Abstract
Each year, thousands of new protistan 18S rRNA sequences are detected in environmental samples. Many of these sequences are molecular signatures of new protistan species, classes, and/or kingdoms that have never been seen before. The main goal of this study was to enable visualization of these novel organisms and to conduct quality ultrastructural examination. We achieved this goal by modifying standard procedures for cell fixation, fluorescence in situ hybridization, and scanning electron microscopy (SEM) and by making these methodologies work in concert. As a result, the same individual cell can now be detected by 18S rRNA-targeted fluorochrome-labeled probes and then viewed by SEM to reveal its diagnostic morphological characteristics. The method was successfully tested on a wide range of protists (alveolates, stramenopiles, kinetoplastids, and cryptomonads). The new methodology thus opens a way for fine microscopy studies of many organisms previously known exclusively by their 18S rRNA sequences.
Research on protistan diversity started two centuries ago (5) but still remains in its infancy. The application of molecular tools to environmental samples is producing a seemingly endless list of organisms that are apparently missed by classical approaches (3, 4, 12, 14, 15, 20). The high rate of acquiring novel protistan rRNA sequences suggests that the global protistan inventory is utterly incomplete and that the captured diversity is but the “tip of an iceberg.” The novelty of some of these sequences indicates discovery of protistan lineages at all levels of taxonomic hierarchy, including new kingdoms (3). There is little doubt that entirely new classes of Protista remain undetected, unseen, and unexplored.
The rRNA approach (2) has proved to be uniquely suited for the initial discovery of novel organisms but provides little information beyond the fact of their existence, abundance in nature, and molecular phylogeny (13). The organisms detected remain bewildering, and their basic biology stays secret. Only direct access to the novel organisms would validate their assignment to high taxonomic levels (3), arguments about the ecological significance of their diversity (13), and their use in reconstruction of early eukaryotic evolution (17, 18). Gaining access to the novel organisms is therefore a natural next step after the discovery of their molecular signatures.
Unlike prokaryotes, microscopic eukaryotes exhibit a very high level of morphological diversity. Because of this, light and electron microscopy studies of the newly discovered organisms would be helpful for confirming their uniqueness, assessing their relatedness to other known protists, and unraveling certain aspects of their lifestyles. However, currently available methods do not allow such studies.
This paper describes a new combination of the modified 18S rRNA, fluorescence in situ hybridization (FISH), and scanning electron microscopy (SEM) methodologies. This combination allows identification of target cells by FISH to be followed by fine-quality SEM studies, with both performed on the same microscopic preparations. This opens a way for researchers to conduct proper morphological studies of organisms known today only from their 18S rRNA signatures in the environment.
MATERIALS AND METHODS
Test organisms.
The test species used in this study included representatives of several major protistan taxa, such as alveolates, stramenopiles, kinetoplastids, and cryptomonads (Table 1). Cultures of all species were obtained from Carolina Biological Supply (Burlington, N.C.), except the ciliate Euplotes sp., which was isolated from the Sippewisset salt marsh, Cape Cod, Mass.
TABLE 1.
Classification of strains used for this study
| Strain no.a | Taxonomy | Species | Isolation sourceb | Isolation source identification no. |
|---|---|---|---|---|
| 131480 | Alveolate (ciliate) | Euplotes eurystomus | Pond at CBS | NAc |
| Our isolate | Alveolate (ciliate) | Euplotes sp. | Great Sippewisset salt marsh | NA |
| 131466 | Alveolate (ciliate) | Dileptus cygnus | Pond at CBS | NA |
| 131452 | Alveolate (ciliate) | Colpidium striatum | Pond at CBS | NA |
| 131554 | Alveolate (ciliate) | Paramecium caudatum | Pond at CBS | NA |
| Our isolate | Alveolate (scuticociliate) | Aspidisca sp. | Great Sippewisset salt marsh | NA |
| 153300 | Alveolate (dinoflagellate) | Prorocentrum micans | The Culture Collection of Algae, University of Texas, Austin | IUCC 1003 |
| 153290 | Alveolate (dinoflagellate) | Peridinium cinctum | The Culture Collection of Algae, University of Texas, Austin | IUCC 1336 |
| 152800 | Euglenozoa (kinetoplastid) | Euglena gracilis | The Culture Collection of Algae, University of Texas, Austin | IUCC 753 |
| 153200 | Stramenopile (heterokonta) | Ochromonas danica | The Culture Collection of Algae, University of Texas, Austin | IUCC 1298 |
| 153210 | Stramenopile (heterokonta) | Synura petersenii | The Culture Collection of Algae, University of Texas, Austin | IUCC 239 |
| 131734 | Cryptomonad | Chilomonas paramecium | Pond at CBS | NA |
Carolina Biological Supply strain number.
CBS, Carolina Biological Supply.
NA, not applicable.
Fixation.
We tested several standard and modified fixative mixtures (11) for the ability to impart sufficient rigidity to the test cells. The list included paraformaldehyde (tested at 2, 4, and 10%; also tested at 2% supplemented with 0.2% glutaraldehyde), formalin (tested at the same concentrations and combinations as described for paraformaldehyde), Schaudinn's fixative (2 parts saturated aqueous HgCl2, 1 part 100% ethanol, and 1% [final concentration] glacial acetic acid; 1 part of this fixative was diluted with 3 parts distilled water and added to the sample at a ratio of 1:1 [vol/vol]), Steve's fixative (76% saturated aqueous HgCl2, 20% formaldehyde, and 4% glacial acetic acid mixed with 3 parts distilled water and added to the sample at a ratio of 1:1 [vol/vol]), and Bouin's fixative mixture (buffered formaldehyde saturated with picric acid and 2% glacial acetic acid added immediately before fixation) tested at 50% with or without the addition of 0.1 to 0.2% glutaraldehyde. The fixation time in all cases was 45 min at 4°C.
FISH probes and probe design.
FISH was used to visualize the marine ciliate Euplotes sp. both in environmental samples and mixed with other test protists (Table 1) as negative controls. We designed two oligonucleotide probes to target different regions of the 18S rRNA. A set of probes (nucleotides, 18 to 22; GC contents, 50 to 60%; nucleotide-nucleotide Tms, ≥57°C) was designed for each Euplotes small-subunit (SSU) rRNA sequence retrieved from GenBank by use of the online tool Primer3 (19). Generated probes were checked against the GenBank sequence collection by a standard nucleotide-nucleotide BLAST search (1) and were compared to an accessibility map of the SSU rRNA of Escherichia coli for hints of probe target sites with promising high signal intensities (8). The potential for hairpin and dimer formation of selected oligonucleotides was assessed by use of the program mfold, v. 3.0 (21). From the original 54 probe candidates, two oligonucleotides were chosen that fulfilled the general criteria of potentially successful probes (10, 16). The probe Eupl240 (5′-TCATCTCAGTAGACCTTGCG-3′) had no mismatches with the SSU rRNA of all but one Euplotes species (E. raikovi) but exhibited a 3-bp mismatch with the next closest sequence in GenBank. The probe Eupl1780 (5′-GACAGTCCAAAGAGGTTCAC-3′) matched all 33 Euplotes sp. SSU rRNA sequences deposited in GenBank and had a mismatch of 3 bp with the next closest GenBank sequence. The probes used were purified by high-performance liquid chromatography and labeled with Cy-3 at the 5′ end by the manufacturer (Integrated DNA Technologies, Coralville, Iowa).
Other probes used included the universal Cy-3-labeled eukaryotic probe Euk1209R (5′-GGGCATCACAGACCTG-3′) (9) and its Cy-3-labeled complement as a nonsense probe.
FISH staining.
We used a standard protocol for in situ hybridization (16), with several important modifications. Preserved samples were placed in wells with bottoms made of polycarbonate membranes (25-mm diameter; 0.4-μm pore size) (Costar Transwells; Corning Inc., Corning, N.Y.). The wells were placed on the base of standard 25-mm-diameter glass filtration units (Millipore, Bedford, Mass.) (Fig. 1). To ensure an even cell distribution on the well's membrane, a 5.0-μm-pore-size 25-mm-diameter cellulose nitrate filter (Whatman, Newton, Mass.) was inserted between the well's bottom and the glass base. The excess of fixative was removed by gentle filtration (<200 mm Hg), always leaving a small amount of liquid covering the membrane. The cells were washed by gradually replacing the remains of the fixative with 1× phosphate-buffered saline (PBS) (0.14 M NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.4). This was achieved in three to five cycles (in the case of Bouin's fixative, washing was continued until the yellow coloration disappeared), each consisting of adding 2 ml of fresh PBS and gently removing it by applying a weak vacuum. We found it essential for the preservation of cell shape to never expose the membrane surface (and thus cells) to drying. The washing buffer was then gradually replaced with a hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 8], 30% formamide [the optimal concentration was found in preliminary experiments], and 0.01% sodium dodecyl sulfate). This was done in two cycles, each consisting of adding and then removing 2 ml of the fresh buffer. The wells, with approximately 300 μl of hybridization buffer, were transferred to a six-well Transwell tissue culture tray, and 50 μl of probe solution (30 ng μl−1 in molecular-grade distilled water) was added to each well. The wells received either a universal eukaryotic, nonsense, or Euplotes-specific probe (separately). The wells with probes added were transferred into glass inserts (stacking Stender dish; 27-mm inner diameter; volume, 8 ml), which were placed in a tightly sealed incubation chamber (plastic jar; 45-mm inner diameter; volume, 70 ml) containing a piece of hybridization buffer-saturated tissue paper at the bottom of the chamber. The incubation chambers with the wells were incubated for 2 h at 46°C. After incubation, 2 ml of a preheated (48°C) washing buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 8], 5 mM EDTA, 0.01% sodium dodecyl sulfate) was added to the wells, which were further incubated for 10 min at 48°C. The wells were then placed back on the base of the filtration units, and their contents were washed by several cycles of addition and then removal of distilled water. As before, special care was taken not to expose the filter to the air. With approximately 300 μl of the last wash still in the well, 500 μl of a DAPI (4′,6′-diamidino-2-phenylindole) solution (1.5 μg ml−1; Sigma-Aldrich, St. Louis, Mo.) was added and incubated in the dark for 5 min at room temperature. The DAPI solution was removed by washing the filter three times with 1 ml of distilled water using the vacuum filtration unit as described above.
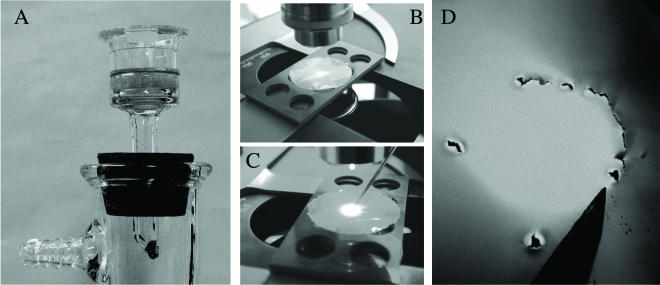
FIG. 1.
Several essential steps for localization of target cells in the combined FISH-SEM method. The images show the Transwell filtration system used to collect and process cells (A), marking of the circular area with the target cell(s) in the middle (B and C), and cutting of the marked area under the dissecting microscope (D).
Preparing cells for SEM.
The probe-stained and DAPI-counterstained cells were dehydrated as follows. The distilled water remaining in the well after DAPI staining was gradually replaced by washing the filter two times with 30% ethanol, using the vacuum filtration system as described above. The wells were transferred to the tissue culture tray, a fresh portion (2 ml) of 30% ethanol was added, and the wells were left in the dark for 12 min. This process was sequentially repeated with 50, 70, 85, 95, and 100% ethanol (the last step was repeated three times for 15 min each). As before, it was essential that the filter not be exposed to air at any point during the procedure. After the last portion of 100% ethanol was removed by use of a vacuum (except for a small amount that was just enough to cover the membrane), the wells were quickly placed into a tissue culture tray, covered with 2 ml of a mixture of 100% ethanol-hexamethyldisilazane (Electron Microscopy Science, Ft. Washington, Pa.) (1:1 [vol/vol]), and allowed to be infiltrated for 20 min in the dark. This was followed by two 20-min infiltration steps with 100% hexamethyldisilazane. At this point, cells were allowed to air dry, as this caused no significant cell distortion. In this state, the filter can be stored in a dark and dry place at room temperature for at least 1 week with no losses in fluorescence intensity.
Fluorescence microscopy.
The air-dried filters were cut out of the wells with a dissecting knife and mounted on an adhesive silicone spacer (Schleicher & Schuell MicroScience, Riviera Beach, Fla.) affixed to a glass microscope slide. The filters were scanned under appropriate epifluorescent illumination on Zeiss Axioplan 2MOT and Axioskop 50 compound microscopes equipped with an HBO 100-W mercury lamp; ×10 Neofluar, ×40 (dry) Neofluar, ×40 (oil) F-Fluar, and ×100 Apo objectives; DAPI- and Cy-3-specific filter sets; and an Hitachi ORCA cooled charge-coupled device camera (Hamamatsu, Hamamatsu City, Japan) operated by the OpenLab software package (Improvision Inc., Lexington, Mass.). Once the positively Cy-3-stained target cell was located by use of the ×10 and ×40 (dry) objectives, a filter area of approximately 5 mm in diameter around the cell was marked with a needle; these and other steps are illustrated in Fig. 1. The marked area was cut out under a dissection microscope (Zeiss Stemi 2000-C) with a dissecting knife. The cut-out piece was checked once again under the fluorescence microscope (×10 and ×40 dry objectives) to confirm the presence and location of the target cell(s). At this point, a digital photograph of the target cell(s) and surrounding areas was taken, with special care taken to capture small landmarks (atypical cell aggregations, unusually shaped particles, etc.). This facilitated locating the cell under SEM. The cut-out piece was then attached to a carbon adhesive tab, mounted on an SEM specimen holder, and sputter coated with 10 to 15 nm of gold-palladium (60:40) by use of a Tousimis Samsputter 2A. SEM was performed on an Amray AMR-1000 scanning electron microscope. The target cells were located at a low SEM magnification, using photographs taken with the epifluorescence microscope.
Some filters were used to examine the Cy-3-labeled cells by epifluorescence rather than SEM. These were mounted on a glass slide, a mixture of 1 part VectaShield (Vector Laboratories, Burlingame, Calif.) and 4 parts Citifluor AF1 (Citifluor, London, United Kingdom) (vol:vol) was added, and a coverslip was gently applied at an angle. This allowed the use of high-numerical-aperture (NA) lenses (×40 oil and ×100 oil) to obtain high-quality epifluorescence images of the target cells. Throughout the project, the principal charge-coupled device camera parameters remained constant (the exposure time was 0.2 s, and all other parameters were at their default settings), except in control preparations, in which cases the exposure time was increased to 2.0 s to avoid featureless blank photos and achieve at least some visualization of the control specimens.
Target cell recovery.
We evaluated the usefulness of our FISH protocol to detect and quantify the target cells in complex species mixes and in environmental samples. The species mixes were prepared using the strains listed in Table 1. Known numbers of Euplotes sp. cells were added to vials with 3 ml of artificial protistan mixes to achieve concentrations of 2, 4, and 8 Euplotes sp. cells/ml. The contents of five vials for each Euplotes sp. concentration were fixed and hybridized to Euplotes-specific probes as well as to the universal and nonsense probes as described above. The negative control contained no probe. All filters were counterstained with DAPI (see above) to obtain the total protistan count. The resulting filters were scanned to examine the specific and nonspecific staining of target and nontarget cells and to count the Cy-3-labeled Euplotes spp.
In addition, 10 Euplotes sp. cells were added to four samples (10 ml each) obtained from Sippewisset salt marsh, which were subsequently stained with the two Euplotes-specific probes. Controls included environmental samples with naturally occurring Euplotes spp. only. For each probe and control treatment, duplicate samples were processed as described above, and duplicate filters were prepared and scanned to detect the Cy-3-labeled target and nontarget cells.
RESULTS AND DISCUSSION
Fixation.
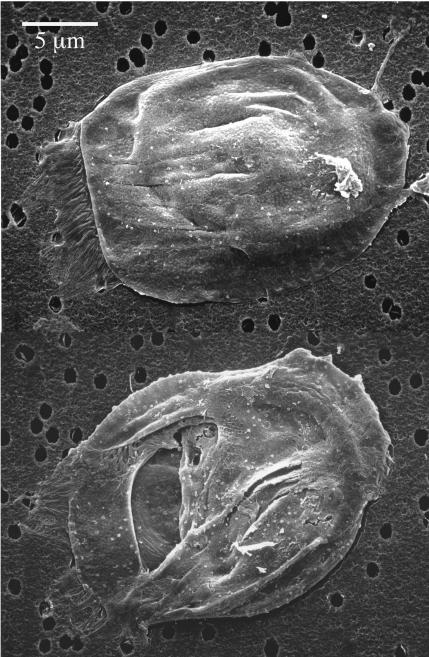
The quality of fixation emerged as one of the most important factors determining the quality of SEM images. The majority of trials resulted in collapse of most cells and clearly substandard SEM images (a typical result is illustrated in Fig. 2). We examined how the cell morphology was changed with each step of the FISH protocol and found that most damage occurred during the dehydration procedure. We addressed this both by modifying the way the cells were handled during FISH and by identifying the fixative that provided sufficient preservation of the cells. Of the nine different fixatives and their combinations tested, only the Bouin's fixative mixture supplemented with glutaraldehyde imparted enough rigidity to the cells such that they retained their shape during FISH and SEM. The qualitatively best combination of cell rigidity, strength of specific fluorescence signal, and low nonspecific background fluorescence was achieved at 50% Bouin's fixative (full-strength solution mixed with sample of equal volume) supplemented with 0.1 to 0.2% glutaraldehyde (see Fig. 3 to 5). Interestingly, Bouin's fixative mixture (at a different concentration and with no glutaraldehyde added) was also found to be optimal for combining FISH and silver staining protocols for ciliate identification (7).
FIG. 2.
Typical SEM images of collapsed Euplotes eurystomus. Heavy damage to the cell morphology was observed with all fixatives except Bouin's fixative supplemented with glutaraldehyde.
FIG. 3.
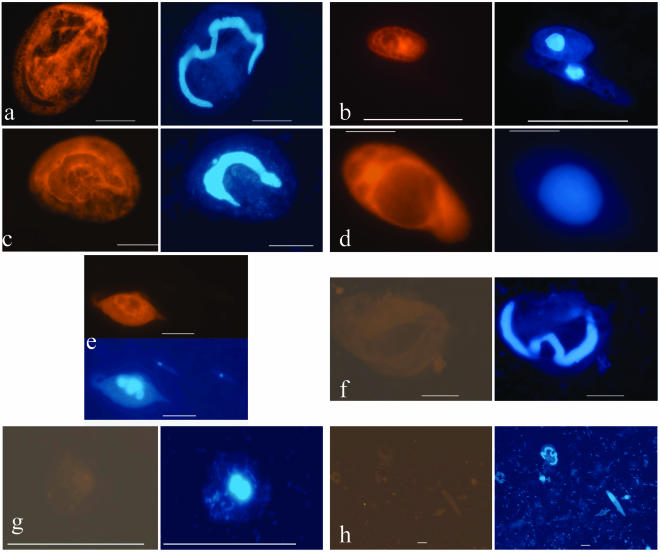
Simultaneous FISH and DAPI staining of several test microeukaryotes in mixed samples. (a to e) Universal eukaryotic probe Euk1209R/DAPI. (f to h) Negative controls with no probe or DAPI (f and g) and with a nonsense probe and DAPI (h). (a) Euplotes eurystomus; (b) Prorocentrum micans and Euglena gracilis (note the lack of probe binding to the latter); (c) Aspidisca sp.; (d) Colpidium striatum; (e) Dileptus cygnus; (f) Euplotes eurystomus; (g) Ochromonas danica; (h) diverse organisms in mixed sample. Bar = 10 μm.
FIG. 5.
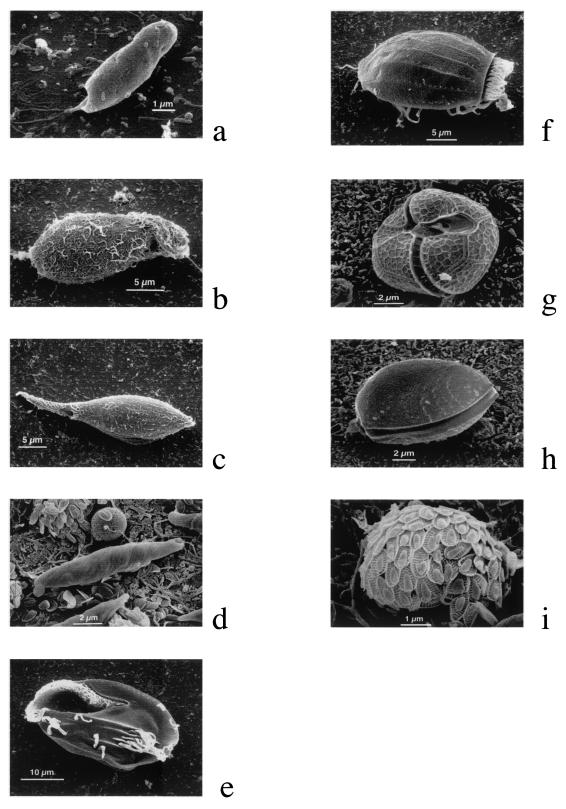
SEM images of organisms retrieved from mixed cultures by use of the combined FISH-SEM protocol. (a) Chilomonas paramecium; (b) Colpidium striatum; (c) Dileptus cygnus; (d) Euglena gracilis; (e) Euplotes eurystomus (ventral surface); (f) Euplotes eurystomus (dorsal surface); (g) Peridinium cinctum; (h) Prorocentrum micans; (i) Synura uvella.
FISH staining.
The commonly used FISH protocol (16) includes a filter-drying step, which serves two main purposes. First, it is a means to attach the target cells to the membrane and prevent excessive cell losses during washing steps. Second, it enhances probe penetration into the target sites in the cell (ribosomes). In our observations, most wall-less organisms collapsed during air drying and became largely unrecognizable. This called for a modification of the standard FISH protocols. We discovered that the cell collapse could be easily prevented if the target cells were kept in solution at all times during FISH. To minimize cell losses, we collected cells in plastic wells with a polycarbonate membrane bottom, as described in a previous protocol (6). When such a well is placed on the base of a typical filtration unit connected to a pump (Fig. 1), reagents can be added to and removed from the well in a controlled fashion. Since the cells stay inside the well during the entire procedure, their losses become minimal (6) (also see below). After the FISH protocol is completed, the membranes can be cut out of the well and used for epifluorescence observations similarly to the standard protocols.
Both Euplotes sp.-specific probes provided satisfactory detection of this ciliate (Fig. 3 and 4). After we optimized one of the principal FISH protocol parameters (i.e., concentration of formamide; data from preliminary trials are not shown), the specificity of staining became 100%. Neither of the two Euplotes sp.-specific probes stained any of the nontarget cells. The nonsense probe did not stain any of the DAPI-counterstained protistan cells, and the level of background fluorescence was sufficiently low (Fig. 3). The universal probe hybridized to most but not all microeukaryotes, which indicated that there were mismatches in the target sequences (SSU rRNA gene) for at least some protists (Fig. 3). Overall, the fluorescence signal was stable for at least 1 week for sample storage in the dark at room temperature.
FIG. 4.
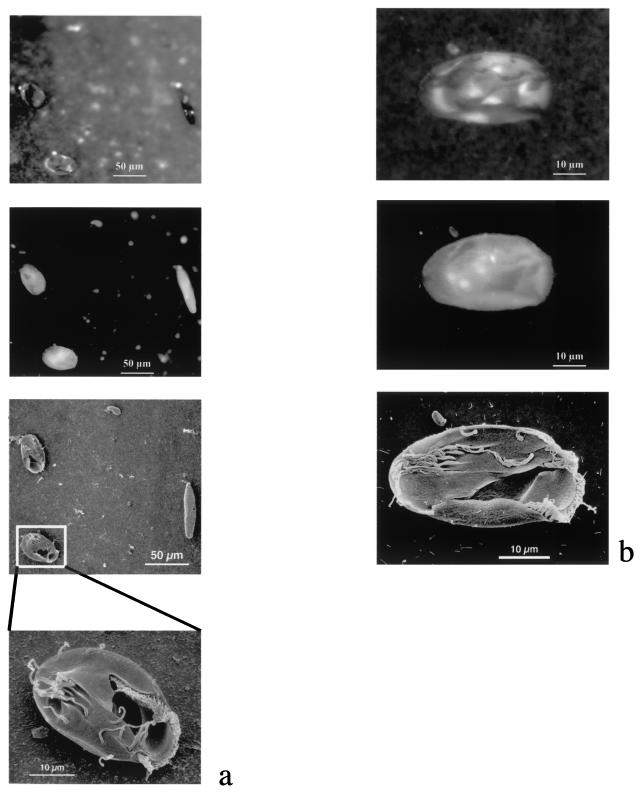
Images obtained at different stages of the combined FISH-SEM protocol. (a) Euplotes eurystomus, Paramecium caudatum, and Colpidium striatum retrieved from a culture mix (10 strains) by use of probe Euk1209R. (b) Euplotes spp. retrieved from an environmental sample by use of probe Eupl240. Top panels, DAPI staining in UV channel; second panels from top, FISH staining in the red channel; third and bottom panels, the same areas under SEM.
We concluded that the modified FISH protocol used was a robust tool for visualization of target cells with an epifluorescence microscope at mid-range magnification (×40 dry objective).
We checked the degree of cell recovery by the modified FISH protocol. We added a known number of Euplotes sp. cells to environmental samples, quantified the increase in the Euplotes sp. count, and compared this increase with the number of added cells (Table 2). The recovery rate was 92% ± 21% for the Euplotes-240 probe and 112% ± 21% for the Euplotes-1780 probe. We also attempted to quantify the recovery of Euplotes spp. from a mix of 11 different protistan cultures (species are indicated in Table 1). The recovery values (for three independent trials) ranged from 93% ± 9% to 97% ± 5%.
TABLE 2.
FISH-aided recovery of Euplotes sp. cells added to environmental samples containing naturally occurring Euplotes
| Probe | No. of cells recovered
|
|||||
|---|---|---|---|---|---|---|
| After the addition of 10 Euplotes cells
|
Control (no addition of Euplotes cells)
|
|||||
| Expt 1 | Expt 2 | Avg ± SD | Expt 1 | Expt 2 | Avg ± SD | |
| Eupl-240 | 9 | 12 | 10.5 ± 2.1 | 3 | 1 | 1.3 ± 1.3 |
| Eupl-1780 | 11 | 14 | 12.5 ± 2.1 | 0 | 1 | 1.3 ± 1.3 |
Bridging FISH and SEM.
Once the target cell is located on the filter by use of the epifluorescence microscope, its position has to be registered to facilitate the search for the same individual cell under SEM. We developed a simple approach to keep track of the cells of interest. This technique consists of several steps: centering the target (stained) cell in the microscopic field of view of the epifluorescence microscope, turning on the transmitted light source to create a light circle around the cell, closing the field diaphragm to bring the diameter of the circle to approximately 3 to 5 mm (Fig. 1B), dotting or puncturing the membrane along the edges of the illuminated circle (Fig. 1C), and cutting a small piece of the membrane containing the circle and the cell of interest in its center (the last step was performed under a dissecting microscope; Fig. 1D). The resulting piece of the membrane can be easily mounted on an SEM specimen holder and scanned under SEM at low magnification to locate the target cell.
SEM.
Once the details of fixation, FISH, and cell handling were worked out, preparation for SEM and SEM observations were done according to standard protocols. We experienced no difficulties in locating the target cells by SEM (Fig. 4), and the quality of material observed was high (Fig. 5). We used a wide selection of organisms across the Protista (cryptomonads, kinetoplastids, stramenopiles, and alveolates) and found all the test species exhibiting their typical morphological characteristics, including those that are important taxonomically (Fig. 5). The images obtained from these preparations are comparable to those obtained by routine SEM techniques alone. The ciliates Dileptus cygnus and Colpidium striatum retained their characteristic shapes and morphological details (Fig. 5b). Euplotes spp. exhibited well-preserved frontoventral, transverse, and caudal cirri as well as the adoral zone of membranelles (Fig. 5e and f). The dinoflagellate Prorocentrum micans (Fig. 5h) showed the expected bilateral compression and two thecal plates typical of the genus (and the order Prorocentrales in general). Images of dinoflagellate Peridinium cinctum revealed this organism's diagnostic tabulation pattern and pronounced ornamentation of thecal plates (Fig. 5g). Representative cell shape, helical striations, and position of the single flagellum were exhibited by the kinetoplastid Euglena gracilis (Fig. 5d). Stramenopile Synura petersenii was seen with its distinctive silica scales (Fig. 5i), and Chilomonas paramecium displayed the normal cryptomonad cell architecture (Fig. 5a). We concluded that the method developed allowed visualization of important diagnostic morphological characteristics across a wide range of protistan diversity and is therefore an adequate tool for studying the morphologies of novel organisms.
Application in the field.
One specific aim of this research was to ensure that our FISH-SEM approach could be used in field-oriented research. In the field, the use of FISH is often impractical or impossible, and so some form of prolonged cell storage must be found. We discovered that Bouin's fixative-glutaraldehyde-fixed cells inside wells could be stored for at least several weeks at −20°C if covered with a mix of 3 parts 1× PBS and 2 parts 100% ethanol. The wells should be placed into tissue culture trays and sealed with Parafilm. Such storage did not appear to compromise the ability of cells to hybridize well with FISH probes.
Conclusions.
We have developed a methodological approach that combines FISH and SEM for studies of protists. It consists of original and modified protocols of standard fixation, FISH, and SEM. The individual components of this approach work together seamlessly and are compatible with the demands of field-based research. The approach provides high-quality SEM images of target cells that are first identified by FISH. It may therefore serve as a convenient tool to conduct ultrastructural studies of organisms known exclusively from their 18S rRNA signatures.
Acknowledgments
We thank Klaus Hausmann (University of Berlin), Denis Lynn (University of Guelph), and Eugene Small (University of Maryland) for their advice on SEM and Christine Beardsley (MPI for Marine Microbiology, Bremen, Germany), Johannes Fried (Technical University, Munich, Germany), and Michael Schweikert (Technical University, Stuttgart, Germany) for helpful discussions on FISH. Ed Jarroll (Northeastern University, Boston, Mass.) was instrumental in providing the institutional support for this project.
This work was funded by Deutsche Forschungsgemeinschaft grant STO414/2-1 to T.S. and by U.S. National Science Foundation grants to S.S.E. (OCE-9618135 and DEB-0103599).
This is contribution 248 of the Marine Science Center of Northeastern University, Nahant, Mass.
REFERENCES
- 1.Altschul, S. F., T. L. Madde, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgcomb, V. P., D. T. Kysela, A. Teske, A. de Vera Gomez, and M. L. Sogin. 2002. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA 99:7658-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrenberg, C. C. 1838. Die infusionstheirchen als vollkommene organismen. Leopold Voss, Leipzig, Germany.
- 6.Field, K. G., D. Gordon, T. Wright, M. Rappe, E. Urback, K. Vergin, and S. J. Giovannoni. 1997. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl. Environ. Microbiol. 63:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fried, J., W. Ludwig, R. Psenner, and K. H. Schleifer. 2002. Improvement of ciliate identification and quantification: a new protocol for fluorescence in situ hybridization (FISH) in combination with silver stain techniques. Syst. Appl. Microbiol. 25:555-571. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovannoni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humason, G. L. 1972. Animal tissue techniques. W. H. Freeman and Company, San Francisco, Calif.
- 12.Lopez-Garcia, P., F. Rodriguez-Valera, C. Pedros-Alio, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 13.Massana, R., L. Guillou, B. Diez, and C. Pedros-Alio. 2002. Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl. Environ. Microbiol. 68:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 15.Moreira, D., and P. Lopez-Garcia. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10:31-38. [DOI] [PubMed] [Google Scholar]
- 16.Pernthaler, J. G. F., W. Schoenhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes, p. 207-226. In J. Paul (ed.), Methods in microbiology: marine microbiology, vol. 30. Academic Press Ltd., London, United Kingdom.
- 17.Philippe, H., and A. Adoutte. 1998. The molecular phylogeny of Eukaryota: solid facts and uncertainties, p. 25-56. In K. V. G Coombs, M. Sleigh, and A Warren (ed.), Evolutionary relationships among protozoa. Chapman and Hall, London, United Kingdom.
- 18.Philippe, H., P. Lopez, H. Brinkmann, K. Budin, A. Germot, J. Laurent, D. Moreira, M. Muller, and H. Le Guyader. 2000. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc. R. Soc. Lond. B Biol. Sci. 267:1213-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 20.Stoeck, T., and S. S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. NATO ASI Series. Kluwer Academic Publishers, New York, N.Y.