Abstract
The aims of this study were to determine the power of discrimination of the real-time PCR assay for monitoring fluctuations in microbial populations within activated sludge and to identify sample processing points where methodological changes are needed to minimize the variability in target quantification. DNA was extracted using a commercially available kit from mixed liquor samples taken from the aeration tank of four bench-scale activated-sludge reactors operating at 2-, 5-, 10-, and 20-day solid retention times, with mixed-liquor volatile suspended solid (MLVSS) values ranging from 260 to 2,610 mg/liter. Real-time PCR assays for bacterial and Nitrospira 16S rRNA genes were chosen because they represent, respectively, a highly abundant and a less-abundant bacterial target subject to clustering within the activated sludge matrix. The mean coefficient of variation in DNA yields (measured as microgram of DNA per milligram of MLVSS) in triplicate extractions of 12 different samples was 12.2%. Based on power analyses, the variability associated with DNA extraction had a small impact on the overall variability of the real-time PCR assay. Instead, a larger variability was associated with the PCR assay. The less-abundant target (Nitrospira 16S rRNA gene) had more variability than the highly abundant target (bacterial 16S rRNA gene), and samples from the lower-biomass reactors had more variability than samples from the higher-biomass reactors. Power analysis of real-time PCR assays indicated that three to five samples were necessary to detect a twofold increase in bacterial 16S rRNA genes, whereas three to five samples were required to detect a fivefold increase in Nitrospira 16S rRNA genes.
The PCR represents a sensitive molecular detection method due to its ability to exponentially amplify a target gene. Although traditional PCR is not quantitative, real-time PCR modifications of the technique allow the rapid quantification of the amount of template present at the start of the amplification process (for recent reviews, see references 6, 8, and 17). The application of real-time PCR for the quantification of particular microbial populations in environmental samples, such as soil, water, sediments, or activated sludge, has increased recently (2, 5, 16, 17, 19, 20, 21, 23, 24, 35). Although this technique is powerful and robust, accurate quantification of specific microbial populations strongly depends on the quality and the yield of DNA extracted from the environmental sample and the inherent variability associated with the PCR amplification.
Bacteria are the major component of activated sludge and are responsible for the oxidation of organic matter and nutrient transformations, as well as the production of polysaccharides and other polymeric materials that aid in the flocculation of the microbial biomass (3). Although the activated sludge system is the most widely used biological process for the treatment of wastewaters, the achievement of reliable high-quality effluent is still elusive, as upsets in plant performance are frequent. Changes in the microbial population of the mixed liquor can lead to significant process problems, such as bulking, foaming, or failure in nitrification (25, 32). Yuan and Blackall (38) recently proposed optimization of the sludge population for control of biological wastewater treatment systems to ensure consistent long-term performance. Real-time PCR provides the opportunity to quantify such changes in individual populations within the sludge under different operational conditions or influent compositions. However, it is important to understand the limitations of real-time PCR assays, particularly the ability to discriminate changes in the microbial population, given the inherent variability in the real-time PCR assay and DNA extraction and recovery between samples.
DNA extraction from activated sludge has been performed for two decades for DNA probing, PCR amplification of specific targets, and the construction of clonal libraries. For these purposes, sample lysis needs to be complete without excessive shearing of the DNA, which can increase artifact formation during the PCR (31). The extracted nucleic acid must be free of contaminants that can inhibit enzymes used in subsequent molecular techniques, such as restriction endonucleases or Taq polymerase (1, 34). For quantitative studies of mixed-liquor population dynamics using real-time PCR, the DNA extraction method also needs to provide uniform DNA recovery and high throughput. Real-time PCR allows the simultaneous quantification of at least 24 samples in 1 to 2 h (in a 96-well format, considering triplicate measurements plus standards and controls). In this study, the performance of a commercially available DNA extraction kit with a rapid vertical angular motion, matrices, and chaotropic reagents to lyse the cells, followed by a GENECLEAN procedure for DNA purification, was tested in mixed-liquor samples and applied towards real-time PCR analysis.
The PCR aspects of the real-time PCR assay are also sources of variability in the quantification of a specific target in an environmental sample. PCR variability may be higher for a less-abundant target than a more-abundant target. Therefore, in this study the variability of a highly abundant biomass target (bacterial 16S rRNA gene) and a less-abundant target of bacteria subject to clustering within the activated sludge matrix (Nitrospira 16S rRNA gene [12, 26, 33]) was determined. All factors contributing to the final assay variability will influence the number of samples necessary to achieve a certain statistical power (18). Power analysis was performed using SamplePower 2.0 to determine the minimum number of samples required to differentiate between 2-, 5-, and 10-fold changes in microbial populations within activated sludge with low and high biomass (MLVSS).
MATERIALS AND METHODS
Samples.
Mixed-liquor samples were collected from the aeration basins of four 10-liter complete-mix bench-scale reactors operating at 2-, 5-, 10- and 20-day solids retention times (SRTs) (14). The bench-scale reactors were fed at a rate of 19 ml/min with primary clarifier effluent collected from a municipal wastewater treatment plant. The reactor dissolved oxygen concentration, during the time period mixed liquor samples were collected, was maintained at 2 mg/liter, and temperature was maintained at 20°C. Both ammonia oxidation and nitrite oxidation were 100% in the 20- and 10-day SRT reactors. Ammonia oxidation was 100% and nitrite oxidation was 97% in the 5-day reactor, and ammonia and nitrite oxidations were incomplete at 76 and 85% in the 2-day reactor (14). MLVSS values were determined following standard method 2540 E (11).
DNA extractions.
Total DNA was extracted in triplicate from mixed-liquor suspended solids using a FastDNA kit (Q-BIOgene, Carlsbad, Calif.) with minor modifications (13). The volume of mixed liquor used for each extraction was 1 ml for 10- and 20-day SRT samples, 2.5 ml for 5-day SRT samples, and 5 ml for 2-day SRT samples. These volumes corresponded to 1.2 to 2.6 mg of MLVSS per extraction for all analyzed samples. CLS-TC cell lysis-DNA solubilizing solution for bacteria, a 1/4-inch ceramic sphere, and the garnet matrix (Q-BIOgene) were used for cell lysis. The sample was homogenized using a 20-s pulse at a speed of 4.0 (FastPrep FP120; Q-BIOgene). After centrifugation to pellet debris, the lysate supernatant was combined with the binding matrix (600 μl) and gently mixed at room temperature for 5 min. SPIN modules (Q-BIOgene) were used for washing and eluting the DNA. Two washes of the binding matrix-DNA complex using 80% (vol/vol) ethanol were included after the recommended salt-ethanol wash step. For elution, the matrix-DNA complex was resuspended in 100 μl of 10 mM Tris-HCl (pH 8.0) buffer, incubated for 5 min at room temperature, and centrifuged for 1 min at 14,000 × g. The elution was repeated for a final volume of 200 μl.
Purity and yield of DNA.
The purity of the extracted DNA was assessed spectrophotometrically by calculating A260/A280 ratios in a Beckman DU640B spectrophotometer (Beckman Instruments Inc., Fullerton, Calif.). DNA was quantified using a PicoGreen double-stranded-DNA quantitation kit (Molecular Probes, Eugene, Ore.) with lambda DNA as the standard, following the manufacturer's instructions. DNA yields were calculated in micrograms of DNA extracted per liter of mixed liquor or per milligram of MLVSS.
Bacterial 16S ribosomal DNA and Nitrospira 16S ribosomal DNA real-time PCR.
Bacterial and Nitrospira 16S rRNA gene quantifications were performed in a total volume of 25 μl with Platinum Quantitative PCR SuperMix-UDG (Life Technologies, Inc., Gaithersburg, Md.) and 5 mM MgCl2 as previously described (21). The Nitrospira primers and probe have no mismatches to a range of Nitrospira 16S rRNA genes isolated from activated sludge including clones in the clade 2 group (7), clones A-4 and A-11 (26), and clones in the b2-b37 cluster (33). The Nitrospira 16S rRNA gene assay contained 15 pmol of primers NSR1113f (5′-CCTGCTTTCAGTTGCTACCG-3′) and NSR1264r (5′-GTTTGCAGCGCTTTGTACCG-3′), 6.25 pmol of TaqMan probe NSR1143fTaq [5′-(6-carboxyfluorescein)-AGCACTCTGAAAGGACTGCCCAGG-(carboxytetramethylrhodamine)-3′], and 0.32 to 1.54 ng of sample DNA or 30 to 3 × 107 copies of the standard (151-bp fragment of Nitrospira 16S rRNA gene; GenBank accession number AF420301). PCR amplification consisted of 2 min at 50°C, 10 min at 95°C, and 55 cycles at 95°C for 30 s and 63°C for 60 s. For the total bacterial 16S rRNA gene assay, the general bacterial primers and probes were derived from broad-specificity 16S rRNA gene primers (30). This assay contained 15 pmol of primers 1055f (5′-ATGGCTGTCGTCAGCT-3′) and 1392r (5′-ACGGGCGGTGTGTAC-3′), 6.25 pmol of TaqMan probe 16STaq1115 [5′-(6-carboxyfluorescein)-CAACGAGCGCAACCC-(carboxytetramethylrhodamine)-3′], and 0.32 to 1.54 ng of sample DNA or 4.5 × 103 to 4.5 × 108 copies of the same plasmid used for the Nitrospira 16S rRNA gene assay. The PCR program consisted of 2 min at 50°C, 10 min at 95°C, and 45 cycles at 95°C for 30 s, 50°C for 60 s, and 72°C for 20 s. All samples were measured three to six times during each assay, and negative controls without template were included in each PCR run. Amplification and detection were performed using the DNA Engine Opticon continuous fluorescence detection system (MJ Research, Waltham, Mass.) or the Bio-Rad iCycler with the iQ fluorescence detector and software version 2.3 (Bio-Rad, Hercules, Calif.). Gene copies per PCR for both assays were calculated using the same universal standard curve generated from 33 individual standard curves (gene copies = power {10, [(−0.24 × CT) + 10.7]}) (CT, threshold cycle). Copies of both targets per liter of mixed liquor and per microgram of DNA were calculated considering the volume of mixed liquor used for each DNA extraction, the volume of DNA added to each PCR, and the concentration of DNA.
Statistical analysis.
All statistical analyses were performed using SPSS version 11.0 and SamplePower 2.0 (SPSS Inc., Chicago, Ill.). Data were normally distributed, and a one-way analysis of variance (ANOVA) test was performed after log10 transformation of the data to meet the assumption of equal variance between groups. In ANOVA tests, the mean difference was significant at the 0.05 level. Multiple comparisons were performed with the Tukey honestly significant difference and Bonferroni tests. The correlation between variables was analyzed using the bivariate two-tailed Pearson correlation. SamplePower 2.0 was used for power analysis to determine the number of samples necessary in each of the two groups being compared. Power analyses were conducted using one-way ANOVA comparisons, a significance level of 0.05, and a power of 80%.
RESULTS
DNA extraction yields.
Twelve mixed-liquor samples from the aeration tank of four bench-scale activated-sludge reactors operating at 2-, 5-, 10-, and 20-day SRTs were extracted in triplicate to determine the variability in DNA yields associated with the DNA extraction. Due to differences in SRT, reactor MLVSS values varied widely: 355 ± 83 mg/liter in 2-day SRT samples, 627 ± 81 mg/liter in 5-day SRT samples, while 10- and 20-day SRT samples had 1,280 ± 89 and 2,127 ± 437 mg/liter, respectively. Therefore, to maintain similar amounts of biomass, between 1 and 5 ml of mixed liquor was used for each DNA extraction. The average A260/A280 ratio for all 36 DNA extractions was 1.93 ± 0.14, suggesting a high level of purity of the extracted nucleic acid.
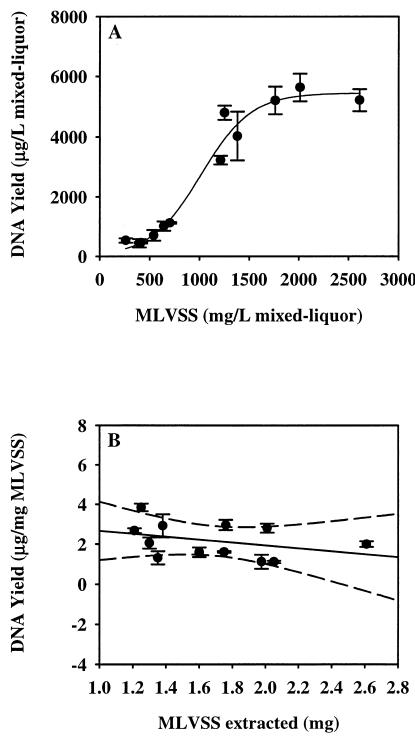
DNA yields (micrograms of DNA/liter of mixed liquor) and MLVSS values were related with an excellent fit to a sigmoid curve (Fig. 1A) (r2 = 0.973). Using mixed-liquor extraction volumes of 5 ml, samples from the 2-day SRT reactor yielded 2.39 ± 0.25 μg of DNA. A volume of 2.5 ml was used in each DNA extraction for 5-day SRT reactor samples, with a yield of 2.37 ± 0.54 μg of DNA. One milliliter of sample from 10- and 20-day SRT reactors yielded an average of 4.02 ± 0.78 and 5.35 ± 0.25 μg of DNA, respectively. No significant differences in micrograms of DNA per milligram of MLVSS were found in pairwise comparisons of 2- and 5-day reactor samples (low biomass) or 10- and 20-day SRT samples (high biomass) (one-way ANOVA, posthoc multiple comparisons; P > 0.05), although differences in DNA yield between the low- and high-biomass reactor samples were significant.
FIG. 1.
(A) Influence of MLVSS values on DNA yields (expressed in micrograms of DNA per liter [L] of mixed liquor). DNA yields from mixed-liquor samples obtained from bench scale reactors operating at different SRTs were fit to a three-parameter sigmoid curve (f = a/{1 + exp[−(X − X0)]/b}) (r2 = 0.973). (B) Influence of the amount of biosolids used in each extraction on DNA yields (expressed in micrograms of DNA per milligram of MLVSS) (y = 3.393 − 0.73x) (r2 = 0.126). The dashed lines represent the 99% confidence interval. Error bars indicate the standard deviation of DNA yields for each triplicate extraction.
Another potential source of variability in the DNA yield and reduction in the DNA recovery is saturation of the binding matrix when excess sample is used in the extraction. An insignificant correlation was found between DNA yields (micrograms of DNA/milligram of MLVSS) and the amount of biomass used in each extraction (between 1.2 and 2.6 mg of MLVSS) (Fig. 1B) (r = 0.354), indicating that saturation of the DNA binding matrix during the extraction protocol was not significant. These combined results show that DNA yield (micrograms per milligram of solids) was slightly higher for the 10- and 20-day SRT reactors (high biomass) at 2.66 ± 0.78 than for the 2- and 5-day SRT reactors (low biomass) at 1.46 ± 0.40, even though similar amounts of total biomass were extracted.
Quantification of bacterial and Nitrospira 16S rRNA genes.
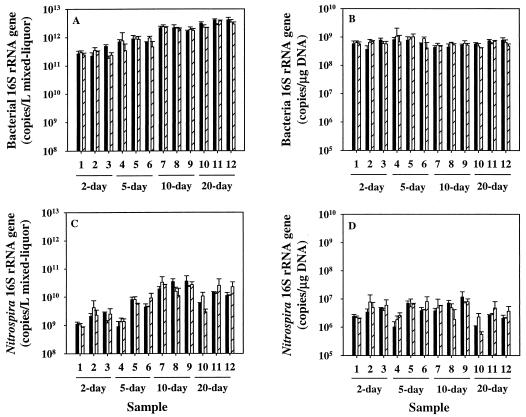
Real-time PCR assays were used to quantify bacterial and Nitrospira 16S rRNA genes in DNA extracts from mixed-liquor samples. Bacterial 16S rRNA genes were used as a measurement of total biomass, since a large fraction of the biosolids in mixed liquor are bacteria. As found with DNA yields, bacterial 16S rRNA genes per liter increased with longer retention times in the bench-scale reactors, from approximately 2.9 × 1011 to 3.2 × 1012 copies/liter of mixed liquor for 2- and 20-day reactor samples, respectively (Fig. 2A). One-way ANOVA indicated significant differences in pairwise comparisons of bacterial 16S rRNA genes per liter in all four reactors (P < 0.05). In addition, MLVSS values and bacterial 16S rRNA genes per liter of mixed liquor were significantly correlated (r = 0.951). When normalized to micrograms of DNA, the mean bacterial 16S rRNA gene copy number remained stable through all 36 extractions at 6.24 (±1.64) × 108 (Fig. 2B). No significant differences were observed between reactors in pairwise comparisons of bacterial 16S rRNA genes per microgram of DNA (P > 0.05), with the exception of samples from 5- and 10-day reactors.
FIG. 2.
Quantification of total bacterial 16S rRNA genes (A and B) and Nitrospira 16S rRNA genes (C and D) using real-time PCR in reactors operating at 2-, 5-, 10-, and 20-day SRTs. Separate extractions are indicated with black, white, and diagonal bars (Average ± standard deviation of three determinations). Targets are expressed in copies/liter (L) of mixed-liquor (A and C) or copies/microgram of DNA (B and D). The detection limits for the bacterial 16S rRNA and Nitrospira 16S rRNA genes were 3.6 × 109 copies/liter and 1.0 × 107 copies/liter, respectively, and 1.5 × 107 copies/μg of DNA and 4.2 × 104 copies/μg of DNA, respectively.
The amount of Nitrospira 16S rRNA genes per liter of mixed liquor ranged from 1.0 × 109 in a 2-day SRT sample to 3.6 × 1010 in a 10-day SRT sample (Fig. 2C). Fewer Nitrospira 16S rRNA gene copies would be expected in the 2-day SRT reactor because it had less total biomass (MLVSS) and lower nitrite oxidation efficiency (85%). In contrast, both the 10- and 20- day SRT reactors had higher biomass and 100% nitrite oxidation efficiency. Therefore, an approximately 10-fold-higher number of Nitrospira 16S rRNA genes per liter in the 10- and 20-day SRT reactors is consistent with better nitrification efficiency. Pairwise comparisons of copies per liter of this target showed three different homogeneous subsets, one containing 2- and 5-day SRT reactor samples, a second containing 5- and 20-day SRT reactor samples, and a third containing 10- and 20-day SRT samples.
When Nitrospira 16S rRNA genes were normalized to micrograms of DNA (Fig. 2D), no significant differences were found between the reactors (one-way ANOVA; F = 1.675), suggesting that differences in the number of Nitrospira 16S rRNA genes per liter between the reactors were biomass dependent. The correlation between biomass (MLVSS) and Nitrospira 16S rRNA genes per liter of mixed liquor (r = 0.525) was less than that for the total bacterial 16S rRNA genes (r = 0.951).
The percentage of Nitrospira cells in the total bacterial population was estimated assuming an average of 3.6 ribosomal operons per bacterial cell and 1 ribosomal operon copy per Nitrospira cell (21). Based on these assumptions, Nitrospira was estimated to be 2.5% ± 1.1% of the total bacterial population in the 2-day reactor, 2.4% ± 2.0% in the 5-day reactor, 4.3% ± 2.0% in the 10-day reactor, and 1.5% ± 0.77% in the 20-day reactor.
Variability and power analyses.
The variability in DNA extraction yields and in bacterial and Nitrospira 16S rRNA gene quantification assays were evaluated to identify the main sources of error for estimating bacterial and Nitrospira 16S rRNA genes in activated sludge. In real-time PCR assays, the threshold cycle (CT), or the PCR cycle at which the fluorescent signal is statistically significant above background, is measured (8). The CT value is compared to a standard curve generated with known target concentrations in order to estimate sample copy concentration. The variability of the real-time PCR assays was studied in both steps in the quantification of the target, CT values and copies of the target.
The coefficient of variation (CV) (standard deviation expressed as a percentage of the mean) of DNA yields (micrograms of DNA per milligram of MLVSS) in the 12 triplicate extractions was 12.2% (Table 1). CVs were calculated per sample (variability of average values per sample for each of the 12 samples) and per DNA extraction (CV of triplicate measurements of the real-time PCR assay in each of the 36 DNA extractions). The CVs of the CT values for bacterial and Nitrospira 16S rRNA genes were similar (Table 1). However, the CVs of the Nitrospira 16S rRNA genes per liter of mixed liquor were approximately 1.5 times higher than the corresponding CVs for bacterial 16S rRNA genes per liter of mixed liquor. These results suggest that the observed error is mainly attributable to the real-time PCR rather than differences in DNA extraction efficiencies.
TABLE 1.
Variability in quantification of bacterial and Nitrospira 16S rRNA gene targets in mixed-liquor samples by real-time PCR
| Data points | Avg CVa ± SD
|
||||||
|---|---|---|---|---|---|---|---|
| DNA yield (μg/mg)b | Bacteria (CT) | Nitrospira (CT) | Bacteria (copies/liter)c | Nitrospira (copies/liter)c | Bacteria (copies/μg)d | Nitrospira (copies/μg)d | |
| 12 mixed-liquor samplese | 12.2 ± 9.2 | 1.8 ± 1.4 | 2.1 ± 1.0 | 19.5 ± 15.9 | 36.0 ± 15.3 | 19.1 ± 9.3 | 34.6 ± 17.2 |
| 36 DNA extractionsf | —g | 1.9 ± 1.5 | 1.9 ± 1.4 | 21.9 ± 18.2 | 29.8 ± 21.7 | 22.0 ± 18.2 | 34.1 ± 29.4 |
CV is the standard deviation expressed as a percentage of the mean.
Micrograms of DNA per milligram of MLVSS.
Copies per liter of mixed-liquor.
Copies per microgram of DNA.
Coefficient of variation based on the mean from three extractions per sample in each of the 12 samples.
Coefficient of variation based on the mean from triplicate measurements of the real-time PCR assay in each of the 36 DNA extractions.
No coefficient of variability was determined because there was only one DNA yield per extraction.
Power analyses were performed to calculate the number of samples necessary to allow the detection of 2-, 5-, and 10-fold differences in bacterial and Nitrospira 16S rRNA genes per liter of mixed liquor (Table 2). Reactor data were first placed into two divisions characterized by biomass concentration: low biomass (2- and 5-day SRT reactors) and high biomass (10- and 20-day SRT reactors). Within these divisions, the variability contribution from DNA extractions was evaluated by performing power analyses using standard deviations obtained with ANOVA comparisons of data grouped by reactor. In the first analysis, mean quantification data for all three DNA extractions on each sample date were considered. In the second analysis, real-time PCR data within the first extraction of each sample date were considered. Similar power analysis predictions would then indicate that DNA extraction contributed little to the gene measurement variability in the activated sludge sample. With regard to the bacterial 16S rRNA gene, the minimum sample size needed to detect a twofold change was three per group in either the high- or low-biomass reactors when three extractions were considered. The minimum number of samples needed when only the first DNA extraction data were considered increased to five for the low-biomass reactors but remained at three for the high-biomass reactors. Five- or 10-fold differences in the 16S rRNA genes should be reliably detected with only two to three samples per group when using either single DNA extraction or triplicate DNA extractions regardless of the bioreactor biomass concentration.
TABLE 2.
Power analysis predictions for sample size required to detect 2-, 5-, and 10-fold differences in bacterial and Nitrospira 16S rRNA gene copies per litera
| Reactor typeb | Target | No. of ext.c | SD | No. of samples per group required for effect size
|
||
|---|---|---|---|---|---|---|
| 2-Fold | 5-Fold | 10-Fold | ||||
| High biomass | Bacteria | 3 | 0.0707 | 3 | 2 | 2 |
| Bacteria | 1 | 0.0707 | 3 | 2 | 2 | |
| Nitrospira | 3 | 0.1732 | 7 | 3 | 2 | |
| Nitrospira | 1 | 0.1789 | 7 | 3 | 2 | |
| Low biomass | Bacteria | 3 | 0.632 | 3 | 2 | 2 |
| Bacteria | 1 | 0.1342 | 5 | 3 | 2 | |
| Nitrospira | 3 | 0.3479 | 22 | 5 | 4 | |
| Nitrospira | 1 | 0.3478 | 22 | 5 | 4 | |
Copies of each target per liter of mixed liquor measured using real-time PCR assays.
Power analysis predictions were divided into two groups based on biomass (MLVSS) concentration. High- and low-biomass predictions utilize one-way ANOVA comparisons of 10- and 20-day SRT reactor data and 2- and 5-day SRT reactor data, respectively.
Number of DNA extractions used for power analysis. Three extraction-parameters results were obtained from one-way ANOVA for three extractions for three sample dates within each reactor grouping. One extraction-parameter results were obtained from one-way ANOVA of triplicate real time results for the first extraction only for each sample date within each reactor grouping. Power predictions were for the log10-transformed copies of the target per liter of mixed liquor and met the assumptions of homogeneity of variance between groups and within group normality. The significance level was 0.05, and the power was 80%.
Due to the higher level of variability observed in the Nitrospira 16S rRNA gene data set in the low-biomass reactors than in the high biomass reactors, considerably more samples were required to detect a twofold change in the low-biomass reactors than in the high-biomass reactors, at 22 and 7 samples, respectively (Table 2). This number decreased to five and four samples for detecting 5- and 10-fold changes in Nitrospira 16S rRNA genes in the low-biomass reactors with triplicate DNA extractions and three and two samples in the high-biomass reactors. As with the 16S rRNA gene data set, the number of samples needed with one DNA extraction to detect 2-, 5-, and 10- fold changes were similar to those found when triplicate DNA extractions were considered.
Variability associated with real-time PCR.
In order to confirm the variability in quantification of a low-copy-number target by real-time PCR, aliquots of a single DNA extract from the 10-day SRT reactor were spiked with plasmid DNA containing the Nitrospira 16S rRNA gene to produce samples with approximately 2-, 5-, and 10-times-higher concentrations of Nitrospira 16S rRNA genes. The unspiked and spiked DNA mixtures were assayed for Nitrospira 16S rRNA genes on five separate days. On each day, the samples were run in triplicate to compare variability within a PCR assay and between PCR assays performed on different days (Table 3). The mean coefficient of variation between PCR assays (37.5%) was twice as high as the coefficient of variation within a PCR assay (16.8%). Based on the copies per liter in the unspiked (1×) sample and the volumes of plasmid DNA added to the DNA extract, the increase in Nitrospira 16S rRNA gene copies relative to the unspiked sample were expected to be 2.4, 4.1, and 7.9 fold. The calculated fold increases compared to the 1× sample were 1.41 (±1.25), 3.92 (±2.19) and 6.84 (±4.53). The data in copies per liter were compared with a one-way ANOVA to determine which mixtures were significantly different (P < 0.05). In these analyses, significant P values were obtained in samples differing by 2.8 or more (1× versus 5×, 1× versus 10×, 2× versus 5×, and 2× versus 10×) but not in samples differing by 1.7 or less (1× versus 2× and 5× versus 10×). These results support the power analyses indicating that more than five samples are needed to resolve twofold differences by the Nitrospira 16S rRNA gene assay.
TABLE 3.
Nitrospira 16S rRNA genes per liter in a DNA sample spiked with a plasmid containing a Nitrospira 16S rRNA gene
| Assayb |
Nitrospira 16S rRNA genes per litera
|
|||
|---|---|---|---|---|
| 1 × Sample | 2 × Sample | 5 × Sample | 10 × Sample | |
| 1 | 5.2 × 109 (± 0.90 × 109) | 3.7 × 1010 (± 0.33 × 1010) | 9.2 × 1010 (± 1.0 × 1010) | 1.2 × 1011 (± 0.28 × 1011) |
| 2 | 3.1 × 1010 (± 0.72 × 1010) | 3.5 × 1010 (± 0.41 × 1010) | 9.2 × 1010 (± 1.5 × 1010) | 1.6 × 1011 (± 0.18 × 1011) |
| 3 | 2.2 × 1010 (± 0.54 × 1010) | 7.5 × 1010 (± 0.89 × 1010) | 1.6 × 1011 (± 0.45 × 1011) | 3.0 × 1011 (± 0.47 × 1011) |
| 4 | 4.5 × 1010 (± 0.94 × 1010) | 3.9 × 1010 (± 1.0 × 1010) | 1.6 × 1011 (± 0.12 × 1011) | 2.9 × 1011 (± 0.11 × 1011) |
| 5 | 4.1 (± 0.90 × 1010) | 3.1 × 1010 (± 0.53 × 1010) | 1.3 × 1011 (± 0.09 × 1011) | 1.7 × 1011 (± 0.94 × 1011) |
| Meanc | 3.6 × 1010 (± 0.38 × 1010) | 4.5 × 1011 (± 2.9 × 1011) | 1.4 × 1010 (± 0.46 × 1010) | 2.3 × 1011 (± 0.99 × 1011) |
Nitrospira 16S rRNA genes per liter; mean of three PCR values (± SD).
Nitrospira 16S rRNA gene assay calculated as copies/liter of MLSS.
Mean of Nitrospira 16S rRNA genes per liter of MLSS for five assays.
DISCUSSION
When quantifying targets using real-time PCR, both the variability attributable to DNA yields between extractions and the PCR assay itself need to be considered. Commercially available kits for the isolation of genomic DNA from mixed-liquor samples are desirable due to the high throughput and elimination of phenol, which requires special disposal protocols and can affect PCR. The main disadvantage of kits over traditional DNA extraction protocols is the higher cost. In this study, the average coefficient of variation in DNA yields between 12 samples from 4 different SRT reactors extracted in triplicate was relatively small at 12.2% (Table 1). The small differences in yield may result from differences in the amount of biosolids included in each extraction (due to the high level of heterogeneity of the mixed-liquor samples or differences produced by the sampling event) and differences in DNA recovery during the extraction.
Variations in cell lysis, DNA binding to the matrix, and DNA elution can contribute to differences in the DNA recovery. In order to improve the binding of the DNA to the matrix, the crude lysate was continuously mixed with the matrix during the recommended 5-min incubation, and the elution of the DNA was performed in two steps to increase the DNA recovery. In addition, since the silica matrix binding and elution properties vary with pH (low pH promotes DNA binding, high pH favors dissociation), 10 mM Tris-HCl (pH 8.0) was used for the elution step. This buffer also increases the stability of the extracted DNA and does not interfere in the PCR, compared with water or Tris-EDTA. DNA yields ranged from 443 to 5,633 μg/liter of mixed liquor and, as expected, increased with increasing MLVSS values (Fig. 1A). These yields were approximately 25-fold lower than those found by Bourrain et al. (4) using phenol-chloroform extraction protocols for extracting DNA from activated sludges and threefold lower than those reported by Yu and Mohn (37). However, the difference in the yield may be related to the method used for the quantification of the DNA, since absorbance at 260 nm can easily overestimate the DNA concentration due to its inability to distinguish between DNA and RNA, and the relative insensitivity of this assay compared to the PicoGreen assay.
Overall, DNA extraction had a smaller effect on total assay variability than PCR, as evidenced by the lower CVs in Table 1 and the similar number of samples predicted for three extractions versus one extraction by power analyses in Table 2. This suggests that the overall assay variability may be due to variability in the real-time PCR. The mean CV of the CT values for triplicate measurements of the real-time PCR assay was 1.9% for both bacteria and Nitrospira 16S rRNA genes (Table 1) and is consistent with 2% CVs reported for TaqMan assays with clinical as well as environmental samples (16, 22, 24). However, low CVs in CT values correspond to considerably higher CVs when linear CT values are converted to exponential copies of the target per liter of mixed liquor (Table 1).
The variability associated with the quantification of a target by real-time PCR is of particular concern when low-target concentrations relative to nontarget DNA (10), such as Nitrospira 16S rRNA genes, are considered. In this study, Nitrospira 16S rRNA genes were approximately 100-fold less abundant than total bacterial 16S rRNA genes. In addition, the concentrations of Nitrospira in reactors with different SRTs are of interest because Nitrospira is considered the primary organism responsible for nitrite oxidation in activated sludge. Reported percentages for Nitrospira by in situ hybridization in a sludge from an industrial plant connected to a rendering factory were 9 and 12% (26, 27), 2.5% for a combined activated sludge-rotating biological contactor process (36), and less than 5% for another rotating biological contactor (15). For comparison purposes, the percentages of Nitrospira for these current reactors under study were calculated assuming 1 ribosomal operon copy per Nitrospira cell and 3.6 ribosomal operon copies per general bacterial cell. Based on these assumptions, the percentages of Nitrospira ranged from 1.5 to 4.3% and were lower than those found in a municipal wastewater treatment plant (8.6%) using the same Nitrospira assay (21). However, these percentages can only be considered an estimate, because the number of ribosomal operons in bacteria can range from 1 to 15 (29). In addition, it is hypothesized that bacteria with short doubling times have more ribosomal operons per cell than bacteria with longer doubling times (28). Therefore, bacteria in the 2-day SRT reactor may have a larger number of ribosomal operons per cell than those in the 20-day SRT reactor. Although the calculated percentage of Nitrospira was similar for all four reactors, the total numbers of Nitrospira cells (assuming 1 ribosomal operon copy per cell) were 6- to 10-fold higher in the two reactors (10- and 20-day) with complete nitrification than in the 2-day SRT reactor with incomplete nitrification due to the higher biomass in the 10- and 20-day SRT bioreactors.
The overall variability in this set of extractions was used in power analyses to estimate the number of samples necessary to find statistically significant changes in the microbial population. Essentially, the larger the group sample size, the greater the power of the statistical test and the more sensitive the experiment would be in detecting differences in microbial populations in the activated sludge under different operational conditions (18). Several power analyses were performed to determine the contributions of DNA extraction, PCR assay, and differences in starting biomass to the number of samples needed to detect 2-, 5-, and 10-fold changes (Table 2). With regard to the power analyses, two observations can be made. First, a 2-, 5-, or 10-fold change should be detected by the 16S rRNA gene assay, representing the high target gene, using a sample size of three to five size regardless of the biomass concentration and number of sample extractions performed. In contrast, the number of samples needed to reliably detect a twofold change with the Nitrospira 16S rRNA gene assay, representing the low target gene, was higher and differed between the high-biomass (7 samples) and low-biomass (22 samples) data (Table 2). However, a 5- or a 10-fold difference would be resolvable by the Nitrospira 16S rRNA gene assay using three samples for the high-biomass data or five samples for the low-biomass data. This suggests that for assays with high-variability and low-abundance genes, either more samples need to be analyzed or larger changes need to occur in the population in order to reliably interpret twofold or lower variation in the microbial population.
Second, only a slight increase in the number of samples per group was required to obtain 80% power when one versus three DNA extractions per sample were considered (Table 2). Therefore, increasing the number of real-time PCR assays performed with a single DNA extract may have as large an effect on statistical power as increasing the number of DNA extractions and real-time PCR assays performed with each activated sludge sample. The interassay variability attributable to the PCR assays was further demonstrated by comparing variability within a single PCR run and between PCR runs using a single DNA extract spiked with Nitrospira 16S rRNA genes. In this experiment, the CVs within a PCR run were approximately one-half those between PCR runs, 16.8% versus 37.5%. Five separate PCR runs were sufficient to detect statistically significant differences in the samples differing in Nitrospira 16S rRNA genes by about fourfold and sevenfold but not less than twofold.
The development of real-time PCR assays for critical microbial populations found in wastewater treatment systems may increase our understanding of microbial population dynamics in activated sludge and potentially influence the design and operation of wastewater treatment plants in the future. However, it is important to recognize the limitations of any new technique, particularly when applied to environmental samples. In order to increase the statistical power, for low-template, high-background DNA situations, it is necessary to increase the sample size or to reduce the variability of the real-time PCR assay. The variability in CT values and thus copies per PCR may be reduced by continuing improvements in the design and optimization of the real-time PCR assays, improvements in commercial reagents, automatization of both the DNA extraction and the dispensing of reagents (9), and the use of internal controls to account for differences in lots of probes, master mixes, and sets of standards. Such improvements may ultimately reduce the number of samples needed to resolve small differences (<2-fold) in gene populations within complex mixtures.
Acknowledgments
This work was funded by a research grant from the Water Environment Research Foundation (WERF Project 98-CTS-2) and in part by the Waste Management Research and Education Institute at the University of Tennessee. H.M.D. is a recipient of a postdoctoral fellowship from CONICET.
We thank the Knoxville Utility Board for access to the wastewater treatment plant and Mike Newman from the statistical consulting center, University of Tennessee, for his advice on statistical analysis of the data.
REFERENCES
- 1.Al-Soud, W. A., and P. Rådström. 1998. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl. Environ. Microbiol. 64:3748-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bach, H.-J., J. Tomanova, M. Schloter, and J. C. Munch. 2002. Enumeration of total bacteria and bacteria with genes for proteolytic activity in pure cultures and in environmental samples by quantitative PCR mediated amplification. J. Microbiol. Methods. 49:235-245. [DOI] [PubMed] [Google Scholar]
- 3.Bitton, G. 1999. Wastewater microbiology, 2nd ed. Wiley-Liss, New York, N. Y.
- 4.Bourrain, M., W. Achouak, V. Urbain, and T. Heulin. 1999. DNA extraction from activated sludges. Curr. Microbiol. 38:315-319. [DOI] [PubMed] [Google Scholar]
- 5.Bowers, H. A., T. Tengs, H. B. Glasgow, Jr., J. M. Burkholder, P. A. Rublee, and D. W. Oldach. 2000. Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl. Environ. Microbiol. 66:4641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunk, C. F., J. Li, and E. Avaniss-Aghajani. 2002. Analysis of specific bacteria from environmental samples using a quantitative polymerase chain reaction. Curr. Issues Mol. Biol. 4:13-18. [PubMed] [Google Scholar]
- 7.Burrell, P. C., J. Keller, and L. L. Blackall. 1998. Microbiology of a nitrite-oxidizing bioreactor. Appl. Environ. Microbiol. 64:1878-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 9.Bustin, S. A. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29:23-39. [DOI] [PubMed] [Google Scholar]
- 10.Chandler, D. P. 1998. Redefining relativity: quantitative PCR at low template concentrations for industrial and environmental microbiology. J. Ind. Microbiol. Biotechnol. 21:128-140. [Google Scholar]
- 11.Clesceri, L., A. Eaton, and A. Greenberg. 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Port City Press, Baltimore, Md.
- 12.Diams, H. M., J. L. Nielsen, P. H. Nielsen, K.-H. Schleifer, and M. Wagner. 2001. In situ charaterizatoin of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dionisi, H. M., A. C. Layton, G. Harms, I. R. Gregory, K. G. Robinson, and G. S. Sayler. 2002. Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants using competitive PCR. Appl. Environ. Microbiol. 68:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dionisi, H. M., A. C. Layton, K. G. Robinson, J. R. Brown, I. R. Gregory, J. J. Parker, and G. S. Sayler. 2002. Quantification of Nitrosomonas oligotropha and Nitrospira spp. using competitive PCR in bench scale wastewater treatment reactors operating at different solids retention times. Water Environ. Res. 74:462-469. [DOI] [PubMed] [Google Scholar]
- 15.Egli, K., F. Bosshard, C. Werlen, P. Lais, H. Siegrist, A. J. B. Zehnder, and J. R. van der Meer. 2003. Microbial composition and structure of a rotating biological contactor biofilm treating ammonium-rich wastewater without organic carbon. Microb. Ecol. 45:419-432. [DOI] [PubMed] [Google Scholar]
- 16.Gerard, C. J., K. Olsson, R. Ramanathan, C. Reading, and E. G. Hanania. 1998. Improved quantitation of minimal residual disease in multiple myeloma using real-time polymerase chain reaction and plasmid-DNA complementarity determining reaction III standards. Cancer Res. 58:3957-3964. [PubMed] [Google Scholar]
- 17.Ginzinger, D. G. 2002. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp. Hematol. 30:503-512. [DOI] [PubMed] [Google Scholar]
- 18.Goldsmith, L. J. 2002. Power and sample size considerations in molecular biology. Methods Mol. Biol. 184:111-130. [DOI] [PubMed] [Google Scholar]
- 19.Grüntzig, V., S. C. Nold, J. Zhou, and J. M. Tiedje. 2001. Pseudomonas stutzeri nitrite reductase gene abundance in environmental samples measured by real-time PCR. Appl. Environ. Microbiol. 67:760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall, S. J., P. Hugenholtz, N. Siyambalapitiya, J. Keller, and L. L. Blackall. 2002. The development and use of real-time PCR for the quantification of nitrifiers in activated sludge. Water Sci. Technol. 46:267-272. [PubMed] [Google Scholar]
- 21.Harms, G., A. C. Layton, H. M. Dionisi, I. R. Gregory, V. M. Garrett, S. A. Hawkins, K. G. Robinson, and G. S. Sayler. 2003. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37:343-351. [DOI] [PubMed] [Google Scholar]
- 22.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 23.Hermansson, A., and P.-E. Lindgren. 2001. Quantification of ammonia-oxidizing bacteria in arable soil by real-time PCR. Appl. Environ. Microbiol. 67:972-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hristova, K. R., C. M. Lutenegger, and K. M. Scow. 2001. Detection and quantification of methyl tert-butyl ether-degrading strain PM1 by real-time TaqMan PCR. Appl. Environ. Microbiol. 67:5154-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins, D., M. G. Richard, and G. T. Daigger. 1993. Manual on the causes and control of activated sludge bulking and foaming, 2nd ed. Lewis Publishers, Chelsea, Mich.
- 26.Juretschko, S., G. Timmermann, M. Schmid, K.-H. Schleifer, A. Pommerening-Röser, H.-P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 28.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrnbd: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., New York, N.Y.
- 31.Ogram, A. 1998. Isolation of nucleic acids from environmental samples, p. 273-288. In R. S. Burlage, R. Atlas, D. Stahl, G. Geesey, and G. Sayler (ed.), Techniques in microbial ecology. Oxford University Press, New York, N.Y.
- 32.Rittmann, B. E., and P. L. McCarty. 2001. Environmental biotechnology: principles and applications. McGraw Hill, New York, N.Y.
- 33.Schramm, A., D. De Beer, J. C. Van Den Heuvel, S. Ottengraf, and R. Amann. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl. Environ. Microbiol. 65:3690-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steffan, R. J., J. Goksøyr, A. K. Bej, and R. M. Atlas. 1988. Recovery of DNA from soils and sediments. Appl. Environ. Microbiol. 54:2908-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stults, J. R., O. Snoeyenbos-West, B. Methe, D. R. Lovley, and D. P. Chandler. 2001. Application of the 5′ fluorogenic exonuclease assay (TaqMan) for quantitative ribosomal DNA and rRNA analysis in sediments. Appl. Environ. Microbiol. 67:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You, S. J., S. H. Chuang, and C. F. Ouyang. 2003. Nitrification efficiency and nitrifying bacteria abundance in combined AS-RBC and A2O systems. Water Res. 37:2281-2290. [DOI] [PubMed] [Google Scholar]
- 37.Yu, Z., and W. W. Mohn. 1999. Killing two birds with one stone: simultaneous extraction of DNA and RNA from activated sludge biomass. Can. J. Microbiol. 45:269-272. [Google Scholar]
- 38.Yuan, Z., and L. L. Blackall. 2002. Sludge population optimisation: a new dimension for the control of biological wastewater treatment systems. Water Res. 36:482-490. [DOI] [PubMed] [Google Scholar]