Abstract
A molecular method based on restriction fragment length polymorphism (RFLP) of PCR-amplified fragments of the 23S rRNA gene was designed to rapidly identify Listeria strains to the species level. Two fragments (S1, 460 bp, and S2, 890 bp) were amplified from boiled DNA. S2 was cut with the restriction enzymes XmnI or CfoI and, if needed, S1 was digested by either AluI or ClaI. This method was first optimized with six reference strains and then applied to 182 isolates collected from effluents of treatment plants. All isolates were also identified by the API Listeria kit, hemolysis, and phosphatidylinositol-specific phospholipase C production (PI-PLC) on ALOA medium. The PCR-RFLP method unambiguously identified 160 environmental strains, including 131 in concordance with the API system, and revealed that 22 isolates were mixed cultures of Listeria monocytogenes and Listeria innocua. Discrepant results were resolved by a multiplex PCR on the iap gene, which confirmed the PCR-RFLP data for 49 of the 51 discordances, including the 22 mixed cultures. Sequencing of the 16S rRNA gene for 12 selected strains and reconstruction of a phylogenetic tree validated the molecular methods, except for two unclassifiable strains. The 158 single identifiable isolates were 92 L. monocytogenes (including seven nonhemolytic and PI-PLC-negative strains), 61 L. innocua, 4 Listeria seeligeri, and 1 Listeria welshimeri strain. The PCR-RFLP method proposed here provides rapid, easy-to-use, inexpensive, and reliable identification of the six Listeria species. Moreover, it can detect mixtures of Listeria species and thus is particularly adapted to environmental and food microbiology.
The genus Listeria comprises six characterized species: Listeria grayi, Listeria innocua, Listeria ivanovii, Listeria monocytogenes, Listeria seeligeri, and Listeria welshimeri (42). All of these species are saprophytic bacteria that are ubiquitous in nature (42). Among them, only L. monocytogenes acts as an opportunistic pathogen for both humans and animals, mainly causing abortions in pregnant females, neonatal sepsis, and severe infections such as septicemia and meningoencephalitis in susceptible hosts (13, 27, 40). Indeed, from its soil-plant reservoir, this bacterium can infect animals, principally ruminants, and also humans via the ingestion of contaminated foodstuffs of vegetal or animal origin. In fact, L. monocytogenes is now recognized as an important food-borne pathogen (13, 40). L. ivanovii is principally responsible for abortions in sheep (27). Other Listeria species are considered nonpathogenic (42), although L. ivanovii and L. seeligeri have been implicated in human listeriosis and L. innocua has been implicated in animal disease (9, 26, 36, 47).
During a recent investigation on the possible enrichment of the environmental reservoir of Listeria spp. by the effluents of treatment plants (D. Paillard, V. Dubois, F. Nathier, E. Hoogland, P. Caumette, and C. Quentin, unpublished data), we isolated a series of Listeria strains from sludge and sewage waters and sought a rapid and accurate method for identifying them to the species level. Traditional methods such as the CAMP (Christie, Atkins, Munch-Petersen) test, sugar fermentation, and nitrate reduction are laborious and can take up to 1 to 2 weeks to complete (42). Easy-to-use and rapid miniaturized identification systems are now commercially available (2, 3, 15, 35). However, because of the high phenotypic similarity of the six Listeria species, their separation is based on a limited number of biochemical characteristics, and unreliable results for some critical tests have been reported, such as for atypical strains lacking critical features (3, 21, 25, 38). On the other hand, a number of molecular methods have been developed since the late 1980s for Listeria species characterization. However, most of them only aim at detecting the Listeria genus and/or L. monocytogenes in food specimens (32-34, 41, 42). The remaining methods fail to identify every Listeria species (18, 30, 43) and/or provide complex intraspecies discrimination (4, 7, 11, 12, 16, 19, 20, 43). Others may require complex reagents (5) or technology (46) or involve time-consuming extraction of chromosomal DNA (6).
In this paper, we describe a novel genotypic method based on restriction fragment length polymorphism (RFLP) of PCR-amplified fragments of the 23S rRNA gene. This method allows the identification of the six species of Listeria, without strain-to-strain variation, by use of a common DNA-based protocol not requiring preliminary DNA extraction. In addition, this method was able to identify mixed cultures of different species, especially L. monocytogenes and L. innocua, which are often found in combination in food and environmental samples (29, 42). The PCR-RFLP method was first validated on six reference strains of Listeria spp. and then was applied to 182 isolates collected from sludge and sewage waters in comparison with biochemical identification.
MATERIALS AND METHODS
Bacterial strains.
A total of 188 Listeria isolates were examined in this study, including six reference strains (L. grayi ATCC 19120T, L. innocua ATCC 51742, L. monocytogenes ATCC 19115, L. seeligeri ATCC 35967T, L. ivanovii ATCC 19119T, and L. welshimeri ATCC 35897T) and 182 isolates recovered over a 1-year period from sludge and sewage waters from six effluent sites in the Bordeaux area of France. All strains were routinely grown on Mueller Hinton (MH) agar (Bio Mérieux, Marcy l'Etoile, France) supplemented with 5% horse blood and were stored in 15% glycerol-Trypticase soy broth and 5% horse blood at −80°C.
Biochemical identification.
Nonsporulating, regular, gram-positive rods occurring singly or in short chains that were catalase positive and oxidase negative and had growth after enrichment in Fraser broth on PALCAM selective medium (Bio-Rad, Marnes-La-Coquette, France) were presumptively considered Listeria spp. All isolates were identified to the species level by the API Listeria kit (Bio-Mérieux), according to the manufacturer's instructions. Hemolysis on horse blood MH agar and phenotype displayed on ALOA, a chromogenic medium for L. monocytogenes detection in food (Laboratoire AES, Combourg, France), were used as additional tests.
PCR-RFLP identification.
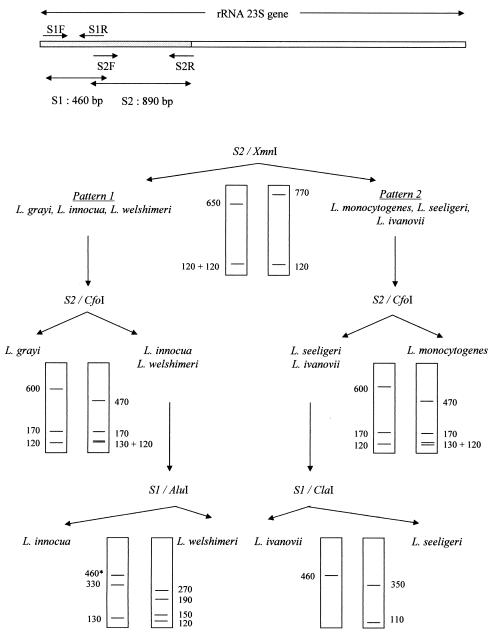
For DNA fragment amplification and restriction endonuclease selection, 23S rRNA gene sequences of Listeria species available in the GenBank database (NC_003210 and Glaser 2001; accession numbers X92948, X92949, X92950, X92951, X92953, and X92954) were aligned and analyzed by use of the Infobiogen restriction program (http:\\www.infobiogen.fr). The theoretical banding patterns revealed a good discrimination between all six Listeria species for two fragments of the gene and four restriction enzymes, following a two- or three-digestion scheme (Fig. 1). Only the typical major bands were taken into account, the minor bands being often nondiscriminative and inconstantly visible at the bottom of the gels (Fig. 2).
FIG. 1.
Scheme used to identify Listeria species by PCR-RFLP on two fragments, S1 (460 bp) and S2 (890 bp), of the 23S rRNA gene, amplified by use of the primer pairs S1R-S1F and S2R-S2F, respectively. S2 was first cut with the restriction enzyme XmnI, giving either pattern 1, for L. grayi, L. innocua, and L. welshimeri, or pattern 2, for L. monocytogenes, L. seeligeri, and L. ivanovii. With pattern 1, restriction of S2 by CfoI individualized L. grayi and digestion of S1 by AluI differentiated L. innocua and L. welshimeri. With pattern 2, restriction of S2 by CfoI characterized L. monocytogenes and digestion of S1 by ClaI discriminated between L. seeligeri and L. ivanovii. The sizes of the bands are indicated in base pairs. *, unrestricted PCR fragment.
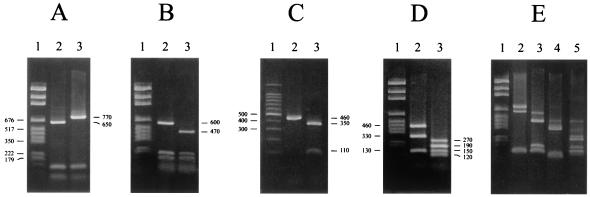
FIG. 2.
DNA restriction patterns of PCR-amplified fragments S1 and S2. The sizes of the fragments are indicated in base pairs. (A) S2 digested by XmnI. Lane 1, size marker; lane 2, pattern 1 (L. grayi, L. innocua, or L. welshimeri); lane 3, pattern 2 (L. monocytogenes, L. seeligeri, or L. ivanovii). (B) S2 digested by CfoI. Lane 1, size marker; lane 2, profile 1 (L. grayi, L. seeligeri, or L. ivanovii); lane 3, profile 2 (L. monocytogenes, L. innocua, or L. welshimeri). (C) S1 digested by ClaI. Lane 1, size marker; lane 2, profile 1 (L. ivanovii); lane 3, profile 2 (L. seeligeri, L. grayi, L. monocytogenes, L. innocua, or L. welshimeri). (D) S1 digested by AluI. Lane 1, size marker; lane 2, profile 1 (L. innocua, L. grayi, or L. monocytogenes); lane 3, profile 2 (L. welshimeri, L. seeligeri, or L. ivanovii). (E) Profiles obtained with mixed cultures. Lane 1, size marker; lane 2, S2 of a mixed culture of L. monocytogenes and L. innocua digested by XmnI; lane 3, S2 of a mixed culture of L. seeligeri and L. monocytogenes digested by CfoI; lane 4, S1 of a mixed culture of L. ivanovii and L. seeligeri digested by ClaI; lane 5, S1 of a mixed culture of L. innocua and L. welshimeri digested by AluI.
For the cell lysis procedure, several colonies were suspended in 100 μl of sterile distilled water. This suspension was immersed in boiling water for 10 min and then placed in cold ice for 5 min. Next, it was centrifuged at 10,000 × g at ambient temperature for 5 min, and the supernatant was kept.
For DNA amplification, a final volume of 50 μl was used, containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.2 mM (each) deoxyribonucleoside triphosphates, 1.5 U of Taq polymerase (Applied Biosystems, Courtabœuf, France), 10 μl of total cell lysate, and 0.5 μM (each) designed laboratory primer for every reaction. Primers S1F (5′-AGTCGGATAGTATCCTTAC-3′) and S1R (5′-GGCTCTAACTACTTGTAGGC-3′) were chosen to amplify a 460-bp DNA fragment (S1) (Fig. 1), and primers S2F (5′-GCCTACAAGTAGTTAGAGCC-3′) and S2R (5′-ACTGGTACAGGAATCTCTAC-3′) were selected to obtain an 890-bp DNA fragment (S2) (Fig. 1). Primers Lis1B, MonoA, Ino2, and Sel1 (6) were used to amplify fragments of the iap gene to confirm the species identification when API and PCR-RFLP data were discordant. Primers hly-a and hly-b (22) were used to amplify the L. monocytogenes hemolysin gene (hlyA). All of these PCR experiments were performed under the following conditions: denaturation at 94°C for 5 min, followed by 35 cycles at 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min and a final cycle at 72°C for 7 min. PCR products were visualized by gel electrophoresis using 1.5% agarose gels containing ethidium bromide.
Endonuclease digestion reactions were carried out separately for each enzyme by incubating 12.5 μl of the amplification products with 0.3 U of the endonuclease XmnI, CfoI, AluI, or ClaI (Promega, Charbonnières, France) per μl (Fig. 1) in a final volume of 15 μl containing the corresponding buffer. The reaction mixture was incubated for 2 h at 37°C. Digestion products were visualized by gel electrophoresis using 2% agarose gels containing ethidium bromide. The molecular sizes of the obtained fragments were estimated by comparison with the 100-bp DNA ladder and pGEM DNA markers (Promega).
16S rRNA gene sequencing.
For 12 strains, DNA amplification was achieved as described above, with 0.5 μM (each) universal primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1489R (5′-TACCTTGTTACGACTTCA-3′), allowing amplification of the entire 16S rRNA gene. PCR experiments were conducted under the following conditions: denaturation at 95°C for 15 min, followed by 35 cycles at 95°C for 1 min, 52°C for 1 min, and 72°C for 1 min 30 s and a final cycle at 72°C for 15 min. PCR products (≈1,400 bp) were visualized by electrophoresis using 1.5% agarose gels containing ethidium bromide.
Sequence reactions using the primers 8F, 1489R, 8F2 (5′-TAACTACGTGCCAGCAGCCG-3′), and 1489R2 (5′-GAAGGGAAAGCTCTGTCTC-3′) were performed by use of the DYEnamic ET Terminator cycle sequencing kit (Amersham, Orsay, France) according to the manufacturer's recommendations. Cycles were as follows: 25 cycles at 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Sequences were analyzed on an automatic ABI 310 sequencer (Perkin Elmer, Courtabœuf, France), using Sequencing Analysis software, and were compared to homologous sequences with Sequence Navigator software.
The 16S ribosomal DNA (rDNA) sequences were aligned from positions 64 to 1459 (according to the Escherichia coli numbering), including gaps, with the CLUSTAL W program. Distance matrices were calculated with the DNADIST program using the algorithm of Jukes and Cantor (23) within the PHYLIP package (J. Felsenstein, University of Washington, Seattle). The phylogenetic tree was inferred from evolutionary distances with the FITCH program of the PHYLIP package (14). The confidence level of the phylogenetic tree topology was evaluated by performing 100 bootstrap replications with the programs SEQBOOT and CONSENSE of the same package.
Nucleotide sequence accession numbers.
The tree sequences obtained in the present work were deposited with EMBL, under the following accession numbers: AJ535693 (strain L169), AJ535694 (L23), AJ535695 (L107), AJ535696 (L151), AJ535697 (L44), AJ535698 (L50), AJ535699 (L52), AJ535700 (L87), and AJ535701 (L97), AJ549928 (L. innocua ATCC 51742), AJ549929 (L. monocytogenes ATCC 19115), and AJ549930 (L. ivanovii subsp. ivanovii ATCC 19119).
RESULTS
Biochemical identification of Listeria species.
The API Listeria system allows a 24-h identification of all Listeria species, based on 10 sugar fermentation reactions and enzymatic reactions in a microtube format, normally without any need for additional tests. All of the ATCC strains were accurately identified to the species level by the kit. Of the environmental strains, 95 were designated L. monocytogenes, 71 were L. innocua, and 1 was L. seeligeri (Table 1). For 15 isolates, no definitive identification was obtained, since several species were proposed for 13 isolates and none for 2 isolates. On horse blood MH agar plates, the ATCC strains of L. monocytogenes, L. ivanovii, and L. seeligeri were hemolytic, while no hemolysis was observed with L. grayi, L. innocua, and L. welshimeri. Of the 182 environmental strains, 92 were hemolytic, 68 were nonhemolytic, and 22 showed a combination of both phenotypes. The ALOA selective medium differentially detects Listeria spp. by their β-glucosidase activity, which translates into bluish colonies, and the pathogenic species L. monocytogenes and L. ivanovii by their phosphatidylinositol-specific phospholipase C (PI-PLC) capacity, leading to an opaque halo around the colonies. PI-PLC production correlated with hemolysis except for four hemolytic, PI-PLC-negative strains.
TABLE 1.
Correlation between RFLP-PCR method and API Listeria kit for identifying 182 environmental strains of Listeria
| Listeria species | No. identified by API Listeria kit | No. identified by RFLP-PCRa
|
||||
|---|---|---|---|---|---|---|
| L. monocytogenes | L. innocua | L. seeligeri | L. welshimeri | Mixture | ||
| L. monocytogenes | 95 | 84 | 5 | 6 | ||
| L. innocua | 71 | 9 | 46 | 1 | 15 | |
| L. seeligeri | 1 | 1 | ||||
| L. welshimeri | ||||||
| Not identified | 15 | 1 | 10 | 3 | 1 | |
| Total | 182 | 94 | 61 | 4 | 1 | 22 |
Concordant identifications between the two methods are indicated in bold.
PCR-RFLP identification of Listeria species.
The Infobiogen program helped us to find two fragments of the 23S rRNA gene with species-specific sites for restriction by four enzymes. A large DNA fragment of 890 bp (S2) was used for XmnI or CfoI restriction, and a small DNA fragment of 460 bp (S1) was used for AluI or ClaI digestion, according to the scheme presented in Fig. 1 and 2. This PCR-RFLP method was first optimized with the six reference strains of Listeria. A large amount of DNA was amplified and no nonspecific bands were obtained with the PCR conditions described here, using chromosomal DNA from boiled bacteria. Since several species of Listeria, principally L. monocytogenes and L. innocua, may coexist in food and environmental samples, an artificial mixture composed of both species was prepared and tested by the PCR-RFLP method. The XmnI restriction enzyme created two bands, of 770 and 650 bp, respectively (Fig. 2E). Identical results were obtained when two species of each pattern were combined. When two species of the same pattern were pooled, the discrimination was achieved at the second or third step, and all mixtures of two species could be detected with the PCR-RFLP method (Fig. 2E). When more than two species were mixed together, many bands were present at the second and third steps and the results were not interpretable.
Of the 182 environmental isolates, 160 were unambiguously identified by PCR-RFLP as 94 L. monocytogenes, 61 L. innocua, 4 L. seeligeri, and 1 L. welshimeri (Table 1). When biochemical and molecular methods gave conflicting data (29 strains), a third method, multiplex PCR of the iap gene (6), was performed. For 27 isolates, the PCR-RFLP identification was confirmed, and for 2 isolates, the multiplex PCR agreed with the API system. The latter isolates (L87 and L151) were designated L. monocytogenes by the PCR-RFLP method and L. innocua by the kit and by amplification of the iap gene. The remaining 22 isolates (12.1%) gave an XmnI PCR-RFLP pattern comprising two fragments, of 770 and 650 bp, consistent with the superimposition of the L. monocytogenes and L. innocua patterns. These data were confirmed by multiplex PCR of the iap gene (6), which showed amplification of both species-specific fragments (data not shown). Isolation on MH blood agar and/or ALOA medium was then performed to separate the strains, and a new PCR-RFLP was carried out. The 22 isolates actually consisted of a combination of the two most frequent species, L. monocytogenes and L. innocua. Therefore, the 182 initial isolates divided into 160 single strains and 22 mixtures.
16S rRNA gene sequences.
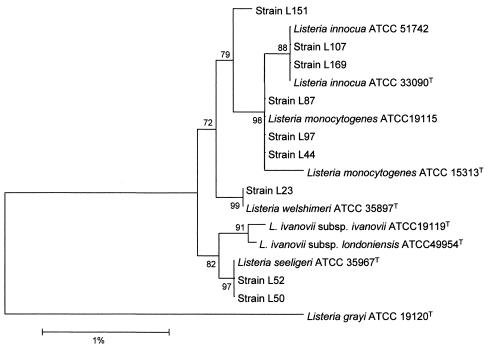
A phylogenetic tree was reconstructed using the 16S rRNA sequences of the seven type strains of Listeria spp. available in GenBank, and the sequences were determined for three reference and nine environmental strains examined in this study (Fig. 3). This analysis showed that all strains exhibited a high level of similarity. Nevertheless, the L. grayi representative strain clearly formed a subline distinct from the other highly interrelated species, in particular from L. monocytogenes and L. innocua. The randomly selected environmental strains (L23, L44, L50, L52, L97, L107, and L169) were very similar to the four corresponding reference strains. According to the 16S rRNA gene sequence analysis, strain L87 was more closely related to the L. monocytogenes cluster than to the L. innocua cluster, consistent with classification by our PCR-RFLP method. However, this strain did not harbor the hlyA gene (data not shown). In contrast, strain L151 possessed this gene (data not shown) but was distant from both clusters. Sequencing of the 23S rRNA region encompassing the XmnI restriction site in L87 and L151 revealed complete homology with the ATCC L. monocytogenes sequences.
FIG. 3.
Phylogenetic tree reconstructed with 16 rDNA sequences of Listeria strains. The bar indicates 1% difference in nucleotide sequence. The numbers in the tree indicate the significance (percent of outcomes) of the branches (bootstrap analysis).
Comparison of the biochemical and PCR-RFLP methods.
Based on the concordant identifications obtained by at least two methods (except for the two unclassifiable strains L87 and L151), the API Listeria kit provided correct identification for 131 of the 158 single and identifiable isolates (82.9%), but the 22 mixed cultures remained mostly unsuspected since they did not yield any species name in one instance and they were positively identified as L. innocua or L. monocytogenes in the 21 other cases (Table 1). Of the 180 environmental isolates, 88 L. monocytogenes strains were hemolytic and PI-PLC positive and 66 strains (59 L. innocua, 6 L. monocytogenes, and 1 L. welshimeri) were nonhemolytic and PI-PLC negative, while the 22 mixtures produced both types of colonies; the 4 L. seeligeri isolates were hemolytic and PI-PLC negative. PCR-RFLP correctly identified all of the single isolates and the mixtures. From a practical point of view, the PCR-RFLP method took about 6 h, owing to the use of DNA from boiled bacteria, compared to more than one working day for multiplex PCR of the iap gene because of the DNA extraction step required.
DISCUSSION
Polymorphism of rRNA genes is commonly used to characterize bacterial species (28). However, for the Listeria genus, such differentiation is difficult because of the highly conserved nature of these genes. Indeed, in the 16S rRNA sequence, only two domains appear to vary among Listeria species (8, 10), while only four signature regions have been identified (39) in the 23S rRNA, although it is twice as large (ca. 2.9 kb). Nevertheless, like DNA-DNA hybridization results (17, 37), 16S and 23S rRNA sequence data have provided evidence that the originally monospecific genus Listeria is composed of six very closely related species, divided into two lines of descent as follows: (i) L. grayi and (ii) the other Listeria spp., the latter subgroup being in turn formed by two distinct evolutionary branches, the first including L. monocytogenes and L. innocua and the second encompassing L. ivanovii, L. seeligeri, and L. welshimeri (8, 39). Since most phylogenetic investigations have been done at the 16S rDNA level, during preliminary studies we performed computer-assisted alignment and restriction analysis of the corresponding available sequences in Listeria spp. to select fragments for amplification and enzymes for their species-specific restriction. However, large amounts of extracted DNA were necessary and many nonspecific bands were obtained (data not shown). In contrast, similar comparison of the 23S rDNA sequences led to a PCR-RFLP assay on boiled DNA, which allowed the precise identification of each Listeria species.
The PCR-RFLP method was first validated on six reference strains, representative of each Listeria species, and then applied to 182 Listeria isolates collected from effluents of treatment plants, in comparison with the API Listeria system. The PCR-RFLP method unequivocally identified 160 environmental strains, including 131 in conformity with the kit, and revealed that 22 isolates were mixed cultures of L. monocytogenes and L. innocua, mostly undetected by the API kit. Discordances between the PCR-RFLP method and the kit were resolved by a multiplex PCR on the invasion-associated protein p60 (iap) gene common to all Listeria species but with internal species-specific portions (6). This method differentiates L. monocytogenes, L. innocua, and L. grayi at the species level, but the L. ivanovii-L. seeligeri-L. welshimeri group has to be further differentiated by additional PCRs. This method also enables coidentification of two Listeria species if they belong to the four different groups (6). Multiplex PCR on the iap gene confirmed the PCR-RFLP data for 49 of the 51 discordances, including the 22 mixtures of L. monocytogenes and L. innocua. In contrast, the multiplex PCR agreed with the API system for two strains of L. innocua, designated L. monocytogenes by the PCR-RFLP method. Sequencing of the 16S rRNA gene for 12 selected strains further supported these findings. Indeed, the phylogenetic tree reconstructed with nine collection strains had a topology consistent with those previously published (17, 39, 44). Moreover, the seven randomly selected environmental strains shared the highest homology with the corresponding reference strains, thus confirming the validity of the PCR-RFLP method. Of the two strains with conflicting identification, strain L87 was more closely related to the L. monocytogenes than to the L. innocua cluster but did not carry the hlyA gene as L. monocytogenes strains do. Strain L151 contained this gene, but could not definitively be identified at the species level on a phylogenetic basis. Other investigations, such as a DNA-DNA hybridization, would be necessary to determine the species affiliation of both strains. Apart from these two atypical strains, the 158 single isolates were 92 L. monocytogenes, 61 L. innocua, 4 L. seeligeri, and 1 L. welshimeri. In the absence of a “gold standard” method, concordant identification obtained by at least two methods (L87 and L151 being omitted) was taken into account. On this basis, the PCR-RFLP exhibited 100% accuracy. Other molecular methods claiming to identify Listeria strains to the species level either target differently conserved chromosomal regions (11, 16, 30, 43), including the iap gene (6, 7, 46), or belong to the epidemiological typing armamentarium, i.e., are usually utilized to type individual strains belonging to a single species (ribotyping, randomly amplified polymorphic DNA, REP- and ERIC-PCR, pulsed-field gel electrophoresis, and multilocus enzyme electrophoresis) (4, 5, 12, 18-20). However, depending on their discriminatory power, either they fail to distinguish all species (18, 30, 43) or they give puzzling intraspecies (often serotype-specific) differentiation (4, 7, 11, 12, 16, 19, 20, 43). Multilocus enzyme electrophoresis involves species-specific reagents (5), and the microarray-based assay using multiple oligoprobes for virulence factor genes is intended for industrial use (46). The multiplex iap PCR method (6) escapes these pitfalls but requires chromosomal DNA extraction and thus more than one working day for completion.
The API Listeria system is the most routinely used biochemical method for identifying Listeria species. In this study, the kit correctly identified 82.9% of the single isolates, gave 8.2% misidentifications, and gave no definitive identification for 8.9% of the strains. In its earliest evaluation (3), the kit accurately identified 85% of 646 Listeria strains: there was one misidentification of an L. ivanovii strain, three unidentified isolates (one L. ivanovii and two L. welshimeri), and 94 isolates needing additional tests to be precisely identified. In another work (21), the API system properly identified to the species level 88.6% of 44 Listeria strains; 3 L. monocytogenes, 1 L. innocua, and 1 L. welshimeri strain gave multiple-species, ambiguous, or no identification. For other investigators (31), the gallery exactly characterized 97% of 350 Listeria strains, with no misidentifications, but provided equivocal profiles for 6 L. monocytogenes and 3 L. innocua isolates. In our study, the high proportion of confusions between L. monocytogenes and L. innocua with the API Listeria system was related in 12 of 14 cases to the arylamidase DIM differentiation innocua-monocytogenes) reaction. Indeed, differentiation between both species relies on this unique characteristic (3), and a positive or negative result depends on the visual appreciation of a color shade. As a consequence, the kit misidentified nine strains of the most pathogenic species L. monocytogenes as the nonpathogenic L. innocua and did the reverse for three strains. In addition, the color strip missed the presence of L. monocytogenes in 16 mixtures, which may be frequent in polymicrobial food and environmental samples (24, 29, 42).
Hemolysis and PI-PLC activity are distinctive criteria for L. monocytogenes and L. innocua identification and might represent supplementary tests when the commercial kits give inconclusive data. Indeed, hemolysin is recognized as a major virulence factor of L. monocytogenes (44). However, other Listeria species (i.e., L. ivanovii and L. seeligeri) are hemolytic, and hemolysin expression in L. monocytogenes is highly variable depending on the strain and the technique employed (13, 15, 21, 25, 31). Accordingly, in our study, seven L. monocytogenes strains were nonhemolytic. ALOA medium differentiates L. monocytogenes and L. ivanovii from the remainder of the genus by their PI-PLC production (1, 24, 35, 45), like other recently proposed chromogenic media (24). Although easier to visualize, the resulting opaque halo was, as expected (24, 44), correlated with hemolysis except for the L. seeligeri strains.
In conclusion, the present PCR-RFLP method allows rapid and accurate identification of the six species of the Listeria genus. Furthermore, in contrast with the API Listeria kit, it can detect different Listeria species contaminating the same sample. Thus, it is particularly useful for identifying Listeria spp. isolated from food products and environment samples but might also be of benefit in clinical and veterinary microbiology for confirming identification of rare species or atypical strains of Listeria spp.
Acknowledgments
This work was supported by a grant from the Agence De l'Environnement et de la Maîtrise de l'Energie (ADEME) and a grant from the Ministère de l'Education Nationale et de la Recherche (EA 525).
We thank Laure Coulange for her technical assistance.
REFERENCES
- 1.Bauwens, L., F. Vercammen, and A. Hertsens. 2003. Detection of pathogenic Listeria spp. in zoo animal faeces: use of immunomagnetic separation and a chromogenic isolation medium. Vet. Microbiol. 91:115-123. [DOI] [PubMed] [Google Scholar]
- 2.Beumer, R. R., M. C. te Giffel, M. T. Kok, and F. M. Rombouts. 1996. Confirmation and identification of Listeria spp. Lett. Appl. Microbiol. 22:448-452. [DOI] [PubMed] [Google Scholar]
- 3.Bille, J., B. Catimel, E. Bannerman, C. Jacquet, M. N. Yersin, I. Caniaux, D. Monget, and J. Rocourt. 1992. API Listeria, a new and promising one-day system to identify Listeria isolates. Appl. Environ. Microbiol. 58:1857-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, S. F., D. R. Gray, D. R. Fenlon, and R. G. Kroll. 1995. Rapid RAPD analysis to distinguish Listeria species and Listeria monocytogenes serotypes using a capillary air thermal cycler. Lett. Appl. Microbiol. 20:188-190. [DOI] [PubMed] [Google Scholar]
- 5.Boerlin, P., J. Rocourt, and J. C. Piffaretti. 1991. Taxonomy of the genus Listeria by using multilocus enzyme electrophoresis. Int. J. Syst. Bacteriol. 41:59-64. [DOI] [PubMed] [Google Scholar]
- 6.Bubert, A., I. Hein, M. Rauch, A. Lehner, B. Yoon, W. Goebel, and M. Wagner. 1999. Detection and differentiation of Listeria spp. by a single reaction based on multiplex PCR. Appl. Environ. Microbiol. 65:4688-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocolin, L., K. Rantsiou, L. Iacumin, C. Cantoni, and G. Comi. 2002. Direct identification in food samples of Listeria spp. and Listeria monocytogenes by molecular methods. Appl. Environ. Microbiol. 68:6273-6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, M. D., S. Wallbanks, D. J. Lane, J. Shah, R. Nietupski, J. Smida, M. Dorsch, and E. Stackebrandt. 1991. Phylogenetic analysis of the genus Listeria based on reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:240-246. [DOI] [PubMed] [Google Scholar]
- 9.Cummins, A. J., A. K. Fielding, and J. McLauchlin. 1994. Listeria ivanovii infection in a patient with AIDS. J. Infect. 28:89-91. [DOI] [PubMed] [Google Scholar]
- 10.Czajka, J., N. Bsat, M. Piani, W. Russ, K. Sultana, M. Wiedmann, R. Whitaker, and C. A. Batt. 1993. Differentiation of Listeria monocytogenes and Listeria innocua by 16S rRNA genes and intraspecies discrimination of Listeria monocytogenes strains by random amplified polymorphic DNA polymorphisms. Appl. Environ. Microbiol. 59:304-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drebot, M., S. Neal, W. Schlech, and K. Rozee. 1996. Differentiation of Listeria isolates by PCR amplicon profiling and sequence analysis of 16S-23S rRNA internal transcribed spacer loci. J. Appl. Bacteriol. 80:174-178. [DOI] [PubMed] [Google Scholar]
- 12.Farber, J. M., and C. J. Addison. 1994. RAPD typing for distinguishing species and strains in the genus Listeria. J. Appl. Bacteriol. 77:242-250. [DOI] [PubMed] [Google Scholar]
- 13.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:479-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 15.Fujisawa, T., and M. Mori. 1994. Evaluation of media for determining hemolytic activity and that of API Listeria system for identifying strains of Listeria monocytogenes. J. Clin. Microbiol. 32:1127-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham, T. A., E. J. Golsteyn-Thomas, J. E. Thomas, and V. P. Gannon. 1997. Inter- and intraspecies comparison of the 16S-23S rRNA operon intergenic spacer regions of six Listeria spp. Int. J. Syst. Bacteriol. 47:863-869. [DOI] [PubMed] [Google Scholar]
- 17.Hartford, T., and P. H. Sneath. 1993. Optical DNA-DNA homology in the genus Listeria. Int. J. Syst. Bacteriol. 43:26-31. [DOI] [PubMed] [Google Scholar]
- 18.Howard, P. J., K. D. Harsono, and J. B. Luchansky. 1992. Differentiation of Listeria monocytogenes, Listeria innocua, Listeria ivanovii, and Listeria seeligeri by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 58:709-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacquet, C., S. Aubert, N. El Solh, and J. Rocourt. 1992. Use of rRNA gene restriction patterns for the identification of Listeria species. Syst. Appl. Microbiol. 15:42-46. [Google Scholar]
- 20.Jersek, B., E. Tcherneva, N. Rijpens, and L. Herman. 1996. Repetitive element sequence-based PCR for species and strain discrimination in the genus Listeria. Lett. Appl. Microbiol. 23:55-60. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, J. L., and C. P. Lattuada. 1993. Comparison of nucleic acid hybridization assays and biochemical characterization tests for the confirmation of Listeria monocytogenes. J. Food Prot. 56:834-840. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, W. M., S. D. Tyler, E. P. Ewan, F. E. Ashton, G. Wang, and K. R. Rozee. 1992. Detection of genes coding for listeriolysin and Listeria monocytogenes antigen A (ImaA) in Listeria spp. by the polymerase chain reaction. Microb. Path. 12:79-86. [DOI] [PubMed] [Google Scholar]
- 23.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. E. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, New York, N.Y.
- 24.Karpiskova, R., M. Pejchalova, J. Mokrosova, J. Vytrasova, P. Smuharova, and J. Ruprich. 2000. Application of a chromogenic medium and the PCR method for the rapid confirmation of L. monocytogenes in foodstuffs. J. Microbiol. Methods 41:267-271. [DOI] [PubMed] [Google Scholar]
- 25.Lachica, R. V. 1996. Hemolytic activity reevaluation of putative nonpathogenic Listeria monocytogenes strains. Appl. Environ. Microbiol. 62:4293-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lessing, M. P., G. D. Curtis, and I. C. Bowler. 1994. Listeria ivanovii infection. J. Infect. 29:230-231. [DOI] [PubMed] [Google Scholar]
- 27.Low, J. C., and W. Donachie. 1997. A review of Listeria monocytogenes and listeriosis. Vet. J. 153:9-29. [DOI] [PubMed] [Google Scholar]
- 28.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 29.MacGowan, A. P., K. Bowker, J. McLauchlin, P. M. Bennett, and D. S. Reeves. 1994. The occurrence and seasonal changes in the isolation of Listeria spp. in shop bought food stuffs, human faeces, sewage and soil from urban sources. Int. J. Food Microbiol. 21:325-334. [DOI] [PubMed] [Google Scholar]
- 30.Manzano, M., L. Cocolin, C. Cantoni, and G. Comi. 2000. Temperature gradient gel electrophoresis of the amplified product of a small 16S rRNA gene fragment for the identification of Listeria species isolated from food. J. Food Prot. 63:659-661. [DOI] [PubMed] [Google Scholar]
- 31.McLauchlin, J. 1997. The identification of Listeria species. Int. J. Food Microbiol. 38:77-81. [DOI] [PubMed] [Google Scholar]
- 32.Meer, R. R., and D. L. Park. 1995. Immunochemical detection methods for Salmonella spp., Escherichia coli O157:H7, and Listeria monocytogenes in foods. Rev. Environ. Contam. Toxicol. 142:1-12. [DOI] [PubMed] [Google Scholar]
- 33.Norton, D. M. 2002. Polymerase chain reaction-based methods for detection of Listeria monocytogenes: toward real-time screening for food and environmental samples. J. Assoc. Off. Anal. Chem. Int. 85:505-515. [PubMed] [Google Scholar]
- 34.Olsen, J. E., S. Aabo, W. Hill, S. Notermans, K. Wernars, P. E. Granum, T. Popovic, H. N. Rasmussen, and O. Olsvik. 1995. Probes and polymerase chain reaction for detection of food-borne bacterial pathogens. Int. J. Food Microbiol. 28:1-78. [DOI] [PubMed] [Google Scholar]
- 35.Paziak-Domanska, B., E. Boguslawska, M. Wieckowska-Szakiel, R. Kotlowski, B. Rozalska, M. Chmiela, J. Kur, W. Dabrowski, and W. Rudnicka. 1999. Evaluation of the API test, phosphatidylinositol-specific phospholipase C activity and PCR method in identification of Listeria monocytogenes in meat foods. FEMS Microbiol. Lett. 171:209-214. [DOI] [PubMed] [Google Scholar]
- 36.Rocourt, J., H. Hof, A. Schrettenbrunner, R. Malinverni, and J. Bille. 1986. Acute purulent Listeria seeligeri meningitis in an immunocompetent adult. Schweiz. Med. Wochenschr. 116:248-251. [PubMed] [Google Scholar]
- 37.Rocourt, J., F. Grimont, P. A. D. Grimont, and H. P. Seeliger. 1982. DNA relatedness among serovars of Listeria monocytogenes sensu lato. Curr. Microbiol. 7:383-388. [Google Scholar]
- 38.Sado, P. N., K. C. Jinneman, G. J. Husby, S. M. Sorg, and C. J. Omiecinski. 1998. Identification of Listeria monocytogenes from unpasteurized apple juice using rapid test kits. J. Food Prot. 61:1199-1202. [DOI] [PubMed] [Google Scholar]
- 39.Sallen, B., A. Rajoharison, S. Desvarenne, F. Quinn, and C. Mabilat. 1996. Comparative analysis of 16S and 23S rRNA sequences of Listeria species. Int. J. Syst. Bacteriol. 46:669-674. [DOI] [PubMed] [Google Scholar]
- 40.Schuchat, A., B. Swaminathan, and C. V. Broome. 1991. Epidemiology of human listeriosis. Clin. Microbiol. Rev. 4:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swaminathan, B., and P. Feng. 1994. Rapid detection of food-borne pathogenic bacteria. Annu. Rev. Microbiol. 48:401-426. [DOI] [PubMed] [Google Scholar]
- 42.Swaminathan, B., J. Rocourt, and J. Bille. 1995. Listeria, p. 341-348. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. ASM Press, Washington, D.C.
- 43.Vaneechoutte, M., P. Boerlin, H. V. Tichy, E. Bannerman, B. Jager, and J. Bille. 1998. Comparison of PCR-based DNA fingerprinting techniques for the identification of Listeria species and their use for atypical Listeria isolates. Int. J. Syst. Bacteriol. 48:127-139. [DOI] [PubMed] [Google Scholar]
- 44.Vazquez-Boland, J. A., G. Dominguez-Bernal, B. Gonzalez-Zorn, J. Kreft, and W. Goebel. 2001. Pathogenicity islands and virulence evolution in Listeria. Microb. Infect. 3:571-584. [DOI] [PubMed] [Google Scholar]
- 45.Vlaemynck, G., V. Lafarge, and S. Scotter. 2000. Improvement of the detection of Listeria monocytogenes by the application of ALOA, a diagnostic, chromogenic isolation medium. J. Appl. Microbiol. 88:430-441. [DOI] [PubMed] [Google Scholar]
- 46.Volokhov, D., A. Rasooly, K. Chumakov, and V. Chizhikov. 2002. Identification of Listeria species by microarray-based assay. J. Clin. Microbiol. 40:4720-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker, J. K., J. H. Morgan, J. McLauchlin, K. A. Grant, and J. A. Shallcross. 1994. Listeria innocua isolated from a case of ovine meningoencephalitis. Vet. Microbiol. 42:245-253. [DOI] [PubMed] [Google Scholar]