Abstract
Purpose
To evaluate the individual and interactive effects of polymorphisms in the myocilin (MYOC),optineurin (OPTN), WD repeat domain 36 (WDR36), and apolipoprotein E (APOE) genes on primary open-angle glaucoma (POAG) in northern Chinese.
Methods
Northern Chinese study subjects, 176 POAG patients and 200 controls, were recruited for screening of the coding exons and splicing regions of MYOC. Five single nucleotide polymorphisms (SNPs) in OPTN (M98K, R545Q, IVS5+38T>G, IVS8–53T>C, and IVS15+10G>A), one SNP in WDR36 (IVS5+30C>T) as well as the APOE promoter and ε2/ε3/ε4 polymorphisms were also examined. Association analysis was performed by using χ2 analysis. High-order gene-gene interaction was also analyzed using the multifactor dimensionality reduction (MDR) method.
Results
In MYOC, 22 variants were identified. Four of them were novel but found in controls only. The missense mutation, Val53Ala, is likely a glaucoma causing mutation, accounting for 0.6% of cases. No individual polymorphism in OPTN, WDR36, or APOE was associated with POAG. MDR analysis identified a best 6-factor model for POAG: MYOC IVS2+35A>G, OPTN Met98Lys, OPTN IVS5+38T>G, OPTN IVS8–53T>C, WDR36 IVS5+30C>T, and APOE −491A>T.
Conclusions
The association pattern between the genes, MYOC, OPTN, WDR36, and APOE, and POAG in northern Chinese is different from that of southern Chinese. Disease-causing mutations in MYOC accounted for a small proportion of northern Chinese POAG patients. Common polymorphisms in these genes were not associated with POAG individually but might interactively contribute to the disorder, supporting a polygenic etiology.
Introduction
Glaucoma is a heterogeneous group of optic neuropathies characterized by progressive degeneration of the optic nerve that leads to irreversible loss of vision. It is the second leading cause of blindness worldwide, estimated to affect about 70 million people [1]. Primary open-angle glaucoma (POAG, OMIM 137760) is the major type of primary glaucoma in most populations. It is evidenced as a complex disorder with multiple risk factors, and genetic factors may play an important role in the etiology of this disorder.
So far, at least 22 genetic loci had been linked to POAG [2-4], and three genes have been identified for POAG from the reported loci, myocilin (MYOC, OMIM 601652) [5,6], optineurin (OPTN, OMIM 602432) [7,8], and WD repeat-domain 36 (WDR36, OMIM 609669) [9]. However, mutations in these three genes account for less than 10% of POAG cases. Therefore, it had been assumed that only a portion of POAG follows the classical Mendelian inheritance while others are caused by variants in several genes, each of which contributes minor effects to disease onset and pathogenesis [10-12]. Presumably, many different genes, each with allelic variations, contribute to the observed variability in a trait, with no particular gene having a single dominant effect [13]. It is likely that single nucleotide polymorphisms (SNPs) in a single gene contributes only a small effect to disease [14] while some genetic variations do not cause diseases individually but act through interaction with other genes. Recently, Park’s study [15] had revealed that OPTN could regulate the expression of MYOC primarily through the control of mRNA stability, indicating that interaction exists between the two glaucoma genes. In addition, apolipoprotein E (APOE; OMIM 107741) has also been reported to be a potent modifier gene and may interactively contribute to POAG [10,11]. These findings provided evidence for the multigenic characteristics of POAG.
To date, the reported findings on glaucoma genetics in Chinese are mostly based on southern Chinese of Hong Kong [2-4,11,16,17], while relevant genetic information from the northern Chinese was limited. Recently, however, a genomic analysis revealed that the genetic ancestries of Chinese can be divided into a southern and a northern group [18]. Therefore, in this present study, we evaluated the genetic association of MYOC, OPTN, WDR36, and APOE polymorphisms with POAG in a group of northern Chinese. The distribution patterns of the gene variants between the northern and southern Han populations were also compared to discern whether any differential distributions of gene variants exist.
Moreover, possible gene-gene interactions among the variants in these genes were also evaluated. Statistically, for gene-gene interaction analysis, logistic regression (LR) is a commonly used method in case-control studies. However, it had been suggested that LR is less powerful if individual variables did not have significant main effects. Only variables that contain an independent main effect will be included in the final model [19]. Moreover, LR is less suitable for a large number of polymorphisms as more analyzed parameters may lead to higher likelihood of false positive and false negative results [20]. In contrast, multifactor dimensionality reduction (MDR) is a nonparametric method, which is less problematic in dimensionality, and is useful for analyzing the interactions among a large number of polymorphisms [20-23]. Therefore, LR was used to analyze the pairwise gene-gene interaction if any two gene variants are found to have significant main effects on POAG. Otherwise, MDR was applied to explore the high-order gene-gene interaction that may be involved in the genetic architecture of POAG in the northern Chinese population.
Methods
Case and control study subjects
Unrelated POAG patients and control subjects were recruited from the Eye Center of Beijing Tongren Hospital (Beijing, China). All the subjects are Han Chinese. They came from Beijing or nearby areas, representing a northern Chinese group. Diagnosis of POAG was based on exclusion of congenital glaucoma and secondary causes (trauma, uveitis, or steroid-induced glaucoma), anterior chamber angle open (grade III or IV gonioscopy), optic disc changes (vertical cup-to-disc ratio greater than 0.5, disc hemorrhage, or thin or notched neuroretinal rim), and visual field changes according to Anderson’s criteria [24]. Visual acuity was determined by the Snellen eye chart, intraocular pressure (IOP) by applanation tonometry, and visual field by a perimeter (Humphrey Field Analyzer; Carl Zeiss Meditec, Dublin, CA) with the Glaucoma Hemifield test. Unrelated control subjects were recruited from people attending the Tongren Eye Center for conditions such as senile cataract, floaters, and itchy eyes. They were given the same ophthalmic examinations and were diagnosed not to have glaucoma or other major eye diseases.
In our study, we included subjects with juvenile onset open-angle glaucoma (JOAG) and adult onset POAG. Totally, there were 176 sporadic patients with POAG, 138 males and 38 females, with age at diagnosis ranging from 10 to 82 years (mean±SD: 38.92±16.33) and with the highest recorded IOP before treatment being higher than 21 mmHg. We included control subjects aged 60 or above as they are less likely to develop POAG later in their lives. There were 200 control subjects, 150 males and 50 females, with age ranging from 61 to 85 years (mean±SD: 69.41±5.97) and with the highest recorded IOP of less than 18 mmHg. The study protocol was approved by the Ethics Committee for Human Research of Tongren Hospital and Capital Medical University (Beijing, China) and adhered to the tenets of the Declaration of Helsinki. Informed consent was obtained from all study subjects after explanation of the nature and possible consequences of the study. Venous blood was obtained from the subjects and stored at −20 °C for less than two months before DNA extraction.
Analysis of the MYOC sequences and single nucleotide polymorphisms in OPTN, WDR36, and APOE
Genomic DNA was extracted from 200 μl of whole blood using a QIAmp Blood Kit (Qiagen, Hilden, Germany). To detect any possible novel disease-causing mutations in MYOC, the three coding exons and adjacent sequences of MYOC were screened by polymerase chain reaction (PCR) followed by direct DNA sequencing with a BigDye Terminator DNA sequencing kit on a 3130XL analyzer (Applied Biosystems, Foster City, CA), using the same set of primers that were used in our previous study [17].
As for the other three genes, only polymorphisms that might be informative in POAG genetics were selected for this study. If any significant association was detected, further fine mapping or re-sequencing of that gene will be performed. Accordingly, five SNPs in OPTN (c.603T>A [M98K], c.1944G>A [R545Q], IVS5+38T>G, IVS8–53T>C, and IVS15+10G>A) were selected, as they had been found to have statistical association with POAG or have interaction with other variants of the disease. They were genotyped by direct sequencing according to the protocol of our previous studies [3,16]. In WDR36, IVS5+30C>T had been found to be significantly associated with POAG in Hong Kong Chinese (unpublished data), and therefore was investigated by direct DNA sequencing, using the same pair of primers (primer for the forward strand: 5′-TAG ATT AGT ATC TAA GTC TGT GG-3′ and primer for the reverse strand: 5′-TGT TAT TTA TAG ACA ACC CTC CA-3′). In APOE, the promoter polymorphisms (−491A>T, −427T>C, and −219T>G) and the ε2/ε3/ε4 polymorphisms in exon 4 (c.526C>T for ε2 and c.388T>C for ε4) were investigated by the TaqMan genotyping assays in an ABI Prism 7000 Sequence Detection System (Applied Biosystems) according to the manufacturer’s instructions. The accuracy of genotyping with TaqMan was evaluated by direct sequencing in about one quarter of randomly selected samples according to the protocol of our previous study [11]. Complete matching of results was obtained.
Statistical analysis
Hardy–Weinberg equilibrium (HWE) was tested for each polymorphism by the χ2 test. Allele or genotype frequencies between patients and controls were compared by the χ2 test or Fisher’s exact test. Significant levels for multiple comparisons were corrected by the Bonferroni method. SPSS version15.0 software (SPSS Inc., Chicago, IL) was used.
Pairwise linkage disequilibrium (LD) estimation and expectation-maximization (EM)-based haplotype association analysis were performed for the variants in MYOC, OPTN, and APOE, respectively, using Haploview 4.0 [25].
LR analysis was used for gene-gene interactions only if the variants were detected to have significant main effects. The disease status was set as the dependent variable (POAG=1, control=0) and gene polymorphisms as the independent variables (homozygote=2, heterozygote=1, wild type=0). The LR model was built with parameters for the independent effects of both variants. If significant interaction was detected, stratified analysis will be used to verify the gene-gene interactions. The study subjects will be stratified according to the genotype of one gene followed by the analysis of another gene in a different stratum defined by the genotype of the former gene. Homogeneity of the odds ratios (ORs) in different strata will be tested by the Breslow-Day test.
High order gene-gene interaction models among all the polymorphisms in the four candidate genes were detected and characterized using the multifactor dimensionality reduction (MDR) method as proposed by Ritchie et al. [23]. A detailed explanation on the MDR method has been described elsewhere [23,26,27]. Among the set of best multifactor models, the combination of genetic factors that maximizes the testing accuracy and/or the highest cross-validation consistency (CVC) is selected and further evaluated using permutation testing. p values associated with each prediction error and cross-validation consistency were determined by the Sign Test (a nonparametric test implemented in the MDR software) and corrected by a permutation test. For the best model, the interaction dendrogram, which was generated by MDR, was used to confirm, visualize, and interpret the interaction model. The MDR analysis was performed by using the open-source MDR software package (version 1.0.0, freely available online at Computation Genetics Laboratory).
Results
Univariate analysis of individual polymorphisms in MYOC
The allele and genotype frequencies of the MYOC variants were summarized in Table 1. In total, 22 polymorphisms had been identified. All of them followed HWE. Association analysis showed that none of these polymorphisms was statistically associated with POAG (p>0.0023, Bonferroni corrected significance level). Four variants (Gln161Arg, Gly183Ser, IVS1+16G>T, and Asn428Ser) were novel. They were detected in controls only. In contrast, variants Gly12Arg, Val53Ala, and Thr353Ile were found only in POAG cases with Gly12Arg in three POAG patients and the other two each in one patient. All were heterozygous. According to Alward’s criteria [28], they were likely disease-causing mutations (DCMs).
Table 1. MYOC polymorphisms identified in this study.
| Location | Sequence change | Codon Change |
Allele frequency (%) |
Genotype frequency |
||
|---|---|---|---|---|---|---|
| POAG (n=352) | Control (n=400) | POAG (n=176) | Control (n=200) | |||
| Promoter |
−126 T>C |
- |
0 (0) |
1 (0.25) |
0/0/176 |
0/1/199 |
| Promoter |
−83 G>A |
- |
16 (4.5) |
30 (7.5) |
2/12/162 |
3/24/173 |
| Exon 1 |
c. 34 G>C |
G12R* |
3(0.85) |
0 (0) |
0/3/173 |
0/0/200 |
| Exon 1 |
c. 57 G>T |
Q19H |
0 (0) |
1 (0.25) |
0/0/176 |
0/1//199 |
| Exon 1 |
c. 136 C>T |
R46X |
2 (0.57) |
3 (0.75) |
0/2/174 |
0/3/197 |
| Exon 1 |
c. 158 T>C |
V53A* |
1 (0.28) |
0 (0) |
0/1/175 |
0/0/200 |
| Exon 1 |
c. 227 G>A |
R76K |
13 (3.69) |
28 (7) |
0/13/163 |
1/26/173 |
| Exon 1 |
c. 369 C>T |
T123T |
2 (0.57) |
1 (0.25) |
0/2/174 |
0/1/199 |
| Exon 1 |
c. 482 A>G |
Q161R |
0 (0) |
1 (0.25) |
0/0/176 |
0/1/199 |
| Exon 1 |
c. 547 G>A |
G183S |
0 (0) |
1 (0.25) |
0/0/176 |
0/1/199 |
| Intron 1 |
IVS1+16 G>T |
- |
0 (0) |
1 (0.25) |
0/0/176 |
0/1/199 |
| Exon 2 |
c. 611 C>T |
T204M |
0 (0) |
2 (0.5) |
0/0/176 |
0/2/198 |
| Exon 2 |
c. 624 C>G |
D208E |
1 (0.28) |
3 (0.75) |
0/1/175 |
0/3/197 |
| Intron 2 |
IVS 2+35 A>G |
- |
69 (19.6) |
84 (21) |
9/51/116 |
7/70/123 |
| Intron 2 |
IVS 2+172 C>A |
- |
4 (1.14) |
1 (0.25) |
0/4/172 |
0/1/199 |
| Exon 3 |
c. 864 C>T |
I288I |
1 (0.28) |
1 (0.25) |
0/1/175 |
0/1/199 |
| Exon 3 |
c. 927 G>A |
Q309Q |
0 (0) |
1 (0.25) |
0/0/176 |
0/1/199 |
| Exon 3 |
c. 1041 T>C |
Y347Y |
0 (0) |
1 (0.25) |
0/0/176 |
0/1/199 |
| Exon 3 |
c. 1058 C>T |
T353I* |
1 (0.28) |
0 (0) |
0/1/175 |
0/0/200 |
| Exon 3 |
c. 1283 A>G |
N428S |
0 (0) |
1 (0.25) |
0/0/176 |
0/1/199 |
| Exon 3 |
c. 1464 C>T |
A488A |
0 (0) |
2 (0.5) |
0/0/176 |
0/2/198 |
| 3′UTR | 1515+73 G>C | - | 3 (0.85) | 1 (0.25) | 0/3/173 | 0/1/199 |
MYOC polymorphisms identified by direct DNA sequencing in this study were shown it the table. The asterisk indicates that the change is a disease-causing mutation and was found in POAG patients. The ‘hyphen’ represents non-coding variation. The numbers under “Genotype frequency” are the counts of homozygotes, heterozygotes, and wild type.
Distribution of single nucleotide polymorphisms in OPTN and WDR36
Genotypic frequencies of OPTN (Met98Lys, Arg545Gln, IVS5+38T>G, IVS8–53T>C, and IVS15+10G>A) and WDR36 (IVS5+30C>T) followed HWE in cases and controls. No significant difference was detected between POAG cases and controls in the genotypic or allelic frequencies (Bonferroni corrected significance level, p>0.01; Table 2).
Table 2. OPTN and WDR36 polymorphisms investigated in the present study.
| Gene | Sequence change | Codon change |
Allele frequency (%) |
Genotype frequency |
||
|---|---|---|---|---|---|---|
| POAG (n=352) | Control (n=400) | POAG (n=176) | Control (n=200) | |||
|
OPTN |
c.603 T>A |
M98K |
39 (11.08) |
48 (12) |
3/33/140 |
3/42/155 |
|
OPTN |
c.1944 G>A |
R545Q |
12 (3) |
13 (3.25) |
0/12/164 |
1/11/188 |
|
OPTN |
IVS5+38 T>G |
- |
118 (33.5) |
121 (30.25) |
17/84/75 |
14/93/93 |
|
OPTN |
IVS8 −53 T>C |
- |
27 (7.67) |
24 (6) |
1/25/150 |
1/22/177 |
|
OPTN |
IVS15+10G>A |
- |
8 (2.27) |
7 (1.75) |
0/8/168 |
0/7/193 |
| WDR36 | IVS 5+30 C>T | - | 134 (38.1) | 170 (42.5) | 22/90/64 | 33/104/63 |
Allelic and genotypic frequencies of the five candidate SNPs in the present study were shown in the table. As only one WDR36 SNP was selected, this SNP was also presented in this table. The “-” represents non-coding polymorphism. The numbers under “Genotype frequency” are the counts of homozygotes, heterozygotes, and wildtype.
Univariate analysis of individual polymorphisms in APOE
Genotypes of all the APOE polymorphisms, i.e., −491A>T, −427T>C, −219T>G, and the ε2/ε3/ε4 polymorphism, followed the HWE in both study groups. Their genotypic or allelic distributions were not statistically different between POAG patients and controls (p>0.0125, Bonferroni corrected significance level, Table 3).
Table 3. APOE polymorphisms investigated in the present study.
| Polymorphism |
Allele frequency (%) |
Genotype frequency (%) |
||||
|---|---|---|---|---|---|---|
| Allele | POAG (n=176) | Controls (n=400) | Genotype | POAG (n=176) | Controls (n=200) | |
| −491 A>T |
T |
8 (2.3) |
13 (3.25) |
TT |
0 (0) |
0 (0) |
| |
A |
344 (97.7) |
387 (96.75) |
TA |
8 (4.5) |
13 (6.5) |
| |
|
|
|
AA |
168 (95.5) |
187 (93.5) |
| −427 T>C |
C |
25 (7.1) |
36 (9.0) |
CC |
1 (0.6) |
1 0.5 (0) |
| |
T |
327 (92.9) |
364 (91.0) |
CT |
23 (13.1) |
34 (17.0) |
| |
|
|
|
TT |
152 (86.3) |
165 (82.5) |
| −219 T>G |
G |
93 (26.4) |
117 (29.25) |
GG |
10 (5.7) |
15 (7.5) |
| |
T |
259 (73.6) |
283 (70.75) |
GT |
73 (41.5) |
87 (43.5) |
| |
|
|
|
TT |
73 (52.8) |
98 (49.0) |
| ε2/ε3/ε4 |
ε4 |
38 (10.8) |
36 (9.0) |
ε4/ε4 |
3 (1.7) |
2 (1.0) |
| |
ε2 |
34 (9.7) |
35 (8.75) |
ε2/ε4 |
5 (2.8) |
4 (2.0) |
| |
ε3 |
280 (79.5) |
329 (82.25) |
ε3/ε4 |
29 (16.5) |
28 (14.0) |
| |
|
|
|
ε2/ε3 |
25 (14.2) |
29 (14.5) |
| |
|
|
|
ε2/ε2 |
2 (1.1) |
1 (0.5) |
| ε3/ε3 | 112 (63.6) | 136 (68.0) | ||||
Allelic and genotypic frequencies of the four APOE polymorphisms in case and control subjects were shown in the table. None of them was significantly associated with POAG.
Haplotype association analysis for the variants in MYOC, OPTN, and APOE
For MYOC, LD analysis revealed one LD block, spanning from the promoter (−83G>A) to exon 1 (c.227G>A). Within this block, polymorphisms −83G>A and c.227G>A were the only two variants that had a minor allele frequency (MAF) greater than 1%. They were in strong LD (D’=0.974). The haplotype G-G, defined by the major alleles of these two SNPs, presented in 95.5% of cases and 92.2% of controls. It was not statistically associated with POAG. The minor haplotype A-A was not associated with glaucoma. IVS2+35A>G was the only other MYOC polymorphism that had a MAF greater than 1%. When it was included in haplotype analysis, none of the haplotypes, which were defined by the three SNPs, showed any significant association with POAG (data not shown).
For OPTN, only five common variants were genotyped, and no LD block was detected. We had tried every possible combination of these five SNPs to evaluate haplotype association but found no haplotype significantly associated with glaucoma (data not shown).
For APOE, only the three promoter SNPs and the ε2/ε3/ε4 polymorphism in exon 4 were genotyped, and no LD block was detected. Haplotype analysis showed that the common haplotypes (frequency>5%), which are defined by the three promoter SNPs (−491A>T, −427T>C, and −219T>G), were not associated with POAG. When the ε2/ε3/ε4 polymorphisms (c.526C>T and c.388T>C) were included in haplotype analysis, no common haplotype was found to be significantly associated with glaucoma.
Gene–gene interaction analysis
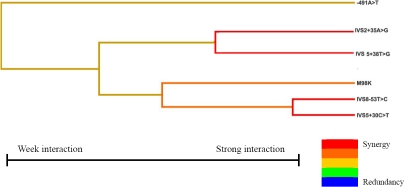
Since we found no variant having strong significant association with POAG, we did not use logistic regression to build the interaction model. By using MDR analysis, six models were formed (Table 4), a best model that included six SNPs from all four genes was identified, including IVS2+35 A>G in MYOC; Met98Lys, IVS5+38T>G, and IVS8–53T>C in OPTN; IVS5+30C>T in WDR36; and −491A>T in APOE. The combination of these possibly interactive polymorphisms in the model gave a maximum CVC of 10/10 and a maximum testing accuracy of 0.5514. The p value associated with the prediction error and CVC was 0.0107 (p<0.001 after corrected by permutation test). Figure 1 illustrates the interaction dendrogram for this model. The hierarchical cluster analysis placed IVS5+30C>T (WDR36), IVS8–53T>C (OPTN), and Met98Lys (OPTN) on the same branch, and their closer position in the diagram clearly showed that the three SNPs may have a synergistic interaction effect on modulating risk of POAG. IVS5+38T>G (OPTN) and IVS2+35A>G (MYOC) were on another branch, revealing an interaction between them. −491A>T (APOE) was located on a different remote branch, suggesting that this SNP may have less of a relationship with other SNPs.
Table 4. Multi-locus interactions by the multifactor dimensionality reduction approach.
| Best candidate model* | Testing accuracy (%) | p value (sign test) | CVC |
|---|---|---|---|
| −219T>G |
43.61 |
0.999 |
5/10 |
| IVS8–53T>C and IVS5+30C>T |
52.5 |
0.377 |
7/10 |
| IVS2+35A>G, IVS5+30C>T, and −219T>G |
43.59 |
1.0 |
4/10 |
| IVS2+35A>G, IVS5+30C>T, −219T>G, and 290C>T |
47.53 |
0.945 |
9/10 |
| A488A, IVS5+38, IVS5+30C>T, −427T>C, and −219T>G |
49.16 |
0.623 |
10/10 |
| IVS2+35A>G, M98K, IVS5+38T>G, IVS8–53T>C, IVS5+30C>T, and −491A>T | 55.14 | 0.0107 | 10/10 |
Interaction models with one to six factors generated by the MDR program were shown in the table. The six-factor model with highest CVC (10/10) and maximum testing accuracy (55.14%) was selected as the best model in this study. The asterisk indicates that as the sample numbers in cases and controls are dissimilar in this study, the T value, which is the threshold ratio used to distinguish high-risk genotype combinations from low risk genotype combinations, was set to 0.88 (number of cases/number of controls, i.e., 176/200) while running the MDR program. CVC: cross-validation consistency.
Figure 1.
Interaction dendrogram for the six polymorphisms modeled by MDR. The colors comprise a spectrum representing a continuum from synergy to redundancy. The red color represents a high degree of synergy (positive information gain), orange a lesser degree, and gold represents the midway point between synergy and redundancy. On the redundancy end of the spectrum (not appeared in our model), the highest degree is represented by the blue color with a lesser degree represented by green.
Discussion
For the first time, we have investigated the association between multiple genes and POAG in a northern Chinese population. In MYOC, we have detected four novel variants, Gln161Arg, Gly183Ser, IVS1+16G>T, and Asn428Ser, each in one control subject, but these variants were not found in POAG patients (Table 1). From published reports, MYOC mutations have been attributed to about 1%-4% of POAG cases, and this rate can be as high as 36% in JOAG families [29]. In the Chinese population of Hong Kong, which is southern Chinese, the prevalence of MYOC mutations is about 1.1%–1.8% in POAG cases [2]. In this present study, the non-synonymous polymorphisms, Gly12Arg, Val53Ala, and Thr353Ile, were found only in POAG cases but not in controls. According to Alward’s criteria [28], they were likely DCMs for POAG. However, a previous study on Hong Kong Chinese POAG patients showed that the Gly12Arg and Thr353Ile variants were not disease-causing; each of them was detected in four control subjects (0.7%) [11]. Therefore, the remaining Val53Ala variant might be the only DCM for POAG in the northern Chinese group, accounting for only 0.6% of POAG cases. Such a mutation rate is lower than that reported in Caucasians and southern Chinese [2,29]. However, since the Gly12Arg variant was detected in 3 of the 176 POAG patients but none of the 200 healthy controls, we could not rule out its possibility of being a POAG-related variant in ethnic northern Chinese. A larger sample size is required to confirm its role on POAG. In addition, we found Gln19His in one control subject, which was previously suggested to be a DCM [11]. This subject was a 69-year-old female with normal fundus. She did not have a family history of glaucoma. Therefore, the Gln19His variant is unlikely a disease-causing mutation for POAG in northern Chinese. Apart from the rare variants, the three common polymorphisms (MAF>1%), −83G>A, R76K, and IVS2+35A>G, were not statistically associated with POAG, consistent with findings in southern Chinese in Hong Kong [11]. The allelic frequencies of these SNPs were also found to be similar between the northern Chinese of this study and the southern Chinese of Hong Kong (Table 5).
Table 5. Allele frequencies of MYOC, OPTN, and APOE variants in southern and northern Chinese.
| Location | Sequence change | Codon change |
Allele frequency in HTG patients (%) |
Comparison (p value) |
Allele frequency in controls (%) |
Comparison (p value) | ||
|---|---|---|---|---|---|---|---|---|
| Southern* (n=588) | Northern (n=352) | Southern (n=562) | Northern (n=400) | |||||
|
MYOC | ||||||||
| Promoter |
−83G>A |
- |
37 (6.3) |
16 (4.5) |
0.26 |
50 (8.9) |
30 (7.5) |
0.44 |
| Exon 1 |
c. 227 G>A |
R76K |
38 (6.5) |
13 (3.7) |
0.07 |
51 (9.1) |
28 (7.0) |
0.25 |
| Intron 2 |
IVS2+35A>G |
- |
119 (20.2) |
69 (19.6) |
0.81 |
91 (16.2) |
84 (21.0) |
0.057 |
|
OPTN | ||||||||
| Exon 5 |
c.603 T>A |
M98K |
100 (17.0) |
39 (11.1) |
0.013 |
88 (15.7) |
48 (12) |
0.11 |
| Exon 16 |
c.1944 G>A |
R545Q |
21 (3.6) |
12 (3.0) |
0.9 |
19 (3.4) |
13 (3.25) |
0.91 |
| Intron 5 |
IVS5+38T>G |
- |
73 (12.4) |
118 (33.5) |
3.8×10–15 |
29 (5.2) |
121 (30.25) |
4.0×10−26 |
| Intron 8 |
IVS8–53T>C |
- |
21 (3.6) |
27 (7.7) |
0.006 |
12 (2.1) |
24 (6.0) |
0.002 |
| Intron 15 |
IVS15+10G>A |
- |
9 (1.5) |
8 (2.3) |
0.41 |
8 (1.4) |
7 (1.75) |
0.69 |
|
APOE | ||||||||
| Promoter |
−491A>T |
- |
28 (4.8) |
8 (2.3) |
0.054 |
15 (2.7) |
13 (3.25) |
0.6 |
| Promoter |
−427 T>C |
- |
5 (0.9) |
25 (7.1) |
1.3×10–7 |
5 (0.9) |
36 (9.0) |
8.4×10−10 |
| Promoter |
−219 T>G |
- |
222 (37.8) |
93 (26.4) |
0.00037 |
187 (33.3) |
117 (29.25) |
0.19 |
| Exon 4 | ε2/ε4/ε3 | R158C, C112R | 63/39/486 (10.7/6.6/82.7) | 34/38/280 (9.7/10.8/79.5) | 0.076 | 48/52/462 (8.5/9.3/82.2) | 35/36/329 (8.8/9.0/82.2) | 0.99 |
Allelic frequencies of MYOC, OPTN, and APOE polymorphisms were compared between southern and northern Chinese. The asterisk indicates that the genetic data of the southern Chinese referred to our previous publication [11. In MYOC, only the three common SNPs (MAF>1%) were shown in the table, and only the frequencies of the minor allele of each polymorphism were shown in the table. The allelic frequencies of the polymorphisms were compared with the χ2 test. The p values were corrected by the Bonferroni method (p<0.05/13=0.0038 was considered statistically significant).
OPTN mutations had been reported to account for 1%–1.6% of sporadic Chinese POAG patients [11,16]. However, later studies reported no glaucoma causing mutations in OPTN among Caucasian and Japanese POAG patients [30-32]. In this study, the five SNPs that were selected to be genotyped had been reported to be either associated with POAG individually or act interactively with other genes on the disease [11,16]. After univariate analysis, none of them showed significant association with glaucoma. However, we found that the allelic distribution of the selected OPTN variants among this present study population was different from the southern Chinese population [11]. The allelic frequencies of the genotyped OPTN variants in the two populations are shown in Table 5. The exonic SNP, Met98Lys, presented a bit lower in the northern Chinese than that in the southern Chinese, but the difference was not statistically different (χ2=2.6, p>0.1 when comparing the allelic frequencies of this variant in the control group in the two studies, 12% versus 15.7% [11]). Another exonic variant, Arg545Gln, and the intronic variant, IVS15+10G>A, also showed comparable allelic distributions among the two Chinese populations. However, IVS5+38T>G, which was significantly associated with POAG in the southern Chinese, was found to be distributed drastically differently between the two populations. It occurred in 12.4% POAG and 5.2% controls in the southern Chinese. In contrast, in the northern Chinese, this SNP was detected in 33.5% of cases and 30.25% of controls, significantly higher than that in the southern Chinese after adjustment for multiple comparison (in cases, χ2=60.59, p=7.0×10−15; in controls, χ2=111.8, p=4.0×10−26). Another intronic SNP, IVS8–53T>C, was also found to have higher allelic frequencies in the northern Chinese, although the p values became borderline after Bonferroni correction (Table 5). Such difference in allelic distributions was also found for APOE −427T>C, which presented at significantly higher frequencies in the northern Chinese than the southern Chinese (Table 5). Such discrepancies can probably be explained by the presence of ancestry-related differences in allele frequencies between the northern and southern Chinese. However, only five SNPs in each of the two genes were investigated in the present study. Our data might have revealed only part of a discrepancy in the distribution pattern of OPTN and APOE polymorphisms. A more thorough investigation of the sequences of these genes is warranted to enable a more comprehensive comparison.
Recent studies suggested that WDR36 defects may contribute to the glaucomatous disease process as a glaucoma modifier gene [33,34]. In a group of southern Chinese POAG patients, only one SNP in WDR36, IVS5+30C>T, showed significant association with POAG, and it is a common polymorphism (unpublished data). However, this SNP did not showed significant association with glaucoma in this study on the northern Chinese.
Findings of this present study suggest that the roles of MYOC, OPTN, WDR36, and APOE on the genetic architecture of POAG are different among northern and southern Chinese. To date, although variations in a variety of genes had shown statistical associations with glaucoma, these associations were often not replicable in other populations. Some of these SNPs may be of functional significance, and their frequencies may vary significantly between different ethnic groups. For example, three DCMs at MYOC (Gly252Arg, Gly367Arg, and Pro370Leu) were found in Asians and Caucasians, and three (Thr293Lys, Thr377Met, and Glu352Lys) were found in Africans and Caucasians. No single mutation was shared by all three ethnic populations, suggesting most DCMs exist in a specific ethnicity [29]. It may also suggest that a single gene locus may not cause glaucoma but act interactively with other gene variants [20]. Evidence for this hypothesis has been reported. Ishikawa et al. [35] found the APOE promoter polymorphism, −491A>T, interacted with MYOC −1000C>G (MYOC.mt1) to increase IOP in POAG patients, but the individual effect of MYOC.mt1 is unclear. Further, Funayama et al. [12] found that common polymorphisms in OPTN and olfactomedin 2 (OLFM2) may interactively contribute to the development of OAG in Japanese patients. In the Chinese population of Hong Kong, Fan et al. [11] detected three pairs of statistical interactions for normal tension glaucoma (between MYOC −83G>A and the APOE ε2/ε3/ε4, MYOC IVS2+35A>G and APOE −219T>G, as well as OPTN Arg545Gln and APOE ε2/ε3/ε4)) and two pairs of interactions for POAG (between MYOC Thr353Ile and OPTN IVS15+10G>A and between OPTN IVS5+38T>G and APOE −491A>T). In this present study, a best interaction model involving six variants in MYOC, OPTN, WDR36, and APOE had been identified for POAG in a northern Chinese population by using MDR. This finding suggested that these genes might act in concert to modulate the risk of POAG, although MYOC attributed a more dominant effect. According to the interaction dendrogram, the variants, WDR36 IVS5+30C>T and OPTN IVS8–53T>C, are expected to have a strong synergistic interaction. OPTN Met98Lys has lesser synergistic interaction with both. Similarly, variants OPTN IVS5+38T>G and MYOC IVS2+35A>G also have strong synergistic interaction themselves but having lesser interaction with the three variants in the first branch. Moreover, APOE −491A>T may have the least degree of interaction with other variants. This dendrogram provided preliminary information about the interactive relationships among the variants in the model. However, one limitation of the MDR method is that it is difficult to evaluate the effects of each individual polymorphism in the models and to authenticate the combination of the risk alleles [36]. These issues still need to be addressed. Another limitation of this study is the lack of replication. As the interaction model detected in this study had not been identified in the southern Chinese or other populations, it should be evaluated in other Chinese populations.
In conclusion, we have for the first time investigated the roles of polymorphisms in MYOC, OPTN, WDR36, and APOE in POAG in a group of northern Chinese. The distributions of some variants were drastically different from that in the general southern Chinese population. The possibly disease-causing mutations in MYOC accounted for only a small proportion of northern Chinese POAG patients. The common polymorphisms in the candidate genes were not significantly associated with glaucoma, although a larger sample of northern Chinese should be required to unravel their contributions to POAG if their effects are mild. However, some of the common polymorphisms in these genes might interactively contribute to POAG, supporting a polygenic etiology of this common and complex disease.
Acknowledgments
This work was supported in part by block grants from Mr. Lai Seung Hung and Mrs. Lai Chan Pui Ngong Fund, Hong Kong and Beijing Natural Science Foundation, 7081001, China, respectively, and by a direct grant, KM200610025025, from Scientific Research Common Program of Beijing Municipal Commission of Education of China. We express our greatest appreciation to all the participants in the study.
References
- 1.Quigley HA. Number of people with glaucoma worldwide. Br J Ophthalmol. 1996;80:389–93. doi: 10.1136/bjo.80.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan BJ, Wang DY, Lam DS, Pang CP. Gene mapping for primary open angle glaucoma. Clin Biochem. 2006;39:249–58. doi: 10.1016/j.clinbiochem.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Pang CP, Fan BJ, Canlas O, Wang DY, Dubois S, Tam PO, Lam DS, Raymond V, Ritch R. A genome-wide scan maps a novel juvenile-onset primary open angle glaucoma locus to chromosome 5q. Mol Vis. 2006;12:85–92. [PubMed] [Google Scholar]
- 4.Wang DY, Fan BJ, Chua JK, Tam PO, Leung CK, Lam DS, Pang CP. A genome-wide scan maps a novel juvenile-onset primary open-angle glaucoma locus to 15q. Invest Ophthalmol Vis Sci. 2006;47:5315–21. doi: 10.1167/iovs.06-0179. [DOI] [PubMed] [Google Scholar]
- 5.Sheffield VC, Stone EM, Alward WL, Drack AV, Johnson AT, Streb LM, Nichols BE. Genetic linkage of familial open angle glaucoma to chromosome 1q21-q31. Nat Genet. 1993;4:47–50. doi: 10.1038/ng0593-47. [DOI] [PubMed] [Google Scholar]
- 6.Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–70. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 7.Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, Heon E, Krupin T, Ritch R, Kreutzer D, Crick RP, Sarfarazi M. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–9. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 8.Sarfarazi M, Child A, Stoilova D, Brice G, Desai T, Trifan OC, Poinoosawmy D, Crick RP. Localization of the fourth locus (GLC1E) for adult-onset primary open-angle glaucoma to the 10p15-p14 region. Am J Hum Genet. 1998;62:641–52. doi: 10.1086/301767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monemi S, Spaeth G, DaSilva A, Popinchalk S, Ilitchev E, Liebmann J, Ritch R, Heon E, Crick RP, Child A, Sarfarazi M. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 2005;14:725–33. doi: 10.1093/hmg/ddi068. [DOI] [PubMed] [Google Scholar]
- 10.Copin B, Brezin AP, Valtot F, Dascotte JC, Bechetoille A, Garchon HJ. Apolipoprotein E-promoter single-nucleotide polymorphisms affect the phenotype of primary open-angle glaucoma and demonstrate interaction with the myocilin gene. Am J Hum Genet. 2002;70:1575–81. doi: 10.1086/340733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan BJ, Wang DY, Fan DS, Tam PO, Lam DS, Tham CC, Lam CY, Lau TC, Pang CP. SNPs and interaction analyses of myocilin, optineurin, and apolipoprotein E in primary open angle glaucoma patients. Mol Vis. 2005;11:625–31. [PubMed] [Google Scholar]
- 12.Funayama T, Mashima Y, Ohtake Y, Ishikawa K, Fuse N, Yasuda N, Fukuchi T, Murakami A, Hotta Y, Shimada N, Glaucome Gene Research Group SNPs and interaction analyses of noelin 2, myocilin, and optineurin genes in Japanese patients with open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006;47:5368–75. doi: 10.1167/iovs.06-0196. [DOI] [PubMed] [Google Scholar]
- 13.Culverhouse R, Suarez BK, Lin J, Reich T. A perspective on epistasis: limits of models displaying no main effect. Am J Hum Genet. 2002;70:461–71. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore JH, Williams SM. Traversing the conceptual divide between biological and statistical epistasis: systems biology and a more modern synthesis. Bioessays. 2005;27:637–46. doi: 10.1002/bies.20236. [DOI] [PubMed] [Google Scholar]
- 15.Park BC, Tibudan M, Samaraweera M, Shen X, Yue BY. Interaction between two glaucoma genes, optineurin and myocilin. Genes Cells. 2007;12:969–79. doi: 10.1111/j.1365-2443.2007.01102.x. [DOI] [PubMed] [Google Scholar]
- 16.Leung YF, Fan BJ, Lam DS, Lee WS, Tam PO, Chua JK, Tham CC, Lai JS, Fan DS, Pang CP. Different optineurin mutation pattern in primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2003;44:3880–4. doi: 10.1167/iovs.02-0693. [DOI] [PubMed] [Google Scholar]
- 17.Pang CP, Leung YF, Fan B, Baum L, Tong WC, Lee WS, Chua JK, Fan DS, Liu Y, Lam DS. TIGR/MYOC gene sequence alterations in individuals with and without primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2002;43:3231–5. [PubMed] [Google Scholar]
- 18.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–4. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 19.Motsinger-Reif AA, Reif DM, Fanelli TJ, Ritchie MD. A comparison of analytical methods for genetic association studies. Genet Epidemiol. 2008;32:767–78. doi: 10.1002/gepi.20345. [DOI] [PubMed] [Google Scholar]
- 20.Heidema AG, Feskens EJ, Doevendans PA, Ruven HJ, van Houwelingen HC, Mariman EC, Boer JM. Analysis of multiple SNPs in genetic association studies: comparison of three multi-locus methods to prioritize and select SNPs. Genet Epidemiol. 2007;31:910–21. doi: 10.1002/gepi.20251. [DOI] [PubMed] [Google Scholar]
- 21.Motsinger AA, Ritchie MD. Multifactor dimensionality reduction: an analysis strategy for modelling and detecting gene-gene interactions in human genetics and pharmacogenomics studies. Hum Genomics. 2006;2:318–28. doi: 10.1186/1479-7364-2-5-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24:150–7. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- 23.Ritchie MD, Motsinger AA. Multifactor dimensionality reduction for detecting gene-gene and gene-environment interactions in pharmacogenomics studies. Pharmacogenomics. 2005;6:823–34. doi: 10.2217/14622416.6.8.823. [DOI] [PubMed] [Google Scholar]
- 24.Anderson DR. Automated static perimetry. St. Louis: Mosby-Year Book; 1992. [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–82. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alward WL, Kwon YH, Khanna CL, Johnson AT, Hayreh SS, Zimmerman MB, Narkiewicz J, Andorf JL, Moore PA, Fingert JH, Sheffield VC, Stone EM. Variations in the myocilin gene in patients with open-angle glaucoma. Arch Ophthalmol. 2002;120:1189–97. doi: 10.1001/archopht.120.9.1189. [DOI] [PubMed] [Google Scholar]
- 29.Gong G, Kosoko-Lasaki O, Haynatzki GR, Wilson MR.Geneticdissection of myocilin glaucoma. Hum Mol Genet 200413 Spec No 1R91–102. [DOI] [PubMed] [Google Scholar]
- 30.Alward WL, Kwon YH, Kawase K, Craig JE, Hayreh SS, Johnson AT, Khanna CL, Yamamoto T, Mackey DA, Roos BR, Affatigato LM, Sheffield VC, Stone EM. Evaluation of optineurin sequence variations in 1,048 patients with open-angle glaucoma. Am J Ophthalmol. 2003;136:904–10. doi: 10.1016/s0002-9394(03)00577-4. [DOI] [PubMed] [Google Scholar]
- 31.Tang S, Toda Y, Kashiwagi K, Mabuchi F, Iijima H, Tsukahara S, Yamagata Z. The association between Japanese primary open-angle glaucoma and normal tension glaucoma patients and the optineurin gene. Hum Genet. 2003;113:276–9. doi: 10.1007/s00439-003-0964-y. [DOI] [PubMed] [Google Scholar]
- 32.Wiggs JL, Auguste J, Allingham RR, Flor JD, Pericak-Vance MA, Rogers K, LaRocque KR, Graham FL, Broomer B, Del Bono E, Haines JL, Hauser M. Lack of association of mutations in optineurin with disease in patients with adult-onset primary open-angle glaucoma. Arch Ophthalmol. 2003;121:1181–3. doi: 10.1001/archopht.121.8.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hauser MA, Allingham RR, Linkroum K, Wang J, LaRocque-Abramson K, Figueiredo D, Santiago-Turla C, del Bono EA, Haines JL, Pericak-Vance MA, Wiggs JL. Distribution of WDR36 DNA sequence variants in patients with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006;47:2542–6. doi: 10.1167/iovs.05-1476. [DOI] [PubMed] [Google Scholar]
- 34.Kramer PL, Samples JR, Monemi S, Sykes R, Sarfarazi M, Wirtz MK. The role of the WDR36 gene on chromosome 5q22.1 in a large family with primary open-angle glaucoma mapped to this region. Arch Ophthalmol. 2006;124:1328–31. doi: 10.1001/archopht.124.9.1328. [DOI] [PubMed] [Google Scholar]
- 35.Ishikawa K, Funayama T, Ohtake Y, Kimura I, Ideta H, Nakamoto K, Yasuda N, Fukuchi T, Fujimaki T, Murakami A, Asaoka R, Hotta Y, Kanamoto T, Tanihara H, Miyaki K, Mashima Y. Association between glaucoma and gene polymorphism of endothelin type A receptor. Mol Vis. 2005;11:431–7. [PubMed] [Google Scholar]
- 36.Coffey CS, Hebert PR, Ritchie MD, Krumholz HM, Gaziano JM, Ridker PM, Brown NJ, Vaughan DE, Moore JH. An application of conditional logistic regression and multifactor dimensionality reduction for detecting gene-gene interactions on risk of myocardial infarction: the importance of model validation. BMC Bioinformatics. 2004;5:49. doi: 10.1186/1471-2105-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]