-Thr | 3 | 0 |
| 10 | Pro-Gln-Asn | 5 | 1 |
| 11 | Leu-Trp-Arg | 2 | 3 |
| 12 | Ile-Arg-Lys | 1 | 1 |
| 13 | Pro-Thr-Ala | 3 | 2 |
| Total | 33 | 31 | |
Sequence numbers are the same as in Table 1.
To determine whether shorter peptides can efficiently bind B. subtilis spores, we constructed two recombinant Ph.D.-7 phage that display either the tetrapeptide Asn-His-Phe-Leu or the pentapeptide Asn-His-Phe-Leu-Pro instead of a 7-mer sequence. (Note that the same sequence, Gly-Gly-Gly-Ser, follows the 4-, 5-, and 7-mer sequences.) We then performed two competitive biopanning experiments using a phage mixture containing 99.9% phage from the Ph.D.-7 library and 0.1% phage displaying either the 4- or 5-mer peptide. A sample of this mixture containing 1010 total phage was mixed with 108 spores for the first round of biopanning. Amplified eluted phage (1010) were mixed with 108 spores for two additional rounds of biopanning. Amplified eluted phage from each round were analyzed as a mixture (i.e., no plaque purification). Genomic DNA was extracted from the eluted phage mixtures (each round separately), and the sequences of the peptide-encoding regions were examined as an aggregate. In the sequencing ladder of each round, it was possible to identify and roughly quantitate (within the mixed sequences) phage displaying peptides of different lengths and also to identify dominant phage species.
In the case of the Ph.D-7 and 4-mer-phage mixture, the results showed that phage displaying the shorter peptide were undetectable in round 1 (i.e., only random 7-mer sequences were observed). In rounds 2 and 3, phage displaying 7-mer peptides with the sequence Asn-His-Phe-Leu-(Pro or Xxx)3 emerged as major species, with no indication of 4-mer phage. Thus, it appeared that phage displaying only the Asn-His-Phe-Leu sequence were relatively poor ligands. The results with the Ph.D.-7 and 5-mer-phage mixture were strikingly different. After round 1, eluted phage contained a mixture of phage displaying the 5-mer peptide and random 7-mer sequences. After rounds 2 and 3, phage displaying the 5-mer were the predominant or only phage species. Therefore, the Asn-His-Phe-Leu-Pro peptide appeared to bind spores as well as a 7-mer containing an Asn-His-Phe-Leu-(Pro or Xxx)3 sequence.
Species specificity of peptide binding.
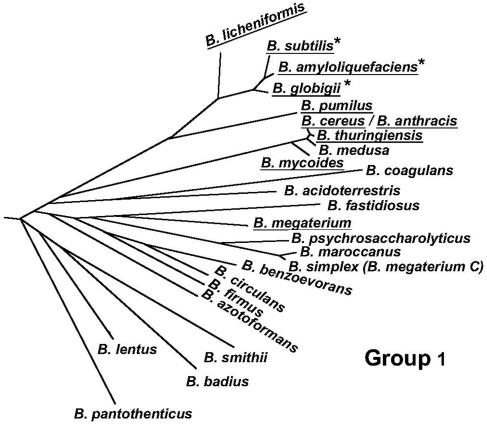
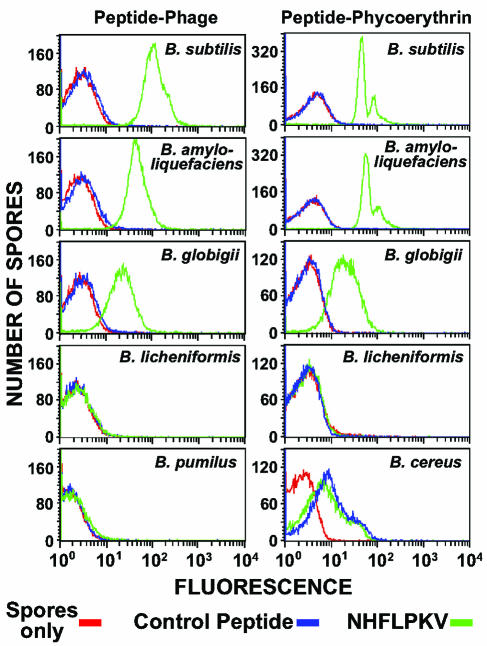
To determine the spore specificity of peptide binding, we first fluorescently labeled M13 phage displaying either 7-mer peptide 4 (Asn-His-Phe-Leu-Pro-Lys-Val) or a control peptide (i.e., Asp-Pro-Leu-Lys-Val-His-Glu). Both phage contained approximately 500 molecules of the fluorochrome Alexa 488 per phage particle. Under standard conditions for assaying peptide binding, spores of B. subtilis and nine other phylogenetically similar Bacillus species (Fig. 1) were individually mixed with fluorescently labeled phage for 10 min, unbound phage were removed by washing, and the spores were analyzed by FACS. This assay measures the increase in spore-associated fluorescence caused by ligand binding. The results show that peptide 4 binds well to spores of B. subtilis, nearly as well to spores of B. amyloliquefaciens, and somewhat weaker to spores of B. globigii (Fig. 2, left panels). No binding of peptide 4 was detected with spores of selected strains of B. licheniformis, B. pumilus, B. thuringiensis, B. cereus, B. anthracis, B. mycoides, and B. megaterium (Fig. 2, left side; only B. licheniformis and B. pumilus results are shown). No binding of the phage displaying the control peptide was detected with any spore species (Fig. 2, left side, and data not shown). In addition, phage displaying peptide 4 did not bind to vegetative cells of B. subtilis (data not shown). Binding of peptide 4 appears to be restricted to spores of species that occupy a single, three-member branch of the Bacillus phylogenetic tree (Fig. 1).
FIG. 1.
Phylogenic tree of group 1 (of five) members of the genus Bacillus based on 16S rRNA sequence analysis. Species of spores that were examined for binding by the Asn-His-Phe-Leu-Pro-Lys-Val peptide are underlined, and those that showed peptide binding are indicated by asterisks. The figure was reproduced with permission from Ash et al. (1) and modified to exclude B. lentimorbus, B. popilliae, and B. lautus, which were reclassified and assigned to the genus Paenibacillus (19; F. G. Priest, personal communication).
FIG. 2.
FACS analysis of peptide binding to Bacillus spores. (Left panels) Spores of the indicated species were mixed with fluorescently labeled M13 phage displaying either the Asn-His-Phe-Leu-Pro-Lys-Val peptide (NHFLPKV) or a peptide with the sequence Asp-Pro-Leu-Lys-Val-His-Glu (control peptide). (Right panels) Spores of the indicated species were mixed with peptide-phycoerythrin conjugates in which the peptide sequence was either Asn-His-Phe-Leu-Pro-Lys-Val (NHFLPKV) or Leu-Phe-Asn-Lys-His-Val-Pro (control peptide). In all analyses, a spores-only control was included.
To demonstrate that peptide binding was independent of the attached fluorochrome (i.e., Alexa-labeled M13), we prepared conjugates of peptide 4 or control peptides (e.g., Leu-Phe-Asn-Lys-His-Val-Pro) with phycoerythrin. Each peptide included a (Gly)3-Cys carboxy-terminal extension, and approximately 10 peptide molecules were attached through the carboxy-terminal Cys residue to the ɛ-amino groups of dispersed lysine residues on one molecule of phycoerythrin (240 kDa). Under standard assay conditions, spores of B. subtilis and seven other Bacillus species (i.e., all of the species described above except B. anthracis and B. mycoides) were individually mixed with a peptide-phycoerythrin conjugate for 60 min, unbound conjugate molecules were removed by washing, and the spores were analyzed by FACS. The results show that peptide 4 binds equally well to spores of B. subtilis and B. amyloliquefaciens and somewhat weaker to spores of B. globigii (Fig. 2, right panels). No binding of peptide 4 was detected with spores of B. licheniformis, B. pumilus, and B. megaterium (Fig. 2, right side; only B. licheniformis results are shown). No binding of the phage displaying the control peptide was detected with spores of B. subtilis, B. amyloliquefaciens, B. globigii, B. licheniformis, B. pumilus, or B. megaterium (Fig. 2, right side, and data not shown). Spores of B. thuringiensis and B. cereus showed slight and equal binding of the peptide 4 and control peptide conjugates, indicating nonspecific association (Fig. 2, right side; only B. cereus results are shown). This nonspecific binding appears to be related to the prominent exosporium present on spores of B. thuringiensis and B. cereus (10, 11). Selective removal of the exosporium from these spores by passage through a French press (27) essentially eliminates this nonspecific binding (data not shown).
A longer binding time (i.e., 60 min) was used with peptide-phycoerythrin conjugates to enhance labeling of spores. Approximately five times more peptide 4 conjugate bound at 60 min than at 10 min of incubation with B. subtilis, B. amyloliquefaciens, and B. globigii spores. The longer incubation time had no effect on the spores. The large and small peaks seen in the histograms of peptide 4 conjugate binding to B. subtilis and B. amyloliquefaciens spores (Fig. 2, right side) apparently represent single spores and pairs (or small aggregates) of spores, respectively, as judged by light-scattering properties analyzed by FACS. The selective spore binding shown in Fig. 2 was confirmed by fluorescence microscopy (data not shown). Finally, the peptide 4-phycoerythrin conjugate was shown to bind germinated spores of B. subtilis prior to outgrowth but not to vegetative cells of B. subtilis (data not shown).
Peptide binding affinity.
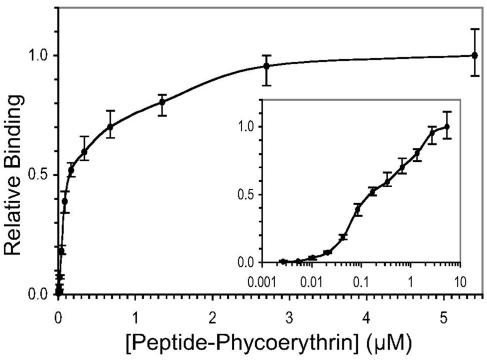
To quantitatively assess the binding of a typical Asn-His-Phe-Leu-Pro-containing peptide, we attempted to measure the Kd for binding of the peptide 4-phycoerythrin conjugate to spores of B. subtilis. Spores (106) were mixed with the conjugate at concentrations ranging from 0.660 μg/ml (2.64 nM) to 1.35 mg/ml (5.40 μM). The mixtures were incubated for 60 min at room temperature, and spore complexes were washed and analyzed by FACS. Conjugate binding was determined from the mean fluorescence associated with spores. (Note that spore aggregates were not detected in this experiment.) A plot of relative fluorescence versus conjugate concentration revealed a binding isotherm with multiple inflection points, which may indicate either binding to nonequivalent sites or positively cooperative binding (Fig. 3). Although the assignment of apparent Kd values for binding was not meaningful under these circumstances, the data indicated an overall half-maximal saturation point (S0.5) of 0.15 μM.
FIG. 3.
Binding affinity of the peptide 4-phycoerythrin conjugate to spores of B. subtilis. At the indicated concentrations, conjugate binding was measured as mean fluorescence per spore as determined by FACS. Each measurement was performed in triplicate, and the average of the three determinations was plotted as relative fluorescence versus the concentration of the conjugate. The error bars indicate the range of the determinations. The same data were plotted linearly and semilogarithmically (inset).
Spore binding of the peptide-phycoerythrin conjugate may have been influenced by the large size of the fluorochrome (i.e., 240 kDa) and the multivalency and structure of the ligand (i.e., approximately 10 peptides randomly attached to phycoerythrin). Therefore, we constructed and attempted to measure the binding affinity of a single peptide attached to a small fluorochrome (e.g., Alexa 488). However, high levels of nonspecific (i.e., independent of peptide sequence) binding or entrapment of this monovalent ligand by spores prevented accurate measurement of the binding affinity (data not shown).
Spore-binding peptides as a developmental targeting sequence.
Conceivably, the spore-binding peptide sequences identified in this study could be used to direct mother cell proteins to a receptor on the surface of the developing forespore. To test this hypothesis, we used the sequence Asn-His-Phe-Leu-Pro as a probe to search the sequenced B. subtilis genome for exact or close matches within known or possible spore surface proteins. The most interesting hits were a perfect match in the SpsC protein and close matches in the SpsA (Asn-His-Phe-Tyr-Pro) and CotA (Thr-His-Phe-Leu-Pro) proteins (mismatches underlined). The SpsC and SpsA proteins are reported to be spore coat proteins involved in polysaccharide biosynthesis (3, 21), and CotA is a pigmented outer spore coat protein with copper-dependent laccase activity (14, 16).
A problem with these matches, however, is that the putative spore binding sequence is not present at the amino terminus of the protein—a requirement indicated by our biopanning results. This is a particularly serious problem for SpsA and CotA, where the putative spore binding sequence is located near the carboxy terminus of the protein. In the case of SpsC, the Asn-His-Phe-Leu-Pro sequence is located near the amino terminus, at positions 6 to 10 (Fig. 4). Interestingly, this sequence is preceded by a basic amino acid (actually two, Lys-Arg), a possible cleavage site for a trypsin-like protease. If cleavage occurred between residues 5 and 6, perhaps as a timing event during spore formation, then presumably the truncated SpsC protein could bind the spore surface and participate in polysaccharide synthesis.
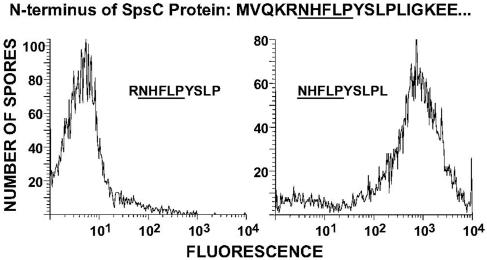
FIG. 4.
FACS analysis of the binding of peptides derived from the amino-terminal region of SpsC to B. subtilis spores. Spores were mixed with fluorescently labeled M13 phage displaying either residues 5 to 14 (left panel) or 6 to 15 (right panel) of SpsC.
To examine more directly the requirements for SpsC binding to the B. subtilis spore surface, we constructed three recombinant M13 phage displaying either SpsC residues 1 to 10, 5 to 14, or 6 to 15 on coat protein pIII. These phage were labeled equally with the fluorochrome Alexa 488 and used to measure spore binding by FACS under standard assay conditions. Phage displaying residues 1 to 10 and 5 to 14 failed to bind spores, while phage displaying residues 6 to 15 (with an amino-terminal Asn-His-Phe-Leu-Pro) readily bound spores (Fig. 4; only results for phage displaying residues 5 to 14 and 6 to 15 are shown). These results confirm that the Asn-His-Phe-Leu-Pro sequence must be present at the amino terminus of a peptide or protein for spore binding. The results also support the hypothesis that SpsC is proteolytically processed prior to participating in spore maturation.
DISCUSSION
From a phage display library, we identified a family of short peptides that bind tightly to spores of B. subtilis. These peptides contain the consensus sequence Asn-His-Phe-Leu-Pro. Using a representative peptide, we demonstrated that binding was restricted to spores of three Bacillus species. We observed nearly equal binding to spores of B. subtilis and its most closely related species, B. amyloliquefaciens, and slightly weaker binding to spores of the closely related species B. globigii. These three species comprise one branch on the Bacillus phylogenetic tree. The representative peptide did not bind to spores of several Bacillus species located on adjacent and nearby branches of the phylogenetic tree or to vegetative cells of B. subtilis. These results show that short peptides can be used as species-specific ligands and suggest that other short peptides can be isolated as specific ligands for different spore species and perhaps for any cell type. Theoretically, the binding site for such ligands can be a fortuitous or physiological receptor on the cell surface.
Our approach of reiteratively biopanning a phage display peptide library yielded a single consensus sequence, presumably the best binding peptide under the conditions employed. Possibly, a different peptide could be recovered by employing different biopanning conditions (e.g., buffer, number of washes, etc.), or by using a library displaying peptides in a different context (e.g., on the M13 major coat protein, pVIII) (24). For all of the peptides isolated in our study, the Asn-His-Phe-Leu sequence is located at the amino terminus, and this location was shown to be essential for spore binding. Typically, the spore-binding 7-mer peptides contain at least one Pro residue at positions 5 to 7. In addition, extension of the nonbinding Asn-His-Phe-Leu peptide by a single Pro residue (when displayed on pIII of phage M13) enables tight spore binding. These results indicate an important but somewhat flexible role for the Pro residue in spore binding. Because of its unique ability to limit polypeptide chain rotation, the Pro residue may stabilize or allow a peptide conformation that permits proper orientation of the four amino-terminal residues with respect to the spore surface receptor.
In the case of B. subtilis, the Asn-His-Phe-Leu-Pro peptide apparently binds to a receptor on the outer surface of the spore. This location for the receptor is based on the fact that molecules as large as the peptide-fluorochrome conjugates used in this study do not penetrate the outer coat of the spore (23). In addition, this receptor may have a physiological role in spore development. This possibility is indicated by the discovery that the spore surface protein SpsC contains the Asn-His-Phe-Leu-Pro sequence near its amino terminus, and this sequence is immediately preceded by a possible cleavage site for a trypsin-like protease. Indeed, trypsin-like proteolytic activity is apparently involved in the temporal processing and activation of mother cell proteins (e.g., the coat proteins CotF and CotT) during the latter stages of sporulation (2, 4, 28). Such a processing event would enable the SpsC protein, directed by its amino-terminal Asn-His-Phe-Leu-Pro sequence, to bind to its forespore receptor at the appropriate time during spore formation and maturation. Once bound to the forespore, the SpsC protein would participate in the synthesis of surface polysaccharides, which gives B. subtilis spores their hydrophilic character (15). At present, little is known about the specific interactions that direct the assembly of the outer layers of the spore. Perhaps, the use of targeting peptides will be shown to be a general mechanism in this process.
Acknowledgments
We thank Dakin Williams, Herb Cheung, and Tibor Bedekovics for assistance and advice.
This work was supported by DARPA grant MDA972-01-1-0030 and ARO grant DAAD19-00-1-0032.
REFERENCES
- 1.Ash, C., J. A. E. Farrow, S. Wallbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 13:202-206. [Google Scholar]
- 2.Bourne, N., P. C. FitzJames, and A. I. Aronson. 1991. Structural and germination defects of Bacillus subtilis spores with altered contents of a spore coat protein. J. Bacteriol. 173:6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charnock, S. J., and G. J. Davies. 1999. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38:6380-6385. [DOI] [PubMed] [Google Scholar]
- 4.Cutting, S., L. B. Zheng, and R. Losick. 1991. Gene encoding two alkali-soluble components of the spore coat from Bacillus subtilis. J. Bacteriol. 173:2915-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Driks, A. 2002. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol. 10:251-254. [DOI] [PubMed] [Google Scholar]
- 6.Driks, A. 2002. Proteins of the spore core and coat, p. 527-535. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. From genes to cells. ASM Press, Washington, D.C.
- 7.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster, S. J., and D. L. Popham. 2002. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. From genes to cells. ASM Press, Washington, D.C.
- 9.Gerhardt, P. 1967. Cytology of Bacillus anthracis. Fed. Proc. 26:1504-1517. [PubMed] [Google Scholar]
- 10.Gerhardt, P., H. S. Pankratz, and R. Scherrer. 1976. Fine structure of the Bacillus thuringiensis spore. Appl. Environ. Microbiol. 32:438-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerhardt, P., and E. Ribi. 1964. Ultrastructure of the exosporium enveloping spores of Bacillus cereus. J. Bacteriol. 88:1774-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 13.Hermanson, G. T. 1995. Bioconjugate techniques. Academic Press, San Diego, Calif.
- 14.Hullo, M.-F., I. Moszer, A. Danchin, and I. Martin-Verstraete. 2001. CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 183:5426-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshikawa, T., M. Yamazaki, M. Yoshimi, S. Ogawa, A. Yamada, K. Watabe, and M. Torii. 1989. Surface hydrophobicity of spores of Bacillus spp. J. Gen. Microbiol. 135:2717-2722. [DOI] [PubMed] [Google Scholar]
- 16.Martins, L. O., C. M. Soares, M. M. Pereira, M. Teixeira, T. Costa, G. H. Jones, and A. O. Henriques. 2002. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J. Biol. Chem. 277:18849-18859. [DOI] [PubMed] [Google Scholar]
- 17.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Ltd., West Sussex, United Kingdom.
- 19.Pettersson, B., K. E. Rippere, A. A. Yousten, and F. G. Priest. 1999. Transfer of Bacillus lentimorbus and Bacillus popilliae to the genus Paenibacillus with emended descriptions of Paenibacillus lentimorbus comb. nov. and Paenibacillus popilliae comb. nov. Int. J. Syst. Bacteriol. 49:531-540. [DOI] [PubMed] [Google Scholar]
- 20.Priest, F. G. 1993. Systematics and ecology of Bacillus, p. 3-16. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. Biochemistry, physiology, and molecular biology. American Society for Microbiology, Washington, D.C.
- 21.Roels, S., and R. Losick. 1995. Adjacent and divergently oriented operons under the control of the sporulation regulatory protein GerE in Bacillus subtilis. J. Bacteriol. 177:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Setlow, P. 1993. Spore structural proteins, p. 801-809. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. Biochemistry, physiology, and molecular biology. American Society for Microbiology, Washington, D.C.
- 24.Smith, G. P., and V. A. Petrenko. 1997. Phage display. Chem. Rev. 97:391-410. [DOI] [PubMed] [Google Scholar]
- 25.Sousa, J. C., M. T. Silva, and G. Balassa. 1976. An exosporium-like outer layer in Bacillus subtilis spores. Nature 263:53-54. [DOI] [PubMed] [Google Scholar]
- 26.Sousa, J. C., M. T. Silva, and G. Balassa. 1978. Ultrastructure and development of an exosporium-like outer spore envelope in Bacillus subtilis. Ann. Microbiol. (Paris) 129B:339-362. [PubMed] [Google Scholar]
- 27.Steichen, C., P. Chen, J. F. Kearney, and C. L. Turnbough, Jr. 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 185:1903-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takamatsu, H., A. Imamura, T. Kodama, K. Asai, N. Ogasawara, and K. Watabe. 2000. The yabG gene of Bacillus subtilis encodes a sporulation specific protease which is involved in the processing of several spore coat proteins. FEMS Microbiol. Lett. 192:33-38. [DOI] [PubMed] [Google Scholar]