Abstract
During aflatoxin biosynthesis, 5′-hydroxyaverantin (HAVN) is converted to averufin (AVR). Although we had previously suggested that this occurs in one enzymatic step, we demonstrate here that this conversion is composed of two enzymatic steps by showing that the two enzyme activities in the cytosol fraction of Aspergillus parasiticus were clearly separated by Mono Q column chromatography. An enzyme, HAVN dehydrogenase, catalyzes the first reaction from HAVN to a novel intermediate, another new enzyme catalyzes the next reaction from the intermediate to AVR, and the intermediate is a novel substance, 5′-oxoaverantin (OAVN), which was determined by physicochemical methods. We also purified both of the enzymes, HAVN dehydrogenase and OAVN cyclase, from the cytosol fraction of A. parasiticus by using ammonium sulfate fractionation and successive chromatographic steps. The HAVN dehydrogenase is a homodimer composed of 28-kDa subunits, and it requires NAD, but not NADP, as a cofactor for its activity. Matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis of tryptic peptides of the purified HAVN dehydrogenase revealed that this enzyme coincides with a protein deduced from the adhA gene in the aflatoxin gene cluster of A. parasiticus. Also, the OAVN cyclase enzyme is a homodimer composed of 79-kDa subunits which does not require any cofactor for its activity. Further characterizations of both enzymes were performed.
Aflatoxins comprise a group of polyketide-derived secondary metabolites mainly produced by certain strains of Aspergillus flavus and A. parasiticus. They are toxic, mutagenic, and carcinogenic to a variety of animal species and humans; thus, their contamination of agricultural commodities has serious deleterious effects on the health of animals and humans (7, 22). Aflatoxin contamination occurs in several crops such as tree-born nuts, peanuts, corn, and cotton. The aflatoxin-producing fungi infect preharvest and postharvest crops (17, 22). Various approaches have been undertaken to reduce or eliminate contamination of food and feed by aflatoxins (18, 20-22, 25).
The pathway for aflatoxin biosynthesis has been extensively studied in an attempt to better understand the molecular regulation of aflatoxin biosynthesis. At least 18 enzymatic reactions are involved in aflatoxin biosynthesis, which is initiated by hexanoate synthesis from acetate (6, 19, 29). Most of these enzymes have been characterized, and their genes have been cloned (reviewed in references 2, 3, 6, 14, 19, 26, 28, and 29). Most of the genes involved in aflatoxin biosynthesis are clustered within a 75-kb DNA fragment in the genomes of A. parasiticus and A. flavus. Furthermore, sterigmatocystin is a late intermediate in aflatoxin biosynthesis and is the final metabolite in the cognate polyketide pathway in A. nidulans. The genes and enzymes for sterigmatocystin production in this species are homologs of the genes and enzymes required for aflatoxin production in A. flavus and A. parasiticus (5, 6, 13, 14).
In the early steps of aflatoxin biosynthesis, norsolorinic acid (NA) is converted to averantin (AVN) by a reductase encoded by nor-1 (27). A monooxygenase encoded by avnA catalyzes the conversion of AVN to 5′-hydroxyaverantin (HAVN) (33). In sterigmatocystin biosynthesis in A. nidulans, the stcF gene is a homolog of the avnA gene (5, 13).
We isolated HAVN from mycelia of Emericella heterothallica IFO 30842 and confirmed that HAVN is a precursor of aflatoxins (31). HAVN was converted to averufin (AVR) in the presence of NAD or NADP in a cell-free system using cell extracts of A. parasiticus, and this reaction preferred NAD to NADP as a coenzyme (31). Consideration of the stereochemistry of HAVN and AVR supported the notion that the 5′-carbon atom of HAVN might be oxidized to a ketone by HAVN dehydrogenase in the presence of NAD, resulting in loss of chirality on the 5′-carbon atom (30). The resultant 5′-oxoaverantin (OAVN) molecule was further assumed to change to (1′S, 5′S) AVR through spontaneous intramolecular acetal formation among the 5′ ketone and two hydroxyl groups, (1′S)-OH and 3-OH (30). [Note that although the absolute configurations of bioactive and inactive AVR were denoted (1′S, 5′R) and (1′R, 5′S), respectively, in a previous paper (30), we have herein corrected them to (1′S, 5′S) and (1′R, 5′R), respectively.] On the other hand, Chang et al. recently reported that disruption of the adhA gene, which is present in the aflatoxin gene cluster of A. parasiticus, caused accumulation of HAVN, indicating that this gene might encode HAVN dehydrogenase (8).
We recently attempted to purify the HAVN dehydrogenase enzyme in order to understand the mechanism of the reaction from HAVN to AVR. We monitored AVR production from HAVN as the HAVN dehydrogenase enzyme activity during purification. Although we attempted complete purification several times, we repeatedly found that the enzyme activity was suddenly and drastically decreased after several purification steps. We thus suspected that more than one enzyme might be involved in the pathway from HAVN to AVR. The goal of this work was to investigate the pathway from HAVN to AVR in detail. For this purpose, we isolated and characterized a new intermediate between HAVN and AVR. Finally, we demonstrated that two enzymes are involved in the pathway from HAVN to AVR by purifying them.
MATERIALS AND METHODS
Microorganisms and growth conditions.
A. parasiticus NIAH-26, a UV-irradiated mutant of A. parasiticus SYS-4 (NRRL-2999) (32), was used as a source of the enzyme. In YES medium (2% yeast extract, 20% sucrose), the fungus produces all the enzymes in the aflatoxin biosynthetic pathway from NA to aflatoxins, although it does not produce aflatoxins, anthraquinone, or xanthone precursors (30, 31).
Preparation of the cytosol fraction.
The cytosol fraction was prepared from the mycelia of A. parasiticus NIAH-26 as previously described (31).
Preparation of HAVN and AVR.
HAVN was isolated from mycelia of E. heterothallica IFO 30842 (31). AVR was prepared from A. versicolor (Vuillemin) (31). The concentrations of the metabolites were determined from UV absorption spectra by using the following absorption coefficients (λmax): for HAVN, 7,100 M−1 cm−1 (466 nm), and for AVR, 10,500 M−1 cm−1 (454 nm).
Preparation and characterization of OAVN.
The crude HAVN dehydrogenase was purified from A. parasiticus NIAH-26 cytosol by ammonium sulfate fractionation and DE 52 column chromatography (see Table 2). To produce OAVN, HAVN (36 mg) was incubated with the crude HAVN dehydrogenase (0.14 mg of protein ml−1) in 375 ml of reaction mixture containing 60 mM Tris-HCl buffer, pH 8.0, and 1.6 mM NAD at 30°C for 120 min. After incubation, the OAVN formed was extracted with ethyl acetate and purified by preparative silica gel thin-layer chromatography (TLC) (silica gel 60 HF254; Merck) with acetone-n-hexane (2:3 [vol/vol]) and Sephadex LH-20 (Amersham Bioscience, Uppsala, Sweden) column chromatography with methanol (MeOH). Eighteen milligrams of OAVN was obtained.
TABLE 2.
Purification of HAVN dehydrogenase
| Purification step | Vol (ml) | Total protein (mg) | Total activity (nmol/min) | Sp act (nmol/mg/min) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Cytosol | 345 | 2,726 | 87 | 0.032 | 1.0 | 100 |
| (NH4)2SO4 (20-55%) | 104 | 1,248 | 42 | 0.034 | 1.1 | 48 |
| DE 52 | 205 | 245 | 62 | 0.25 | 8 | 71 |
| Butyl Toyopearl | 35 | 19 | 11 | 0.57 | 18 | 13 |
| Hydroxyapatite | 40 | 16 | 8.4 | 0.51 | 16 | 10 |
| First Superdex | 11 | 2.1 | 2.2 | 1.06 | 33 | 2.5 |
| First Mono Q | 4.5 | 1.95 | 1.8 | 0.92 | 29 | 2.1 |
| Second Superdex | 1.7 | 0.31 | 0.67 | 2.15 | 67 | 0.8 |
| Resource Q | 3.5 | 0.15 | 0.50 | 3.31 | 104 | 0.6 |
| Second Mono Q | 2.4 | 0.01 | 0.16 | 22.2 | 694 | 0.2 |
The UV spectrum of OAVN was recorded on a Shimadzu UV-2200 UV-VIS recording spectrometer or a Hitachi U-2001 spectrometer, and the infrared (IR) spectrum was recorded on a JASCO FT/IR 7000 spectrometer. Optical rotation was measured with a Horiba SEPA-200 highly sensitive polarimeter. Electrospray ionization mass spectrometry (ESI-MS) was performed with a Thermo Quest LCQ LC/MS system. Nuclear magnetic resonance (NMR) spectra were measured in acetone-d6 with a JEOL JNM-ECP 500 NMR spectrometer or a JEOL Lambda 400 NMR spectrometer. Chemical shifts were referenced to acetone-d6 (δH 2.04, δC 29.8). Solid-state NMR spectra were measured with a JEOL JNM-ECP 300 NMR spectrometer. Chemical shifts were referenced to hexamethyl benzene (δC 17.4). For enzyme assays, the concentration of OAVN was determined by using the molar absorption coefficient, 8,500 M−1 cm−1 (457 nm) (Table 1).
TABLE 1.
Physicochemical properties of OAVN and HAVN
| Property | Value or description |
|---|---|
| OAVN | |
| Form | Orange powder |
| UV (EtOH) λmax (nmɛ) | 221 (27,900), 262 (16,400), 296 (20,900), 314 (22,700), 457 (8,500) |
| IR (KBr) νmax cm−1 | 3,380, 2,930, 1,702, 1,671, 1,622, 1,580, 1,473 |
| [α]D2,4 | +170° (c 0.1 MeOH) |
| ESI MS m/z | 385.0 [M − H]− |
| 1H-NMR δ (acetone-d6) | 1.66-1.87 (4H, m), 2.08 (3H, s), 2.53 (2H, t, J = 7.0 Hz), 5.40 (1H, dd, J = 8.3, 3.9 Hz), 6.62 (1H, d, J = 2.5 Hz), 7.11 (1H, s), 7.22 (1H, d, J = 2.5 Hz), 12.21 (1H, bs), 12.76 (1H, bs) |
| 13C-NMR δ (acetone-d6) | 20.3, 29.0, 36.4, 43.3, 69.8, 109.0, 109.5, 109.6, 110.2, 110.5, 122.0, 134.6, 136.4, 161.7, 165.3, 166.0, 166.0, 181.9, 190.7, 207.9 |
| HAVN | |
| 1H-NMR δ (acetone-d6)a | 1.07 (3H, d, J = 5.9 Hz), 1.20-1.90 (6H, m), 3.69 (1H, tq, J = 5.9, 5.9 Hz), 5.38 (1H, dd, J = 7.8, 4.9 Hz), 6.59 (1H, d, J = 2.7 Hz), 7.08 (1H, s), 7.18 (1H, d, J = 2.7 Hz), 12.16 (1H, s), 12.71 (1H, s) |
| 13C-NMR δ (acetone-d6) | 22.4, 24.1, 37.2, 39.9, 67.5, 70.1, 109.0, 109.4, 109.7, 110.2, 110.7, 122.4, 134.6, 136.5, 161.8, 165.6, 166.1, 166.3, 182.0, 190.6 |
Data are from reference 31.
Mono Q column chromatography of cytosol fraction.
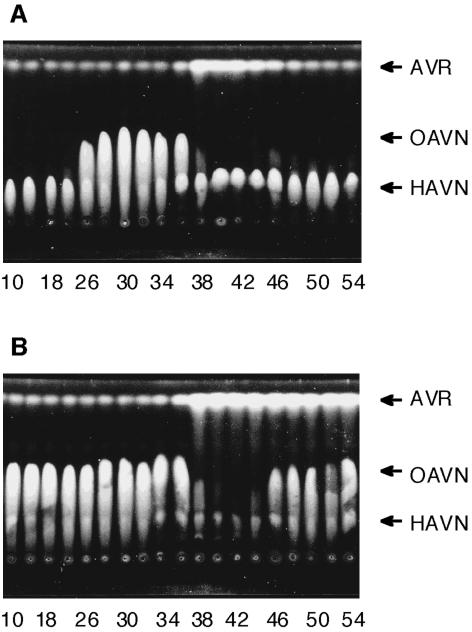
The cytosol fraction from A. parasiticus NIAH-26 was desalted by using a PD-10 column (Amersham Bioscience) which had been equilibrated with buffer A (20 mM Tris-HCl buffer [pH 7.5], 0.4 mM EDTA, 10 mM MgCl2, 1 mM 2-mercaptoethanol, 10% glycerol [vol/vol]). The resultant cytosol (9 ml) was injected onto a Mono Q HR 5/5 column (Amersham Bioscience) equilibrated with buffer A. The column was washed with the same buffer, and then the bound proteins were eluted with a linear gradient of 0 to 0.5 M NaCl in buffer A at a flow rate of 0.4 ml min−1 for 50 min. Each 0.25-ml eluate was collected, and 2 μl of each faction was assayed for enzyme activities involved in the reaction from HAVN to AVR. The reaction products were detected by TLC (silica gel 60; Merck), with ethyl acetate-benzene (1:1 [vol/vol]) used as the developing solvent (Fig. 1A). To detect OAVN cyclase activity, a two-step reaction was used. HAVN was first converted to OAVN by incubating HAVN (0.13 mM) with a portion of the pooled fractions containing HAVN dehydrogenase (fractions 29 to 33 in Fig. 1) in 0.1 M potassium phosphate buffer, pH 7.5 (total volume, 615 μl), containing 10% glycerol (vol/vol) and 2 mM NAD, at 30°C for 30 min. Twenty microliters of the resultant reaction mixture containing OAVN was then added to 5 μl of each fraction eluted. After incubation at 30°C for 15 min, the products were detected by TLC. The reaction products were observed under UV light (365 nm).
FIG. 1.
Mono Q ion-exchange chromatography of the cytosol fraction from A. parasiticus NIAH-26. (A) Each fraction of the Mono Q ion-exchange chromatography step was incubated with HAVN in the presence of NAD. (B) HAVN was first converted to OAVN by incubation of HAVN with the fractions containing HAVN dehydrogenase enzyme activities (fractions 29 to 33 fractions; see panel A). Each fraction of the Mono Q chromatography step was then added to an aliquot of the first reaction mixture and incubated. The reaction products were analyzed by TLC.
Purification of HAVN dehydrogenase.
All purification steps for HAVN dehydrogenase were performed at 0 to 4°C.
(i) Ammonium sulfate precipitation.
Ammonium sulfate was added to the cytosol fraction (345 ml) up to 20% saturation. After removing pellets by centrifugation at 10,000 × g for 15 min, the saturation level of the supernatant was brought up to 55% with ammonium sulfate and then the resultant pellets were collected by centrifugation. The pellets were dissolved in buffer A and dialyzed against 20 mM Tris-HCl buffer, pH 7.5, containing 10% glycerol (vol/vol) and 0.1 mM phenylmethylsulfonyl fluoride.
(ii) DE 52 column chromatography.
The dialyzed material was diluted with buffer A to 104 ml and loaded onto a DE 52 column (3.0 by 12 cm; Whatman) equilibrated with buffer A. The column was washed with buffer A, and the enzyme was eluted with 50 mM NaCl in buffer A (240 ml). Active fractions were combined and used for the next step.
(iii) Butyl Toyopearl column chromatography.
The ammonium sulfate concentration of the combined fractions was adjusted to 0.8 M with 3.2 M ammonium sulfate in buffer A. The solution was loaded onto a Butyl Toyopearl 650 M column (3 by 10 cm; Tosoh) equilibrated with 0.8 M ammonium sulfate in buffer A. The column was washed with 0.8 M ammonium sulfate in buffer A, and then the enzyme was eluted with a linear gradient of 0.8 to 0 M ammonium sulfate in buffer A (200 ml). The enzyme was eluted at about 0.4 M ammonium sulfate, and active fractions were combined.
(iv) Hydroxyapatite column chromatography.
The sample was loaded onto a hydroxyapatite column (1.7 by 9.5 cm; Wako Chemical, Osaka, Japan) equilibrated with buffer A. The proteins that were not bound to the column were collected and concentrated by ultrafiltration with the Ultrafree-15 system (Millipore).
(v) First Superdex column chromatography.
The concentrated sample was filtered through a membrane filter with a pore size of 0.22 μm. One-fifth of the filtrate was loaded onto a Superdex 200 HR 10/30 column (Amersham Bioscience) equilibrated with buffer A. Elution was performed with buffer A at a flow rate of 0.5 ml min−1. The same procedure was repeated for the remaining sample. Active fractions were combined.
(vi) First Mono Q column chromatography.
The combined sample was injected onto a Mono Q HR 5/5 column (Amersham Bioscience) equilibrated with buffer A. The column was washed with the equilibration buffer, and then the enzyme was eluted with a linear gradient of 0 to 0.5 M NaCl in buffer A for 40 min at a flow rate of 0.5 ml/min. Active fractions were combined and concentrated by ultrafiltration with Ultrafree-4.
(vii) Second Superdex column chromatography.
The concentrated sample was loaded onto a Superdex 200 HR 10/30 column equilibrated with buffer A. Elution was performed with buffer A at a flow rate of 0.5 ml min−1. Active fractions were combined.
(viii) Resource Q column chromatography.
The enzyme solution was injected onto a Resource Q column (Amersham Bioscience) equilibrated with buffer A. The column was washed with equilibration buffer, and then the enzyme was eluted with a linear gradient of 0 to 0.5 M NaCl in buffer A for 120 min at a flow rate of 0.5 ml min−1. HAVN dehydrogenase was eluted at 0.14 M NaCl, and active fractions were pooled.
(ix) Second Mono Q column chromatography.
The pooled sample was diluted to double the volume and then was injected onto a Mono Q HR 5/5 column equilibrated with buffer A. The column was washed with buffer A, and then the enzyme was eluted with a linear gradient of 0 to 0.3 M NaCl in buffer A for 40 min at a flow rate of 0.5 ml min−1. Active fractions were pooled as the purified enzyme.
HAVN dehydrogenase assay.
For detection of enzymatically active fractions during enzyme purification, TLC analyses were routinely used. HAVN (0.2 mM) was incubated with each fraction during purification in 25 μl of buffer B (60 mM potassium phosphate buffer [pH 7.5] and 10% glycerol [vol/vol]) containing 1.6 mM NAD at 30°C for 10 min, and the reaction was then terminated by adding 40 μl of water-saturated chloroform and mixing it with a Vortex mixer. After centrifugation at 10,000 × g for 1 min, 23 μl of the lower chloroform layer was spotted onto a TLC plate (silica gel 60; Merck), and the plate was developed with acetone-n-hexane (2:3 [vol/vol]). The reaction products, including OAVN, were observed under long-wave (365 nm) UV light.
To determine HAVN dehydrogenase activity quantitatively, high-performance liquid chromatography (HPLC) was used. HAVN (0.2 mM) was incubated with 2 μl of enzyme solution from each purification step or the purified enzyme solution in buffer B containing 1.6 mM NAD (total volume, 25 μl) at 30°C for 5 min, and the reaction was terminated by adding 50 μl of ethyl acetate and mixing it with a Vortex mixer. After centrifugation at 2,000 × g for 30 s, 30 μl of the upper layer was collected. Fifty microliters of ethyl acetate was mixed with the resulting lower layer, and the upper ethyl acetate layer was collected. The same extraction procedure was repeated three more times. The combined ethyl acetate extracts (110 μl) were left in the dark and dried. The residue was dissolved in 40 μl of MeOH and analyzed by HPLC. An HPLC apparatus (LC-6A; Shimadzu Co., Kyoto, Japan) equipped with a Cosmosil 5Ph column (0.46 by 15 cm; Nacalai Tesque) was used, with 70% MeOH in 1% acetic acid aqueous solution as the eluting solvent, at a flow rate of 0.5 ml/min. The temperature of the column was kept at 30°C, and the UV absorption at 290 nm was monitored.
Purification of OAVN cyclase. (i) Ammonium sulfate precipitation.
Ammonium sulfate fractionation of the cytosol (198 ml) was done by the same procedure as for the purification of HAVN dehydrogenase.
(ii) DEAE-Sepharose column chromatography.
The dialyzed solution (180 ml) was loaded onto a DEAE-Sepharose CL-6B column (2.8 by 10 cm; Amersham Bioscience) which had been equilibrated with buffer A. The column was washed with buffer A, and the proteins bound to the column were eluted with a linear gradient of 0 to 0.5 M KCl in buffer A (200 ml). The enzyme was eluted by ∼0.16 M KCl. Active fractions were combined.
(iii) Phenyl Sepharose column chromatography.
Ammonium sulfate concentration of the combined solution was adjusted to 0.8 M with 3.2 M ammonium sulfate in buffer A. The solution was loaded onto a phenyl Sepharose CL-4B column (1.6 by 8.5 cm; Amersham Bioscience) which had been equilibrated with buffer A containing 0.8 M ammonium sulfate. The column was washed with the same solution, and the proteins were eluted with a linear gradient of 0.8 to 0 M ammonium sulfate in buffer A (160 ml). The enzyme was eluted broadly around 0.46 M ammonium sulfate. The active fractions were combined (96 ml). In order to reduce the volume of the enzyme solution, the enzyme was concentrated by use of a small phenyl Sepharose CL-4B column (0.7 by 15 cm) and one-step elution by changing the ammonium sulfate concentration in buffer A from 0.8 M to 0 M. The enzyme fractions were combined (8.5 ml) and concentrated by ultrafiltration with a Centriprep-10 concentrator (Millipore).
(iv) Sephacryl S-300 column chromatography.
The concentrated sample (0.9 ml) was loaded onto a Sephacryl S-300 column (1.6 by 68 cm; Amersham Bioscience). The active fractions of OAVN cyclase were combined.
(v) First Mono Q column chromatography.
The sample was fractionated by use of a Mono Q HR 5/5 column by the same procedure as that used for the first Mono Q chromatography of HAVN dehydrogenase. The OAVN cyclase activities were eluted at 0.23 M KCl. The active fractions were combined and concentrated by ultrafiltration with an Ultrafree-15 filter (Millipore).
(vi) Superdex column chromatography.
The concentrated sample was loaded onto a Superdex 200 HR 10/30 column as described above. Active fractions were combined.
(vii) Matrex gel Green A column chromatography.
The enzyme solution was loaded onto a Matrex gel Green A column (1.0 by 8 cm; Millipore). The proteins that were not bound to the column were collected.
(viii) Second Mono Q column chromatography.
The fractions collected were filtered through a membrane filter with a pore size of 0.22 μm. The filtrate was injected onto a Mono Q HR 5/5 column as for the first Mono Q chromatography step. The enzyme was eluted with a linear gradient of 0 to 0.1 M KCl in buffer A for 5 min and then with a linear gradient of 0.1 to 0.45 M KCl in buffer A for 55 min at a flow rate of 0.5 ml min−1. The enzyme was eluted by ∼0.21 M KCl. Active fractions were collected.
(ix) Resource PHE column chromatography.
The collected solution was brought up to 0.8 M ammonium sulfate and then injected onto a Resource PHE column (Amersham Bioscience) which had been equilibrated with 0.8 M ammonium sulfate in buffer A. The column was washed with the equilibration buffer, and then the proteins were eluted with a linear gradient of 0.8 to 0 M ammonium sulfate in buffer A for 40 min at a flow rate of 0.5 ml min−1. The OAVN cyclase was eluted by ∼0.26 M ammonium sulfate and was pooled as the purified enzyme.
OAVN cyclase assay.
Since the desired OAVN cyclase behaved together with the HAVN dehydrogenase for the first three purification steps (ammonium sulfate fractionation, DEAE-Sepharose chromatography, and phenyl Sepharose chromatography), the activity of the OAVN cyclase was detected by using HAVN as a substrate, by which HAVN was converted to OAVN by the contaminated HAVN dehydrogenase, and the resultant OAVN served as the substrate for the desired cyclase activity. HAVN (0.87 mM) was incubated with an aliquot of each fraction during purification in 22 μl of buffer B containing 1.8 mM NAD at 30°C for 10 min. The reaction products were extracted with water-saturated chloroform and detected by using TLC. TLC plates were developed with benzene-ethyl acetate (1:1 [vol/vol]).
During the purification process, the residual HAVN dehydrogenase was almost completely separated from the OAVN cyclase at the Sephacryl S-300 column chromatography step. We recently found that the cytosol fraction of Lactobacillus brevis IFO 12005 contains HAVN dehydrogenase activity which can convert HAVN to OAVN (E. Sakuno et al., unpublished data). Therefore, we used a two-step reaction assay for the detection of OAVN cyclase activity after this step. HAVN was first converted to OAVN by the L. brevis enzyme, and then the resultant OAVN was used as the substrate for the OAVN cyclase. Lactobacillus HAVN dehydrogenase (0.12 mg ml−1) was added to buffer B (total volume, 15 μl) containing 0.13 mM HAVN and 2.7 mM NADP, so that NADP, not NAD, was used as a cofactor of the bacterial enzyme. After incubation at 40°C for 90 min, a 3-μl aliquot of each fraction and NAD (final concentration, 4.3 mM) were added to the mixture (final volume, 23 μl). We later confirmed that OAVN cyclase does not require any cofactor for its activity. The resulting mixture was further incubated at 30°C for 20 or 30 min, and the amount of AVR produced was then determined by TLC.
To determine OAVN cyclase activity quantitatively, we used HPLC. OAVN (0.29 mM) was incubated with 2 μl of each enzyme fraction (total volume, 50 μl) in buffer B at 30°C for 5 min. The reaction was terminated by adding 50 μl of ethyl acetate and mixing it with a Vortex mixer. After centrifugation, 30 μl of the upper ethyl acetate layer was directly injected into an HPLC apparatus equipped with a Shimpack CLC-ODS column (0.46 by 15 cm; Shimadzu). The reaction products were eluted from the column with 90% MeOH in water at a flow rate of 1 ml min−1.
Characterization of HAVN dehydrogenase and OAVN cyclase.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (15). The proteins were stained with Coomassie brilliant blue R-250. The molecular mass of the denatured enzyme was determined with a low-molecular-weight calibration kit (Amersham Bioscience). The relative molecular mass of the native enzyme was determined by gel filtration on a Superdex 200 HR 10/30 column as described above. The column was calibrated with the following standard proteins (Bio-Rad Laboratories): thyroglobulin (670 kDa), gamma globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and cyanocobalamin (1.35 kDa). The protein concentration was determined by use of a protein assay solution (Bio-Rad), with bovine serum albumin as the standard (4).
The effect of temperature on HAVN dehydrogenase activity was measured in the range of 20 to 55°C. HAVN (0.2 mM) was incubated with partially purified HAVN dehydrogenase (0.19 mg ml−1) in 25 μl of buffer B containing 2 mM NAD at each temperature for 5 min. The reaction product, OAVN, was extracted with ethyl acetate and measured by HPLC as described above. To determine the pH effect on enzyme activity, 0.1 M sodium phosphate buffer was used for pHs 6.0 to 8.5 and 0.1 M Tris-HCl buffer was used for pHs 8.5 to 10.0. Twenty-five microliters of a mixture of 0.1 mM HAVN, 2 mM NAD, 10% glycerol (vol/vol), and partially purified enzyme (0.19 mg ml−1) in the various buffer solutions described above was incubated at 30°C for 5 min. To extract the reaction product effectively, the pH of the reaction mixture was decreased by adding 10 μl of 0.5 M sodium phosphate buffer, pH 6.5, before extraction with ethyl acetate. OAVN production from HAVN was measured by HPLC as described above. To determine the kinetic values of the enzyme, various concentrations of HAVN were incubated with partially purified HAVN dehydrogenase (0.04 mg ml−1) in 25 μl of buffer B containing 2 mM NAD at 30°C for 8 min. The reaction product was extracted with ethyl acetate and analyzed by HPLC as described above.
To determine the effect of pH on OAVN cyclase activity, 0.1 M sodium acetate buffer was used for pHs 3.5 to 5.5 and 0.1 M sodium phosphate buffer was used for pHs 5.5 to 8.0. A mixture of 80 μM OAVN, 10% glycerol (vol/vol), and partially purified enzyme (0.13 mg ml−1) in the various buffer solutions described above was incubated at 30°C for 15 min. AVR production from OAVN was measured by HPLC as described above. The effect of temperature was determined at pH 7.5 in the range of 20 to 55°C. To determine the kinetic values of the cyclase, various concentrations of OAVN were incubated with the purified enzyme (0.11 mg/ml) after Resource PHE column chromatography in a final volume of 50 μl at 30°C for 11 min. The reaction products were extracted with 75 μl of water-saturated ethyl acetate, and 60 μl of the ethyl acetate extract was directly injected into an ODS column as described above.
Peptide MS fingerprinting analysis.
After SDS-PAGE, a band corresponding to HAVN dehydrogenase was excised from the gel. The gel piece was incubated at 35°C for 20 h in Tris buffer, pH 8.0, containing trypsin. The peptide solution was purified and concentrated by ZipTip C18 concentrators (Millipore). The peptides were analyzed on a Voyager-DE STR machine (Applied Biosystems), with delayed extraction in reflector mode. 2,5-Dihydroxybenzoic acid (2,5-DHB) in 50% acetonitrile and 0.1% trifluoroacetic acid was used for the matrix. Matrix-assisted laser desorption ionization (MALDI) spectra were calibrated by external calibration using a mixture containing 2,5-DHB (monoisotopic mass, 273.040), des-argl-bradykinin (monoisotopic mass, 904.4681), angiotensin 1 (monoisotopic mass, 1,296.6853), adrenocorticotropin (clip 1-17) (monoisotopic mass, 2,093.0867), and adrenocorticotropin (clip 7-38) (monoisotopic mass, 3,657.9294). The mass list from the HAVN dehydrogenase digest was used to search the database.
RESULTS
Involvement of two enzymes in the pathway from HAVN to AVR.
The desalted cytosol fraction of A. parasiticus NIAH-26 was applied onto a Mono Q column, and then the proteins bound to the column were eluted with a linear gradient of 0 to 0.5 M NaCl. When HAVN was incubated with each fraction, the fractions around 0.16 M KCl (no. 26 to 36) produced a novel substance which was neither HAVN nor AVR (Fig. 1A). In contrast, the fractions around 0.24 M KCl (no. 38 to 42) yielded larger amounts of AVR than did the other fractions. Corresponding to the formation of AVR, the amounts of the remaining HAVN decreased remarkably. These results suggested that the cytosol fraction contains two different enzymes, which catalyze the formation of the unknown substance and of AVR, respectively. In order to confirm these results, the following two-step procedure was devised. HAVN was first incubated with pooled fractions 29 to 33 to produce the unknown substance, and then each fraction obtained after Mono Q column chromatography was added to an aliquot of the resultant reaction mixture. AVR was produced significantly when each fraction from no. 38 to 54 was added, whereas other fractions did not show any change (Fig. 1B). These results indicated the following two things. First, fractions 26 to 36 contained HAVN dehydrogenase, which catalyzed the conversion of HAVN to the new substance. Second, fractions 38 to 54 contained another enzyme, which catalyzed conversion of the substance to AVR. Although we previously suggested that the latter AVR formation reaction proceeds spontaneously (30), these data confirmed that a specific cytosolic enzyme catalyzes it.
Characterization of a novel intermediate.
The novel intermediate was very unstable, because it was easily converted to AVR while being kept in solution during drying. However, we succeeded in isolating it from the preparation obtained by incubation of HAVN with purified HAVN dehydrogenase by avoiding use of any drastic procedure, such as evaporation under high temperature or column chromatography using an acidic solvent. The physicochemical properties of the new intermediate and the 13C and 1H NMR data for HAVN are shown in Table 1, for which the 1H NMR data of HAVN have been previously reported (31). The molecular weight of the compound is 386, which is 2 units lower than that of HAVN. Its 1H and 13C NMR spectra are very similar to those of HAVN, except for the following. In its 13C NMR spectrum, the resonance (δC 67.5) assigned to C-5′ of HAVN disappears, and a new resonance at δC 207.9 is observed. In its 1H NMR spectrum, the methyl signal of C-6′ is shifted to δH 2.08 and appears as a singlet. The resonance at δH 3.72 (1H, tq) in the 1H NMR spectrum of HAVN disappears, and a resonance at δH 2.53 (2H, t) is observed in that of the new intermediate. These data indicate that the intermediate is OAVN. Thus, we named the novel enzyme which catalyzes the reaction from OAVN to AVR OAVN cyclase. In the IR spectrum of OAVN, absorption around 1,700 cm−1 is remarkably weak, although a ketone group usually shows strong absorption around 1,700 cm−1. However, the signal of the 5′-ketonic carbon is also observed at δC 207.9, even by the solid-state 13C NMR measurement of OAVN, indicating that OAVN must be in the oxo-form in the solid state and in solvent.
Purification of HAVN dehydrogenase.
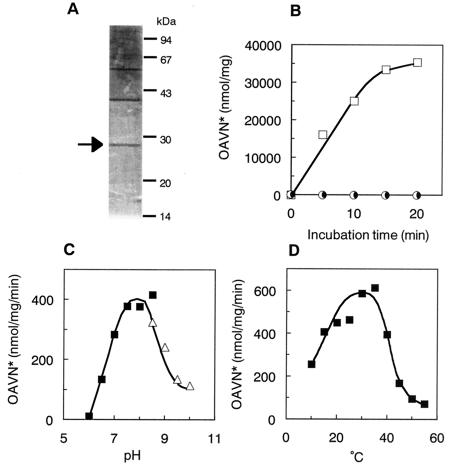
HAVN dehydrogenase was purified from mycelia of A. parasiticus NIAH-26 by monitoring the enzyme activity leading to the formation of OAVN from HAVN. The enzyme purification process is summarized in Table 2. OAVN cyclase was separated from HAVN dehydrogenase by DE 52 column chromatography. After the final Mono Q chromatography step, the enzyme was purified 694-fold over the initial step, with a yield of 0.2%. The highly active fractions 29 to 37 obtained by the final Mono Q chromatography step showed four bands by SDS-PAGE. A protein band (28 kDa) was correlated with the enzyme activity by comparison of the enzyme activity with the protein bands on an SDS-PAGE gel (Fig. 2A). The molecular mass of the enzyme was close to that (29.6 kDa) of the AdhA protein, as deduced from its cDNA sequence (8).
FIG. 2.
Characterization of purified HAVN dehydrogenase. (A) SDS-PAGE of purified HAVN dehydrogenase. The peak fraction of HAVN dehydrogenase activity after the second Mono Q chromatography step was analyzed by SDS-PAGE on a 12% polyacrylamide gel, and proteins were then stained with Coomassie brilliant blue. Arrow, HAVN dehydrogenase. (B) Purified enzyme was incubated with HAVN in the absence (•) or presence of NAD (□) or NADP (○) for various times. The amount of OAVN produced was measured. Symbols on the baseline overlap each other. (C) OAVN formation from HAVN was measured under various pH conditions. (▪), sodium phosphate buffer; (▵), Tris-HCl buffer. (D) OAVN formation from HAVN was measured at various temperatures. OAVN*, total amounts of OAVN and AVR, which was spontaneously produced from OAVN. The proportion of AVR in the total amounts was 7% on average (0 to 35%).
Characterization of HAVN dehydrogenase.
Based on Superdex 200 HR gel filtration chromatography, the native molecular mass of HAVN dehydrogenase was estimated to be 60 kDa. These data suggested that the active HAVN dehydrogenase was a homodimer composed of identical 28-kDa subunits. The purified HAVN dehydrogenase required NAD, but not NADP, for its activity (Fig. 2B). The optimum pH of the enzyme activity was between 7.5 and 8.5 (Fig. 2C). The optimum temperature for the enzyme was around 30 to 35°C, although the range of 20 to 40°C appeared to be suitable (Fig. 2D). From Lineweaver-Burk plots, the Km and Vmax of the enzyme for HAVN were estimated to be 83 μM and 2.3 μmol mg of protein−1 min−1, respectively.
Although we tried to determine the N-terminal sequence of the enzyme, no sequence could be detected, indicating that the N terminus of this enzyme may be blocked. Therefore, the purified HAVN dehydrogenase was digested by trypsin and the products were analyzed by MALDI-time of flight (TOF) MS. Seven peptide fragments were observed, and the molecular masses matched the theoretical fragment masses from the gene product of adhA (Table 3), which was reported by Chang et al. to be the gene encoding HAVN dehydrogenase (8). This result demonstrated that HAVN dehydrogenase was encoded by the adhA gene. Furthermore, the fragment corresponding to the N-terminal 16 amino acids showed the ion peak at a molecular mass of 1,777.9, and its calculated molecular mass is 1,718.8, indicating that the N-terminal methionine of HAVN dehydrogenase is posttranslationally acetylated and oxidized to form acetyl methionine sulfoxide. Methionines at either or both of the 172nd and 179th positions of the amino acid sequence were also oxidized in some, but not all, enzyme molecules.
TABLE 3.
Peptide masses from HAVN dehydrogenase digested with trypsin
| Fragment | Monoisotopic mass
|
Peptide sequence | Amino acid residues | Comment(s) | |
|---|---|---|---|---|---|
| Observed | Predicted | ||||
| 1 | 1,718.88 | MEVLDTTVDLGTLQGK | 1-16 | ||
| 2 | 1,777.91 | 1,777.88 | MEVLDTTVDLGTLQGK | 1-16 | 1 N-Acetyl (protein); 1 oxidation (M) |
| 3 | 1,515.84 | 1,515.84 | SALITGGASGIGLATAR | 17-33 | |
| 4 | 1,222.65 | 1,222.64 | LDVNPSPPDIR | 122-132 | |
| 5 | 1,698.86 | 1,698.84 | SLILMGSIGSYMDSPK | 168-183 | |
| 6 | 1,714.84 | 1,714.83 | SLILMGSIGSYMDSPK | 168-183 | 1 Oxidation (M) |
| 7 | 1,730.82 | 1,730.83 | SLILMGSIGSYMDSPK | 168-183 | 2 Oxidation (M) |
| 8 | 478.27 | 478.27 | FGVR | 192-195 | |
| 9 | 492.29 | 492.29 | GLFR | 196-199 | |
| 10 | 573.34 | 573.33 | ELGVR | 207-211 | |
Purification of OAVN cyclase.
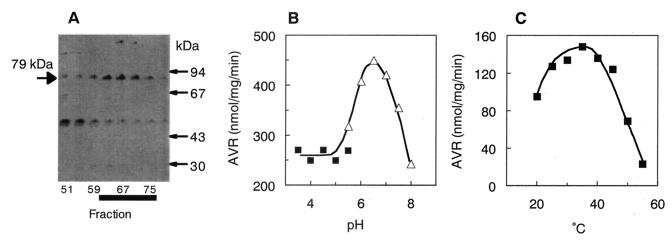
OAVN cyclase was purified from the cytosol fraction of A. parasiticus NIAH-26. The purification procedure is summarized in Table 4. HAVN dehydrogenase was present together with OAVN cyclase in the cytosol fraction, and these enzymes were copurified until the phenyl Sepharose chromatography step. Therefore, HAVN, instead of OAVN, could be used as a substrate to detect OAVN cyclase activities, because HAVN was converted into OAVN by the endogenous HAVN dehydrogenase in the same reaction mixture. HAVN dehydrogenase was mostly separated by Sephacryl S-300 chromatography, because the molecular mass of OAVN cyclase (158 kDa) was rather bigger than that of HAVN dehydrogenase (60 kDa). The enzyme activity after this step was measured by using OAVN, which was prepared from HAVN by HAVN dehydrogenase from L. brevis, because we previously found that some L. brevis strains had stable HAVN dehydrogenase activity (Sakuno et al., unpublished data). After the final Resource PHE chromatography, the enzyme was purified 12.3-fold over the initial step, with a yield of 2.1%. The peak fraction of the enzyme activity showed three protein bands, one major band of 79 kDa and two minor bands, of 115 and 48 kDa, by SDS-PAGE (Fig. 3A). The 79-kDa protein correlated with the enzyme activity in the last three purification steps, indicating that this protein was OAVN cyclase.
TABLE 4.
Purification of OAVN cyclase
| Purification step | Vol (ml) | Total protein (mg) | Total activity (nmol/min) | Sp act (nmol/mg/min) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Cytosol | 198 | 711 | 4,846 | 6.81 | 1.0 | 100.0 |
| (NH4)2SO4, 20-55% | 180 | 422 | 5,048 | 12.0 | 1.8 | 104.2 |
| DEAE-Sepharose | 88 | 247 | 1,271 | 5.14 | 0.8 | 26.2 |
| Phenyl Sepharose | 96 | 82 | 223 | 2.73 | 0.4 | 4.6 |
| Sephacryl S-300 | 17.2 | 24 | 570 | 23.3 | 3.4 | 11.8 |
| First Mono Q | 8.3 | 8.3 | 379 | 45.9 | 6.7 | 7.8 |
| Superdex | 5.1 | 6.2 | 207 | 33.4 | 4.9 | 4.3 |
| Matrex Green A | 7.4 | 4.1 | 181 | 44.0 | 6.5 | 3.7 |
| Second Mono Q | 3.9 | 2.3 | 185 | 81.8 | 12.0 | 3.8 |
| Resource PHE | 11.1 | 1.2 | 103 | 83.6 | 12.3 | 2.1 |
FIG. 3.
Characterization of purified OAVN cyclase. (A) SDS-PAGE of partially purified OAVN cyclase. The proteins in the active fractions eluted from the Resource PHE column were electrophoresed in an SDS-polyacrylamide gel and stained with Coomassie brilliant blue R-250. OAVN cyclase appeared as a 79-kDa band. Fractions that had high OAVN cyclase activity are marked with a thick black line. (B) AVR formation from OAVN was measured under various pH conditions. (▪), sodium acetate buffer; (▵), sodium phosphate buffer. (C) AVR formation from OAVN was measured at various temperatures.
Characterization of OAVN cyclase.
The specific activity of the purified enzyme was 83.6 nmol mg of protein−1 min−1, and neither NADP(H) nor NAD(H) was required for the enzyme activity. The molecular mass of the native enzyme was determined to be 158 kDa by the Superdex 200 gel filtration chromatography step. Since the denatured enzyme was 79 kDa, OAVN cyclase must function as a homodimer. The enzyme showed an optimum pH range for activity between 6.0 and 7.0 when it was measured by using buffers from pHs 3.5 to 8.0 (Fig. 3B). The optimum temperature of the enzyme was broad, from 25 to 45°C (Fig. 3C). The kinetic values Km and Vmax of the enzyme for OAVN were 20 μM and 6.67 nmol mg of protein−1 min−1, respectively.
DISCUSSION
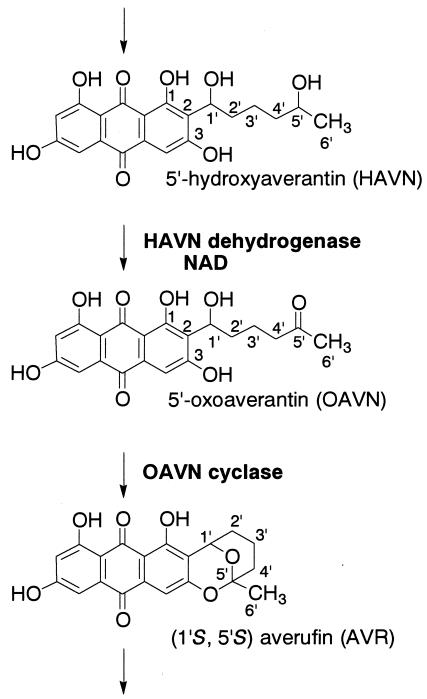
We demonstrated that the pathway of HAVN → OAVN →AVR is included in aflatoxin biosynthesis (Fig. 4). We previously proposed that OAVN would be produced as a transient intermediate between HAVN and AVR and that OAVN would cyclize to form AVR spontaneously, not enzymatically. Since the latter enzyme, OAVN cyclase, did not require any cofactors, the reaction from HAVN to AVR had appeared to be a one-enzyme reaction depending on the presence of NAD (30, 31). However, in this work, we clearly demonstrate that an enzyme catalyzed the reaction from OAVN to AVR.
FIG. 4.
Pathway from HAVN to AVR clarified in this study. HAVN is converted to OAVN by HAVN dehydrogenase, which requires NAD, but not NADP, for its activity. OAVN is then converted to AVR by a novel OAVN cyclase enzyme.
The biosynthetic pathway from HAVN to AVR has been controversial for a long time. McCormick et al. reported that averufanin was converted to AFB1 in a feeding experiment, suggesting that averufanin is involved in the step between AVN and AVR (16). In contrast, we showed that HAVN is a precursor of aflatoxins in a feeding experiment, that HAVN, but not averufanin, is converted to AVR in a cell-free experiment (31), and that aflatoxin is not produced from averufanin in a feeding experiment. We also found that HAVN is an unstable substance which is easily converted to averufanin through spontaneous dehydration. In this study, we demonstrated that HAVN is converted to OAVN by an AdhA dehydrogenase enzyme depending on NAD. The resultant OAVN was converted to AVR through dehydration by a new enzyme activity, that of OAVN cyclase. Therefore, this study brings the long controversy about the pathway from HAVN to AVR to an end. The pathway is composed of two reactions, that is, (i) the dehydrogenation reaction from HAVN to OAVN by AdhA dehydrogenase and (ii) the dehydration (cyclization) reaction from OAVN to AVR by OAVN cyclase. Although averufanin had been found in an AVR-accumulating mutant (23) as well as many fungal species (1, 11, 12), it was most likely produced by nonenzymatic dehydration of HAVN. In fact, gentle manipulation of the metabolites HAVN and OAVN was indispensable for the success of this work.
OAVN was a very unstable substance and was easily converted to AVR. Even mild drying or simply storing in MeOH solution at −20°C caused gradual dehydration to form AVR. Because we had routinely used a formic acid-containing solvent as a developing solvent for TLC analysis, we had not previously noticed the involvement of OAVN in the pathway from HAVN to AVR (30, 31). In fact, the reaction products formed by some eluted fractions were observed as a smeary and broad band on the TLC plate even when we used acid-containing developing solution, the most intensive part of which corresponded to AVR (data not shown). In this work, we could isolate OAVN from the reaction mixture of HAVN and the purified HAVN dehydrogenase by avoiding any drastic procedure such as evaporation under high temperature or column chromatography using an acidic solvent. Finally, we investigated the structure of OAVN (Table 1). In structural analyses of OAVN, 5′-oxo function in the IR spectrum showed weak absorption, which was unexpected. However, we confirmed the presence of the oxo- group at the 5′-C site by both solid and liquid 13C NMR measurement. Therefore, the weak absorption shown on the IR spectrum remains to be studied.
On the other hand, OAVN as well as HAVN and AVR were detected by TLC as smearing spots (Fig. 1) when ethyl acetate-benzene (1:1 [vol/vol]) was used as the developing solvent. Similar smearing patterns were observed when relatively large amounts of other metabolites were developed instead of OAVN. In contrast, more compact spots of OAVN and HAVN were also obtained when another developing solution was used (data not shown), indicating that the smearing pattern on the TLC plate may be caused by low solubility of the metabolites in this solution.
For the first time, we purified HAVN dehydrogenase. The molecular masses of the native enzyme and the denatured enzyme were 60 and 28 kDa, respectively, indicating that this enzyme is a homodimer. The molecular mass of the gene product encoded by adhA is 29.6 kDa (8). We herein confirmed that HAVN dehydrogenase is encoded by the adhA gene by peptide mass fingerprinting by MALDI-TOF MS (Table 3). The masses of the peptides from HAVN dehydrogenase digested with trypsin matched those of theoretical digestion reactions of the adhA gene product. Also, the MALDI-TOF MS analysis revealed that the N terminus of this enzyme is posttranslationally acetylated. Although we have found that the N termini of several enzymes involved in aflatoxin biosynthesis were blocked based on the failure of amino acid sequencing (data not shown), HAVN dehydrogenase is the first enzyme whose blocking structure has been clarified among enzymes in aflatoxin biosynthesis. Also, two methionines of the enzyme protein were found to be oxidized to methionine sulfoxides, as determined by MALDI-TOF MS. Methionine sulfoxidation is known to cause conformational changes of many proteins (9, 10, 24). Reversible oxidation and reduction of methionine have also been suggested as a biological regulatory mechanism (9). Therefore, oxidation of methionine in HAVN dehydrogenase could indicate a regulatory mechanism of aflatoxin biosynthesis.
This work demonstrated that HAVN dehydrogenase enzyme exclusively required NAD, but not NADP, as a cofactor (Fig. 2B), although the crude cytosol fraction showed significant activity even in the presence of NADP instead of NAD (31). This suggests that an oxido-reductase enzyme may be involved in conversion between NADP(H) and NAD(H) in the cytosol. In aflatoxin biosynthesis, NADP(H) is commonly used as a cofactor for many enzyme activities (31). The reversible reaction between NA and AVN uses NAD(H) as well as NADP(H) as a cofactor (31). However, the HAVN dehydrogenase enzyme is the sole enzyme requiring NAD exclusively for its activity among the enzymes involved in aflatoxin biosynthesis.
OAVN cyclase was also purified from the cytosol fraction, which clearly demonstrated that the reaction of OAVN to AVR proceeds enzymatically. OAVN cyclase is a homodimer with a native mass of 158 kDa which is composed of two 79-kDa monomer proteins. The enzyme catalyzes intramolecular acetal formation among 5′-ketone and 3- and 1′-hydroxyl groups in OAVN. Further characterization of this enzyme and the gene encoding the enzyme is now in progress by our group.
Acknowledgments
We thank Y. Ando, National Institute of Animal Health, Japan, for help with taking photographs and M. Nakayama, National Institute of Floricultural Science, Japan, for measuring the ESI-MS.
This work was supported in part by a grant-in-aid (Bio-Design Program) from the Ministry of Forestry and Fisheries, Japan (BMP-03-VI-1-4).
REFERENCES
- 1.Aucamp, P. J., and C. W. Holzapfel. 1970. Polyhydroxyanthraquinones from Aspergillus versicolor, Aspergillus nidulans and Bipolaris sp. Their significance in relation to biogenetic theories of aflatoxin B1. S. Afr. Chem. Inst. J. 23:40-56. [Google Scholar]
- 2.Bennett, J. W., P. K. Chang, and D. Bhatnagar. 1997. One gene to whole pathway: the role of norsolorinic acid in aflatoxin research. Adv. Appl. Microbiol. 45:1-15. [DOI] [PubMed] [Google Scholar]
- 3.Bhatnagar, D., T. E. Cleveland, and P. J. Cotty. 1994. Mycological aspects of aflatoxin formation, p. 327-346. In D. L. Eaton and J. D. Groopman (ed.), Toxicology of aflatoxins: human health, veterinary, and agricultural significance. Academic Press, Inc., New York, N.Y.
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Brown, D. W., J. H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, M. P., C. S. Brouwn-Jenco, and G. A. Payne. 1999. Genetic and molecular analysis of aflatoxin biosynthesis. Fungal Genet. Biol. 26:81-98. [DOI] [PubMed] [Google Scholar]
- 7.Busby, W. F., and G. N. Wogan. 1985. Aflatoxins, p. 945-1136. In C. E. Searle (ed.), Chemical carcinogens, 2nd ed., vol. 2. American Chemical Society, Washington, D.C.
- 8.Chang, P. K., J. Yu, K. C. Ehrlich, S. M. Boue, B. G. Montalbano, D. Bhatnagar, and T. E. Cleveland. 2000. adhA in Aspergillus parasiticus is involved in conversion of 5′-hydroxyaverantin to averufin. Appl. Environ. Microbiol. 66:4715-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciorba, M. A., S. H. Heinemann, H. Weissbach, N. Brot, and T. Hoshi. 1997. Modulation of potassium channel function by methionine oxidation and reduction. Proc. Natl. Acad. Sci. USA 94:9932-9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Härndahl, U., B. P. A. Kokke, N. Gustavsson, S. Linse, K. Berggren, F. Tjerneld, W. C. Boelens, and C. Sundby. 2001. The chaperone-like activity of a small heat shock protein is lost after sulfoxidation of conserved methionines in a surface-exposed amphipathic α-helix. Biochim. Biophys. Acta 1545:227-237. [DOI] [PubMed] [Google Scholar]
- 11.Heathcote, J. G., and M. F. Dutton. 1969. New metabolite of Aspergillus flavus. Tetrahedron 25:1497-1500. [Google Scholar]
- 12.Holker, J. S. E., S. A. Kagal, L. J. Mulheirn, and P. M. White. 1966. Some new metabolites of Aspergillus versicolor and a revised structure of averufin. J. Chem. Soc. Sect. D 24:911-913. [Google Scholar]
- 13.Keller, N. P., C. M. H. Watanabe, H. S. Kelkar, T. H. Adams, and C. A. Townsend. 2000. Requirement of monooxygenase-mediated steps for sterigmatocystin biosynthesis by Aspergillus nidulans. Appl. Environ. Microbiol. 66:359-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.McCormick, S. P., D. Bhatnagar, and T. E. Cleveland. 1987. Averufanin is an aflatoxin B1 precursor between averantin and averufin in the biosynthetic pathway. Appl. Environ. Microbiol. 53:14-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh, S. F., and G. A. Payne. 1984. Preharvest infection of corn silks and kernels by Aspergillus flavus. Phytopathology 74:1284-1289. [Google Scholar]
- 18.Marth, E. H., and M. P. Doyle. 1979. Update on molds: degradation of aflatoxin. Food Technol. 42:81-87. [Google Scholar]
- 19.Minto, R. E., and C. A. Townsend. 1997. Enzymology and molecular biology of aflatoxin biosynthesis. Chem. Rev. 97:2537-2555. [DOI] [PubMed] [Google Scholar]
- 20.Ono, M., S. Sakuda, A. Suzuki, and A. Isogai. 1996. Aflastatin A, a novel inhibitor of aflatoxin production by aflatoxigenic fungi. J. Antibiot. 50:111-118. [DOI] [PubMed] [Google Scholar]
- 21.Park, D. L. 1993. Controlling aflatoxin in food and feed. Food Technol. 47:92-96. [Google Scholar]
- 22.Payne, G. A. 1992. Aflatoxin in maize. Crit. Rev. Plant Sci. 10:423-440. [Google Scholar]
- 23.Prieto, R., G. L. Yousibona, and C. P. Woloshuk. 1996. Identification of aflatoxin biosynthesis genes by genetic complementation in an Aspergillus flavus mutant lacking the aflatoxin gene cluster. Appl. Environ. Microbiol. 62:3567-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy, V. Y., P. E. Desrochers, S. V. Pizzo, S. L. Gonias, J. A. Sahakian, R. L. Levine, and S. J. Weiss. 1994. Oxidative dissociation of human α2-macroglobulin tetramers into dysfunctional dimmers. J. Biol. Chem. 269:4683-4691. [PubMed] [Google Scholar]
- 25.Samarajeewa, U., A. C. Sen, M. D. Cohen, and C. Z. Wei. 1990. Detoxification of aflatoxins in foods and feeds by physical and chemical methods. J. Food Prot. 53:489-501. [DOI] [PubMed] [Google Scholar]
- 26.Trail, F., N. Mahanti, and J. Linz. 1995. Molecular biology of aflatoxin biosynthesis. Microbiology 141:755-765. [DOI] [PubMed] [Google Scholar]
- 27.Trail, F., P. K. Chang, J. Cary, and J. E. Linz. 1994. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus. Appl. Environ. Microbiol. 60:4078-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woloshuk, C. P., and R. Prieto. 1998. Genetic organization and function of the aflatoxin B1 biosynthetic genes. FEMS Microbiol. Lett. 160:169-176. [DOI] [PubMed] [Google Scholar]
- 29.Yabe, K.2002. Pathway and genes of aflatoxin biosynthesis, p. 227-251. In F. Fierro and J. Francisco (ed.), Microbial secondary metabolites: biosynthesis, genetics and regulation. Research Signpost, Kerala, India.
- 30.Yabe, K., Y. Matsuyama, Y. Ando, H. Nakajima, and T. Hamasaki. 1993. Stereochemistry during aflatoxin biosynthesis: conversion of norsolorinic acid to averufin. Appl. Environ. Microbiol. 59:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yabe, K., Y. Nakamura, H. Nakajima, Y. Ando, and T. Hamasaki. 1991. Enzymatic conversion of norsolorinic acid to averufin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 57:1340-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yabe, K., H. Nakamura, Y. Ando, N. Terakado, H. Nakajima, and T. Hamasaki. 1988. Isolation and characterization of Aspergillus parasiticus mutants with impaired aflatoxin production by a novel tip culture method. Appl. Environ. Microbiol. 54:2096-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, J., P. K. Chang, J. W. Cary, D. Bhatnagar, and T. E. Cleveland. 1997. avnA, a gene encoding a cytochrome P-450 monooxygenase, is involved in the conversion of averantin to averufin in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 63:1349-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]