Abstract
For many ecological studies of cyanobacteria, it is essential that closely related species or strains can be discriminated. Since this is often not possible by using morphological features, cyanobacteria are frequently studied by using DNA-based methods. A powerful method for analysis of the diversity and dynamics of microbial populations and for checking the purity and affiliation of cultivated strains is denaturing gradient gel electrophoresis (DGGE). We realized high-resolution discrimination of a variety of cyanobacteria by means of DGGE analysis of sections of the internal transcribed spacer between the 16S and 23S rRNA genes (rRNA-ITS). A forward primer specific for cyanobacteria, targeted at the 3′ end of the 16S rRNA gene, was designed. The combination of this primer and three different reverse primers targeted to the rRNA-ITS or to the 23S rRNA gene yielded PCR products of different sizes from cultures of all 16 cyanobacterial genera that were tested but not from other bacteria. DGGE profiles produced from the shortest section of rRNA-ITS consisted of one band for all but one cyanobacterial genera, and those generated from longer stretches of rRNA-ITS yielded DGGE profiles containing one to four bands. The suitability of DGGE for detecting intrageneric and intraspecific variation was tested by using strains of the genus Microcystis. Many strains could be discriminated by means of rRNA-ITS DGGE, and the resolution of this method was strikingly higher than that obtained with previously described methods. The applicability of the developed DGGE assays for analysis of cyanobacteria in field samples was demonstrated by using samples from freshwater lakes. The advantages and disadvantages associated with the use of each developed primer set are discussed.
Cyanobacteria are a major component of many aquatic and terrestrial ecosystems and can be found in virtually all habitat types, including marine and freshwater environments, deserts, hot springs, hypersaline environments, rocks, and ice. In addition, they can be involved in symbiotic associations with a remarkable range of eukaryotic host organisms. In all these environments, cyanobacteria are major contributors to photosynthesis and nitrogen fixation (38). Cyanobacterial populations may reach high densities during blooms in surface waters. Detrimental effects of such blooms are of growing concern for water managers worldwide (9).
For ecological studies of cyanobacteria, it is essential to be able to differentiate between closely related organisms. Some important features, such as nitrogen fixation and toxicity, are not uniformly dispersed across taxa or morphological groups and may vary between species or even between strains of the same species (9, 41). For example, toxic cyanobacterial blooms often require characterization below the genus level. Both toxin-producing and -nonproducing strains are included in most morphologically distinguishable species of the genera Microcystis (18, 30) and Anabaena (5). Shifts in the ratio of toxic to nontoxic genotypes of Microcystis may (partly) explain the often poor correlations of cell counts with toxin content in many field studies (6).
To study the functions and interactions of cyanobacteria, it is necessary to reveal the composition and dynamics of their populations. Also, it is important to relate isolated strains to their counterparts in nature for extrapolation of findings from physiological experiments carried out on cultures to natural conditions. Linking laboratory cultures and field populations is often problematic due to the selectivity inherent to cultivation and to morphological changes occurring after cultivation (27, 31). Denaturing gradient gel electrophoresis (DGGE), a technique for sequence-dependent separation of PCR products (25), can be used to assess the genotypic diversity in environmental samples and to judge the purity and uniqueness of isolated strains. Isolated cultures can be assigned to field populations based on the comparison of their DGGE profiles, and sequence information from profile bands can be used to characterize the organisms that are present. A section of DNA is suitable for DGGE analysis if it can be specifically amplified from the target organisms, has sufficient sequence heterogeneity for the desired resolution, and, preferably, is part of a gene for which a considerable number of sequences have been deposited in sequence databases. DGGE of hetR, a gene assigned to heterocyst differentiation, has been used to study diversity in isolated strains of the cyanobacterial genera Trichodesmium and Nostoc (28, 32). Also, nifH, a gene encoding dinitrogenase reductase in many microorganisms, including cyanobacteria (42), has been used for DGGE analysis of the very diverse functional group of diazotrophic (N2-fixing) organisms (22). An important drawback for the application of both protein-encoding genes, especially for the analysis of communities of cyanobacteria, is that they are present in only a limited number of cyanobacterial genera. Nübel et al. (27) developed primers for specific amplification of a 16S rRNA gene segment from cyanobacteria and plastids, allowing DGGE analysis of cyanobacterial populations (1, 11, 27). However, the taxonomic resolution offered by 16S rRNA genes is insufficient for discrimination of closely related organisms. As a result, research has increasingly focused on the rRNA 16S to 23S internal transcribed spacer (rRNA-ITS). The greater degree of sequence heterogeneity, as well as a considerable number of published rRNA-ITS sequences, makes rRNA-ITS very suitable for high-resolution analysis of cyanobacteria. Restriction enzyme digestion of rRNA-ITS has been used to resolve closely related cyanobacterial strains (21, 23, 26, 32), and direct sequencing has been used to study subgeneric phylogenetic relationships in genera such as Microcystis (30), Trichodesmium (28), and picocyanobacteria (34). Recently, DGGE has been used for analysis of Synechococcus (4) and Aphanizomenon (20) rRNA-ITS sequences.
In the present work, we combined the relatively high sequence variation in rRNA-ITS with the potential of DGGE to separate even small differences in sequence to introduce methods for high-resolution analysis of cultures and populations of cyanobacteria. We developed and tested primers and protocols for selective amplification and DGGE analysis of cyanobacterial genotypes in the presence of DNA from contaminating microorganisms. Their value for analysis of cyanobacterial strains of various genera and of field samples was examined, and the resolution that could be achieved by different rRNA DGGE methods was compared by using closely related Microcystis strains.
MATERIALS AND METHODS
Sampling and DNA isolation.
Lakes Kinselmeer and Sneekermeer are shallow lakes (maximum depth, approximately 3 m), and Volkerak and Zeegerplas are deeper lakes (maximum depth, 15 and 25 m, respectively). All these lakes have extensive blooms of various cyanobacteria, mostly in spring and summer. Water samples from the lakes were collected 0.5 m below the surfaces in sterile bottles from a boat in the middle of the lakes and stored in the dark at 4°C. Within 4 h after sampling, a volume of 250 ml of water (or less if the filter clogged) was filtered over a 25-mm diameter 0.2-μm-pore-size mixed-esters filter (ME 24; Schleicher & Schuell, Dassel, Germany). The filter was cut in two with a sterile scalpel, and each half was stored in a microcentrifuge tube at −80°C until further processing. DNA from filters containing field samples was isolated as described by Zwart et al. (43). DNA from the pellets of 2-ml cyanobacterial cultures in which growth was visible by eye was isolated by means of the DNA isolation procedure described by Tillett and Neilan (37).
Strain isolation and culture conditions.
The strains of cyanobacteria used in this study are given in Table 1. The cultures isolated in this study (Microcystis and Gloeotrichia spp.) were obtained by picking single colonies from water samples by using a dissecting microscope and sterile glass Pasteur pipettes with narrow openings, followed by repeated washings in a small volume of O2 medium (8). Alternatively, single Microcystis colonies were grown on plates containing O2 medium solidified with agarose (0.3%, wt/vol). The colonies grown on plates were rendered unialgal through repeated plating. None of the cyanobacterial cultures that were used in this study were axenic. All cyanobacteria able to grow in freshwater medium were maintained in our laboratory in O2 medium (5 ml in 30-ml tubes or 30 ml in 100-ml Erlenmeyer flasks) at 20°C at an irradiance of approximately 15 μmol · m−2 · s−1 for a cycle with a light-to-dark ratio of 16:8 h. A Microcoleus isolate was maintained as decribed by Jonkers et al. (16). The bacteria used as negative controls to test the specificity of the primers were Serratia marcescens (DSM 1636), Acinetobacter calcoaceticus strain BD4 (DSM 586), Erwinia carotovora subsp. carotovora (DSM 30168), Escherichia coli (DSM 423), Rhodococcus erythropolis (DSM 43188), Lactobacillus reuteri (DSM 20016), Lactococcus lactis subsp. lactis (NIZO-81), Bacillus polymyxa (DSM 36), Bacillus subtilis (DSM 10), and Pseudomonas stutzeri (DSM 5190). Cultures were obtained from the German Collection of Microorganisms and Cell Cultures (cultures encoded DSM) and from the Dutch Institute for Dairy Research (encoded NIZO).
TABLE 1.
Cultures of cyanobacteria used in this study
| Code | Identity | Origin | Source or referencea |
|---|---|---|---|
| K29, K50 | Microcystis sp. | Kinselmeer, The Netherlands | This study |
| S2 | Microcystis sp. | Sneekermeer, The Netherlands | This study |
| V28, V40, V67, V72, V73, V80, V88, V89, V91 | Microcystis sp. | Volkerak, The Netherlands | This study |
| Z6, Z11 | Microcystis sp. | Zeegerplas, The Netherlands | This study |
| CYA43 | Microcystis aeruginosa Kütz | United States | NIVA |
| CYA140 | Microcystis aeruginosa Kütz | Bendig's Pond, Bruno, Canada | NIVA |
| CYA228 | Microcystis aeruginosa Kütz | Lake Akersvatnet, Vestfold, Norway | NIVA |
| PCC7806 | Microcystis aeruginosa | Braakman Reservoir, The Netherlands | PCC |
| PCC7820 | Microcystis aeruginosa | Loch Balgavies, Scotland | PCC |
| SAG17.85 | Microcystis aeruginosa Kütz | Lake Neusiedler, Austria | SAG |
| PCC6803 | Synechocystis sp. | Freshwater from California | PCC |
| PCC7942 | Synechococcus sp. | Freshwater from California | PCC |
| PCC73110 | Leptolyngbya sp. | PCC | |
| CYA99 | Lyngbya sp. | River Suldalslågen | NIVA |
| Microcoleus sp. | Microbial mat from hypersaline Lake Chirana, Spain | 16 | |
| CYA146 | Pseudanabaena catenata Lauterb. | Canada | NIVA |
| CYA24 | Planktothrix prolifica | Lake Levrasjön, Skåne, Sweden | NIVA |
| CYA126 | Planktothrix aghardii | Lake Långsjön, Finland | NIVA |
| ATCC 29413 | Anabaena variablis | ATCC | |
| CYA135 | Anabaenopisis arnoldii Aptek. | Ferguson Gulf, Lake Turkana, Kenya | NIVA |
| CYA103 | Aphanizomenon flos-aquae f. gracile (Lemm.) | Pond at Vingrom, Lillehammer, Norway | NIVA |
| 1401/7 CCAP | Aphanizomenon flos-aquae | Freshwater from Brantry Lough, N. Ireland | CCAP |
| CYA225 | Cylindrospermopsis raciborskii | Lake Balaton, Hungary | NIVA |
| CYA124 | Nostoc sp. | Lake Steinsfjorden, Buskerud, Norway | NIVA |
| KAC13 | Nodularia spumigena | Baltic Sea | KAC |
| BV16R | Gloeotrichia sp. | Lake Broekvelden, The Netherlands | This study |
| IMS101 | Trichodesmium erythraeum | North Carolina coast | KAC |
| PCC9006 | Prochlorothrix hollandica | Lake Loosdrecht, The Netherlands | PCC |
NIVA, Culture Collection of Algae, Norwegian Institute for Water Research; PCC, Pasteur Culture Collection of Cyanobacteria, Institut Pasteur, Paris, France; KAC, Kalmar Algae Collections, Kalmar University, Sweden; SAG, Culture Collection of Algae at the University of Göttingen, Germany; CCAP, Culture Collection of Algae and Protozoa, CEH, Windermere, United Kingdom; ATCC, American Type Culture Collection, Manassas, Virginia.
Sequence alignment and investigation.
Sequences of the 16S rRNA of bacteria and cyanobacteria were obtained from the ribosomal database project (RDPII) (24) and the GenBank, EMBL, and DDBJ databases. The program Dedicated Comparative Sequence Editor (DCSE) (10) and the program package ARB (http://www.ARB-home.de) were used for sequence alignments based on comparison of secondary structural elements in the rRNA. These alignments, containing most available cyanobacterial sequences, and an alignment of 218 representative bacterial sequences in RDPII were used to search for potential primer sites in the 16S rRNA gene. The number of mismatches of the designed primer with the deposited sequences was investigated by using the probe match option in ARB and ARB's 6spring2001 database, supplemented with 19 sequences (W. Ludwig and A. Ernst, personal communication) containing 8,229 16S rRNA gene sequences of more than 1,400 bp (of which 145 are from cyanobacteria and 77 from chloroplasts). Based on this database, databases were constructed containing either all cyanobacteria, all plastids, or all sequences except cyanobacteria and plastids. The number of sequences with a given number of mismatches was determined by using probe match, followed by a correction for degenerate base positions in the primer sequence. Sequences from the rRNA-ITS region were obtained from GenBank, EMBL, and DDBJ and were aligned by using the programs ClustalW and BioEdit Sequence Alignment Editor (12).
PCR amplification.
Primer sequences and references are given in Table 2. The forward primer for amplification of part of the 16S rRNA gene was slightly modified from that used by Nübel et al. (27) to further improve their theoretical specificity, because the efficiency of amplification and therefore the selectivity of the primer is typically determined by the nature of the 3′-end nucleotides (19, 35). PCR amplification was performed in an MBS 0.5 S thermocycler (ThermoHybaid, Ashford, United Kingdom) in a 25-μl reaction mixture containing approximately 50 ng of DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.01% (wt/vol) of gelatin, 200 μM (each) deoxynucleotide, 1.5 mM MgCl2, 2.5 U of Taq DNA polymerase (Boehringer Mannheim, Mannheim, Germany), and 0.5 μM (each) primer. The temperature cycling conditions for the amplification of part of the 16S rRNA were modified slightly from those of Nübel et al.(27). After preincubation at 94°C for 5 min, a total of 30 cycles were performed at 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. The temperature cycling was concluded with a final step of 5 min at 72°C. The optimized temperature cycling conditions for the amplification of rRNA-ITS (ITSa, ITSb, and ITSc; Results and Table 2) were as follows. After preincubation at 94°C for 5 min, a total of 30 cycles were performed at 94°C for 1 min, annealing temperature (Ta) for 1 min, and 72°C for 1 min. In the first 20 cycles, Ta decreased by 1°C after every second cycle, from 62°C in the first cycle to 52°C in the twentieth. This touch down procedure was followed to reduce nonspecific annealing of the primers. In the last 10 cycles, Ta was 52°C. The temperature cycling was concluded with a final step of 30 min at 72°C.
TABLE 2.
Primers used in this study
| Primera | Sequence (5′ to 3′) | Reference |
|---|---|---|
| 16S rRNA | ||
| (GC-)CYA371F | CCT ACG GGA GGC AGC AGT GGG GAA TTT TCC | 27 |
| CYA783R | GAC TAC (A/T)GG GGT ATC TAA TCC C(A/T) | 27 |
| (GC-)CSIF | G(T/C)C ACG CCC GAA GTC (G/A)TT AC | This study |
| rRNA-ITS | ||
| 373R | CTA ACC ACC TGA GCT AAT | 40 |
| ITS3R | TAT GCA GTT TTC AAG GTT C | 39 |
| 23S rRNA | ||
| ULR | CCT CTG TGT GCC TAG GTA TC | 13, 26 |
F (forward) and R (reverse) refer to the primer orientation in relation to rRNA genes. A 40-nucleotide G-C clamp 5′-CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC C-3′ was added to the 5′ end of the forward primers indicated with (GC-).
DGGE profiling.
Theoretical melting profiles were constructed by using the program MELT94, which can be found on the Internet at http://web.mit.edu/osp/www/melt.html. DGGE was performed essentially as described by Muyzer et al. (25). Briefly, PCR products were separated on a 1.5-mm-thick vertical gel containing 8% (wt/vol) polyacrylamide (at an acrylamide-to-bisacrylamide ratio of 37.5:1) and a linear gradient of the denaturants urea and formamide, increasing from 25 or 30% at the top of the gel to 40% at the bottom. The concentrations depended on the gene products that were analyzed and are given in the figure legends. Here, 100% denaturant is defined as 7 M urea and 40% (vol/vol) formamide. Electrophoresis was performed in a buffer containing 0.04 M Tris-acetate and 0.001 M EDTA (pH 7.6) (0.5× TAE buffer) for 16 h at 75 V. The gel was stained for 1 h in water containing 0.5 μg of ethidium bromide ml−1. An image of the gel was recorded with a charge-coupled device camera system (Imago; B & L Systems, Maarssen, The Netherlands).
Sequencing of DNA from DGGE bands.
A small piece of gel from the middle of the target band was excised from the DGGE gel and incubated in 50 μl of sterile Milli-Q-purified water for 24 h at 4°C. The eluent was reamplified by using the original primer set and run on DGGE to confirm its identity. For sequence analysis, the eluent was reamplified with reverse primers that had M13 priming sites added to the 5′ side of the original primers. The PCR products were purified by using the Concert Rapid PCR Purification System (GibcoBRL Life Technologies, Paisley, United Kingdom), and these products were used as templates for sequencing reactions with the Thermo Sequenase Primer cycle sequencing kit (Amersham Pharmacia Biotech). Sequencing reaction products were analyzed on an ALF Express II sequencer (Amersham Pharmacia Biotech) with CY5 fluorescence-labeled M13 sequence primers or a labeled G-C clamp. The sequences were processed by using the program Sequencher, version 4.0.5 (Gene Codes Corp., Ann Arbor, Mich.), and similarity with sequences deposited in the GenBank, EMBL, and DDBJ databases was checked by using the program BLAST (2) (http://www.ncbi.nlm.nih.gov/BLAST/).
Template mixtures experiment.
ITSa was amplified from the selected cultures, and for each strain the amount of PCR product was estimated from an agarose gel. The PCR products were mixed and diluted in order to obtain template DNA from selected cultures or from mixtures thereof, all in identical final concentrations. This template DNA was used for ITSa amplification, and the PCR products were analyzed on a DGGE gel.
Nucleotide sequence accession numbers.
The sequences were deposited at EMBL and were assigned accession numbers AJ579895 to AJ579906.
RESULTS
Design of primers for rRNA-ITS amplification.
Our aim was to develop a PCR for specific amplification of cyanobacterial rRNA-ITS in the presence of noncyanobacterial DNA to yield PCR products that can be separated by means of DGGE. To ensure specific amplification of the rRNA-ITS gene section, the target area for at least one of the primers had to be exclusive for cyanobacteria. For the design of a specific primer, we focused on the 16S rRNA gene, since ample sequence information is available from this gene and it includes areas with various degrees of sequence conservation. We used alignments of all available 16S sequences from cyanobacteria and of sequences from other representative prokaryotes in the RDPII (24), GenBank, EMBL, and DDBJ databases. The alignments were searched upstream, starting from the 3′ end of the 16S rRNA gene, which resulted in the identification of a 20-nucleotide sequence (CSIF) (Table 2) corresponding to nucleotides 1423 to 1442 in the Synechococcus sp. strain PCC6301 sequence. This sequence was highly conserved in all phylogenetic groups of cyanobacteria from which sequences were available. Out of 145 cyanobacterial sequences, 84 had no mismatches, 46 had one mismatch, 2 had two mismatches, and 13 had three mismatches with the primer sequence, as revealed by a probe search in ARB. Sequences with three mismatches were found in the genera Nostoc, Spirulina, Gloeobacter, and Calothrix. From the first two genera there were also sequences deposited with fewer mismatches. Importantly, the three bases at the 3′ end (TAC), which are the most selective bases when the sequence is used as the forward primer (19, 35), appeared to be conserved almost exclusively among cyanobacteria. Only the few deposited sequences of the genera Gloeobacter and Calothrix and a sequence related to Limnothrix redekei, misnamed Oscillatoria limnetica, did not contain a C at the 3′ end. As expected from their close relationship with cyanobacteria, plastid sequences had relatively few mismatches with the primer sequence. A probe search in ARB revealed that from 77 plastid sequences, 1 had a perfect match, 45 had one or two mismatches, and 31 had three or more mismatches. To confirm the specificity for cyanobacteria of the primer sequence, a combined probe search was performed (using the probe match option) in the RDPII database and in an ARB database containing more than 8,000 full-length 16S rRNA prokaryotic sequences. No sequences for prokaryotes other than cyanobacteria were found with less than three mismatches. Two sequences (from Ammonifex degensii and Thermodesulfobacterium hverag) had three mismatches, 116 sequences had four mismatches, and the remainder had five or more mismatches. A BLAST search (2) of the primer sequence yielded only cyanobacteria in the hits with the highest similarity scores.
Reverse primer sites for the amplification of cyanobacterial rRNA-ITS sequences were gathered from previous research (Table 2). Three reverse primers were used, two (373R and ITS3R) targeted at highly conserved sequence motifs in the ITS and one (ULR) at the 5′ end of the 23S gene. The combination of the forward primer (CSIF) and the reverse primers yields three primer sets, ITSa (primers CSIF plus 373R), ITSb (CSIF plus ITS3R), and ITSc (CSIF plus ULR) (Table 2). The rationale for using more than one primer combination for amplification of the rRNA-ITS was the trade-off that exists between the broad applicability of the shorter fragments and the different melting behavior and higher amount of sequence information associated with longer fragments (see below). Alignment of cyanobacterial rRNA-ITS sequences deposited in the GenBank, EMBL, and DDBJ databases showed that virtually all cyanobacteria had at least one operon containing a perfect match with the reverse primer 373R, which is targeted to the highly conserved tRNAIle gene. Only some Prochlorococcus strains contained one mismatch. Most deposited cyanobacterial sequences also had a perfect match with the ITS3R primer sequence. Only a number of picocyanobacteria did not contain the target sequence. A BLAST search performed on the reverse primer sequences resulted in the highest similarity scores for cyanobacteria. However, it cannot be concluded that the reverse primers are selective for cyanobacteria, since the database of rRNA ITS and 23S sequences is relatively small (compared to that for 16S sequences) and may be biased for cyanobacteria.
Theoretical melting curves calculated for rRNA-ITS sequences of several major genera (using the program MELT94) showed that the G-C clamp positioned at the forward (5′) primer yielded a favorable melting curve, with a lower melting domain at the 3′ end.
In vitro specificity and applicability of the primers for rRNA-ITS amplification.
The forward primer presented in Table 2 had a complete or almost complete match with cyanobacterial sequences, but it also had some sequence similarity with several noncyanobacterial 16S rRNA sequences in the database. To confirm the selectivity of the primers for amplification of cyanobacterial rRNA-ITS, we tried to amplify DNA from bacterial strains with some degree of sequence homology to the forward primer (four to five mismatches) and from a few randomly chosen strains. No PCR products were generated from these bacteria either at the optimized annealing temperature (as described below) or at 10° below this temperature.
The phylogenetic coverage within the cyanobacterial phylum of the primers was investigated by using DNA from strains of a range of different cyanobacterial genera as templates for PCR amplification (Table 3). For each primer combination, the highest temperature at which PCR products were generated for all genera was selected as the optimal PCR amplification temperature. As a consequence, for several genera amplification is possible at higher annealing temperatures than those used in our optimized protocol. The number of PCR products and their sizes and relative intensities varied among the different genera (Table 3). For all strains that were tested, ITSa amplification yielded one band of 275 to 350 bp. ITSb amplification resulted in PCR products between 350 and 800 bp for most tested strains, with the exception of Synechocystis, Leptolyngbya, and Lyngbya. One PCR product was formed from the genera Microcystis, Pseudanabaena, Planktothrix, Trichodesmium, Prochlorothrix, and Synechococcus, and two PCR products were formed from Aphanizomenon, Anabaena, Anabaenopsis, Gloeotrichia, Nodularia, Nostoc, and Cylindrospermopsis. ITSc primers successfully amplified DNA from all the strains that were tested, yielding for most genera PCR products ranging between 450 and 900 bp. Prochlorothrix hollandica yielded one very long product of 1,050 bp. Amplification with ITSb or ITSc primers yielded the same number of bands for most strains; only ITSc amplification of Planktothrix strains produced two bands instead of one.
TABLE 3.
Number and approximate sizes of the PCR products of strains from various cyanobacterial genera amplified with the primers described in this studya
| Strain | ITSa | ITSb | ITSc |
|---|---|---|---|
| Microcystis aeruginosa | 300 | 500 | 550 |
| Synechocystis sp. | 325 | 675 | |
| Synechococcus | 325 | 650 | 750 |
| Leptolyngbya | 275 | 500 | |
| Lyngbya sp. | 300 | 675 | |
| Microcoleus sp. | 300 | ND | 675 |
| Pseudanabaena catenata | 300 | 675 | 750 |
| Planktothrix prolifica | 300 | 550 | 525, 700 |
| Planktothrix aghardi | 300 | 550 | 525, 700 |
| Anabaena variabilis | 300 | 375, 600 | 500, 700 |
| Anabaenopsis arnoldii | 300 | 350, 650 | 450, 725 |
| Aphanizomenon flos-aquae | 300 | 400, 625 | 475, 700 |
| Aphanizomenon flos-aquae f. gracile | 300 | 400, 625 | 475, 700 |
| Cylindrospermopsis raciborskii | 300 | 350, 525 | 425, 600 |
| Nostoc sp. | 300 | 375, 650 | 500, 775 |
| Nodularia spumigena | 300 | 400, 700 | 575, 875 |
| Gloeotrichia sp. | 275 | 375, 675 | 475, 775 |
| Trichodesmium erythraeum | 300 | 550 | 675 |
| Prochlorothrix hollandica | 350 | 800 | 1050 |
Primer combinations are described in the text. The forward primers include a 40-nucleotide GC-clamp. PCR product sizes (in base pairs) were estimated from agarose gels. ND, not determined.
High-resolution separation of Microcystis strains by means of rRNA-ITS DGGE.
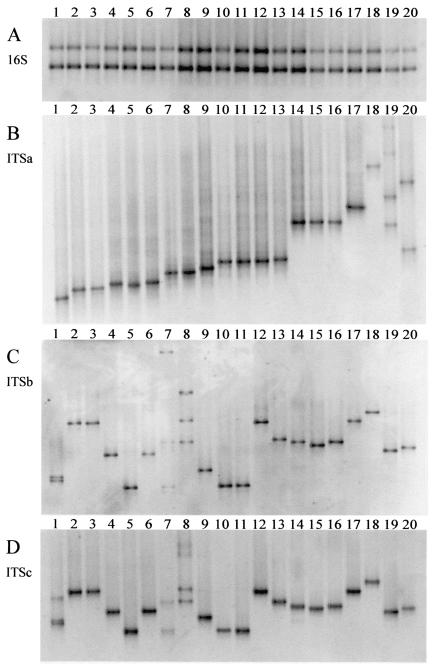
The ability to differentiate related organisms by means of DGGE was examined by using a number of Microcystis strains that had been isolated from Dutch lakes or obtained from culture collections (Table 1). The DGGE gels in Fig. 1 demonstrate the possibility of separating PCR products obtained with the primer set targeted to 16S and the primer sets ITSa, ITSb, and ITSc by using DNA from 20 selected Microcystis strains. The 16S rRNA PCR product of approximately 450 bp gave rise to two bands in the DGGE gel occupying identical positions for all strains (Fig. 1A). The occurrence of two bands can be explained by the presence of degenerate bases in the non-G-C-clamped reverse primer (Table 2). In contrast, ITSa amplicons (approximately 325 bp) resulted in one DGGE band for most of our isolated strains and for all culture collection strains and at a unique position for many strains (Fig. 1B). Examples of strains with more than one band are V40, with three bands (lane 20; the upper band is not on the figure), and V88 (lane 19), with four bands. These profiles containing two or more bands were encountered predominantly in Microcystis cultures that had been established by isolating single colonies. Compared to plating, this isolation strategy is more likely to yield cultures that are not yet unialgal. The upper bands in lanes 19 and 20 of Fig. 1 resulted from heteroduplex formation (15), which was confirmed through excision, reamplification, and DGGE analysis of these bands. The lower two bands in these cultures were both derived from Microcystis strains, which was revealed by a BLAST search of the sequences obtained from these bands.
FIG. 1.
Discrimination of closely related cyanobacteria. DNA from 20 Microcystis strains was amplified with four different primer combinations (A through D) and separated on DGGE gels. The primer sequences and combinations are given in Table 2. The different primer combinations amplified a segment of the 16S gene (A) or different portions of the rRNA-ITS, i.e., ITSa (B), ITSb (C), or ITSc (D). The Microcystis strains are SAG17.85 (lane 1), V80 (lane 2), V72 (lane 3), V73 (lane 4), CYA43 (lane 5), V67 (lane 6), K29 (lane 7), K50 (lane 8), CYA140 (lane 9), CYA228 (lane 10), PCC7820 (lane 11), PCC7806 (lane 12), V91 (lane 13), Z6 (lane 14), Z11 (lane 15), V89 (lane 16), V28 (lane 17), S2 (lane 18), V88 (lane 19), and V40 (lane 20). Information about the Microcystis strains is given in Table 1. The DGGE gels had a 30 to 40% denaturant concentration gradient, and only the part of the gel containing bands is shown.
Results of the DGGE analysis of ITSb and ITSc amplicons from the selected Microcystis strains were largely comparable. Amplification yielded single bands of approximately 500 and 550 bp for ITSb and ITSc, respectively, resulting in one band on DGGE gels (Fig. 1C and D) except for strains SAG17.85 (lane 1), K29 (lane 7), and K50 (lane 8). Positions in the gel were unique for most strains, and importantly, a number of strains that could not or could hardly be distinguished based on ITSa DGGE were separated by using ITSb or ITSc DGGE. For example, ITSa amplicons from strains CYA228 (lane 10), PCC7820 (lane 11), and PCC7806 (lane 12) were undistinguishable and differed only faintly from those of strain V91 (lane 13), whereas ITSb or ITSc amplicons enabled a clear separation and left only CYA228 and PCC7820 undifferentiated (Fig. 1C and D). Three cultures, SAG17.85, K29, and K50, revealed multiple bands when analyzed using ITSb or ITSc DGGE, whereas ITSa DGGE had only one band (Fig.1B through D). Again, the upper bands were shown to result from heteroduplex formation, and all excised lower bands were confirmed to represent Microcystis strains. In contrast to the improved strain differentiation obtained by ITSb or ITSc DGGE compared to that for ITSa DGGE, examples were found for the opposite situation. For instance, strains PCC7806 and V28 (lane 17) could hardly be distinguished when analyzed by using ITSb or ITSc amplicons but occupied clearly different positions in the DGGE gel when ITSa amplicons were used (Fig. 1B through D). The two strains that had yielded two bands on ITSa DGGE, V88 (lane 19) and V40 (lane 20), gave rise to a single band with ITSb or ITSc DGGE (Fig. 1B through D).
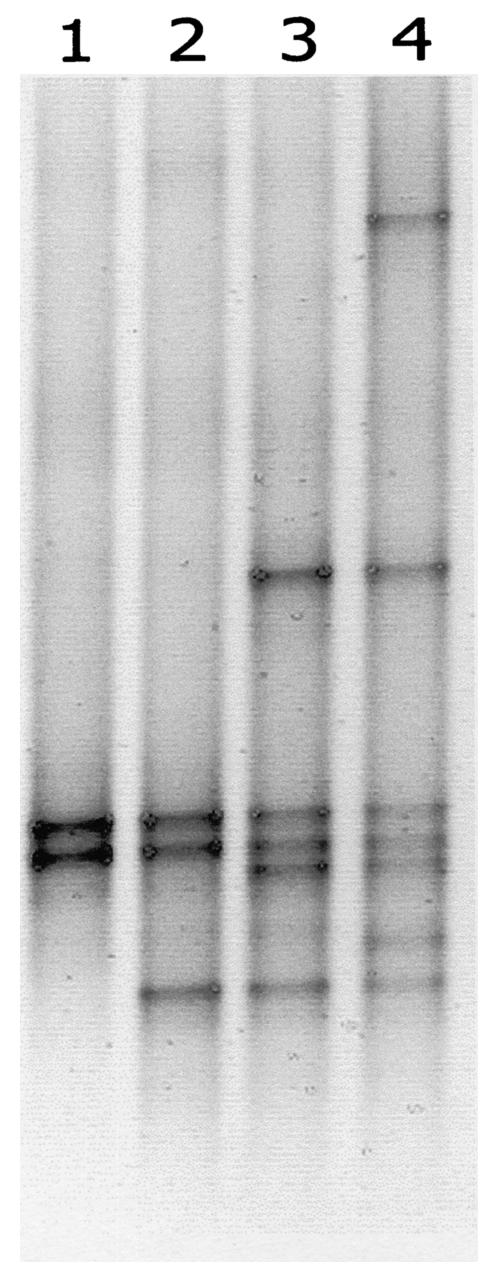
The presence of several homologous DNA templates may affect PCR amplification and result in heteroduplex (see above) and chimera formation. To investigate the occurrence of these phenomena, we mixed ITSa amplification products from seven Microcystis strains that could be separated by using DGGE and used these mixtures as templates for ITSa amplification. Figure 2 shows the DGGE banding patterns generated from template mixtures of two, three, five, or seven Microcystis strains. In spite of the presence of several related sequences, each strain in the mixtures gave rise to one single band of similar intensity and no additional bands were detected.
FIG. 2.
DGGE profiles of ITSa amplified from DNA template mixtures. Lanes 1 through 4 show ITSa PCR products which were amplified from equal concentrations of template mixtures of Microcystis strains PCC7820 plus K29 (lane 1), PCC7820 plus K29 plus SAG17.85 (lane 2), Z6 plus PCC7820 plus CYA140 plus K29 plus SAG17.85 (lane 3), and S2 plus Z6 plus PCC7820 plus CYA140 plus K29 plus CYA43 plus SAG17.85 (lane 4).
DGGE profiles of rRNA-ITS from different cyanobacterial genera.
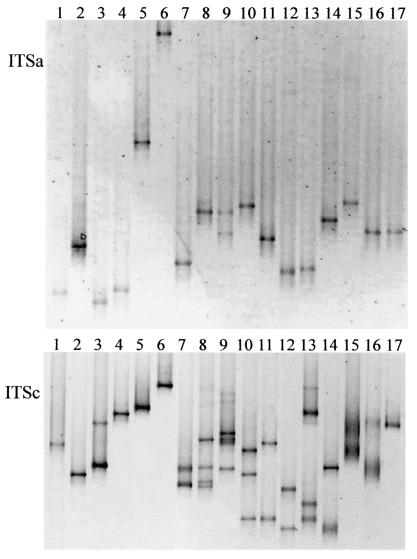
The developed protocols that proved to be effective for Microcystis separation were applied to DGGE analysis of specifically amplified rRNA-ITS of other cyanobacterial genera (Table 3). For almost every strain that was tested, ITSa amplification gave rise to one sharp band, mostly occupying a unique position in the gel (Fig. 3A). Only Aphanizomenon gracile yielded two bands. Also, the longer amplification products of ITSb and ITSc were analyzed using DGGE (Fig. 3B). The patterns for ITSb were mostly similar to those for ITSc and are therefore not shown. Strains from the genera Synechococcus, Synechocystis, Lyngbya, Pseudanabaena, Trichodesmium, and Prochlorothrix yielded one band (Fig. 3). Leptolyngbya had a second band of lower intensity. Strains from the genera Anabaena, Aphanizomenon, Planktothrix, Cylindrospermopsis, and Gloeotrichia yielded two to four clear bands in most cases and a few less-distinct additional bands for Anabaenopsis. Possibly some of the bands resulted from heteroduplex formation. Nostoc and Nodularia strains yielded two very fuzzy bands. In addition to the strains shown in Fig. 3, a Microcoleus sp. isolate was analyzed and yielded sharp, single bands on both ITSa and ITSc DGGE gels (data not shown).
FIG. 3.
DGGE profiles of various cyanobacterial genera amplified with ITSa and ITSc primers. The following strains were analyzed: Synechococcus sp. (lane 1), Synechocystis sp. (lane 2), Leptolyngbya sp. (lane 3), Lyngbya sp. (lane 4), Pseudanabaena catenata (lane 5), Trichodesmium erythraeum (lane 6), Anabaena variablis (lane 7), Aphanizomenon flos-aquae (lane 8), Aphanizomenon gracile (lane 9), Planktothrix aghardii (lane 10), Planktothrix prolifica (lane 11), Cylindrospermopsis raciborskii (lane 12), Anabaenopsis arnoldi (lane 13), Gloeotrichia sp. (lane 14), Nostoc sp. (lane 15), Nodularia spumigena (lane 16), and Prochlorothrix hollandica (lane 17). The gels had a denaturant concentration gradient of 25 to 40%, and only the part of the gel containing bands is shown. The lower gel was assembled from two gels.
Field samples.
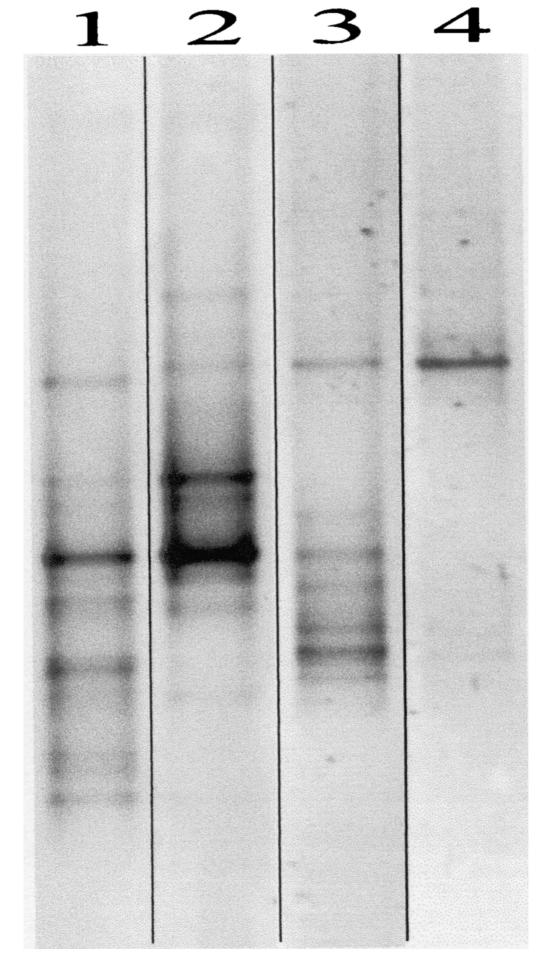
To confirm that the developed methods for high-resolution analysis of cyanobacteria are valid for analysis of the complex microbial communities encountered in the field, we tested two rRNA-ITS primer sets on field samples. Samples from three freshwater lakes were taken in summer, when dense blooms of cyanobacteria of various genera were observed, and from one lake also in winter, when there was no visible presence (including microscopic) of distinctive cyanobacteria. When analyzed on DGGE, ITSa and ITSc PCR products gave rise to complex banding patterns. As shown in the ITSa profiles in Fig. 4, unique as well as (probably) similar genotypes could be identified. The dominant bands were excised from the gel, and the PCR products were reamplified and sequenced. A BLAST search performed on these sequences revealed that the retrieved bands were all derived from cyanobacteria of various genera, including Microcystis, Anabaena, Synechococcus, Oscillatoria, and Nostoc.
FIG. 4.
DGGE profiles generated from ITSa amplification products from freshwater lake samples. The samples were taken from Lake Zeegerplas on July 20 (lane 1) and August 21 (lane 2) when cyanobacteria were blooming and from Lake Kinselmeer on May 5 (lane 3) during a cyanobacterial bloom and on January 16 in the absence of a visible presence of cyanobacteria (lane 4). Several dominant bands were excised, reamplified, and sequenced.
DISCUSSION
There are two main considerations for using primers specific for cyanobacteria in DGGE analyses. First, specific primers prevent amplification of the abundant DNA of noncyanobacterial microbes in field samples. The resulting DGGE profiles are less complex than those generated with general bacterial primers; hence, detection of cyanobacteria that are less abundant or have lower amplification efficiencies is more feasible. Second, characterization of cultures by DGGE, but also by restriction enzyme digestion or sequencing, is not possible when DNA from contaminants is coamplified. Rendering cyanobacteria axenic can be a difficult and time-consuming procedure, and cyanobacteria are often cultivated more easily when accompanied by heterotrophic bacteria (27). The selectivity of our developed primer sets was confirmed through homology searches in databases and through in vitro tests using DNA from cultures of heterotrophic bacteria and from complex microbial communities in freshwater lakes (Fig. 4). The appearance of only one to four distinct bands in DGGE profiles of cultures from various genera (Fig. 1 and 3) may serve as additional support for selectivity of the developed primer sets. None of the cultures used in our study was axenic, yet no unexplainable additional bands were detected on DGGE even when PCR conditions were made less selective (by substantially lowering annealing temperatures).
Primer 322, described by Wilmotte et al. (40), and primer 16CITS, described by Neilan et al. (26), are targeted to sequences just upstream from the target sequence of forward primer CSIF and thus could produce virtually identical amplification products. Primer 322, however, is targeted to highly conserved bacterial sequences and is therefore suitable only for studies of axenic cultures of cyanobacteria (14, 21). Primer 16CITS is specific for cyanobacteria. However, from our alignment of 16S sequences it appears that primer CSIF has more sequence differences with noncyanobacterial sequences, especially at the 3′ site. For primer 16CITS, we found 71 noncyanobacterial sequences with two mismatches (as opposed to none for primer CSIF) and 266 noncyanobacterial sequences with three mismatches (as opposed to two for CSIF). Primer CSIF can therefore be considered more specific for cyanobacteria. Nevertheless, we emphasize that this was based on sequences in the present database and that no experimental comparisons of the primers were made.
Coverage of a broad range of genera for primer CSIF was confirmed by the probe match software tool by using published cyanobacterial sequences and was supported by in vitro tests (Table 3 and Fig. 3). We were able to generate PCR products from Nostoc strain CYA124, even though some deposited sequences from this genus had three mismatches with the primer sequence. Investigations must still be done to determine whether PCR products can be formed from all strains from the genera Nostoc and Spirulina (two or three mismatches in deposited sequences), from Gloeobacter and Calothrix (three mismatches), and from genera without deposited sequences. The generation of amplification products from strains of all tested cyanobacterial genera with primer 373R as the reverse primer and from most genera with primer ITS3R is supported by the universal occurrence of the highly conserved gene for tRNAIle and of certain structural elements in cyanobacteria (7, 13, 34).
The difference in resolution with 16S and ITS DGGE in Microcystis is in agreement with the reported average sequence diversity of less than 1% in 16S (7, 29) compared to up to 7% in rRNA-ITS (26, 30). The high resolution and the single bands that are generated make rRNA-ITS DGGE a valuable method for analysis of this genus. We made the assumption that the multiple bands containing Microcystis sequences in the rRNA-ITS DGGE profiles of some cultures reflect the coisolation of two different Microcystis strains instead of the occurrence of two different rRNA operons. This assumption can be confirmed unequivocally only by repeated plating to make certain that the analyzed cultures are unialgal. This elaborate procedure was not carried out, since we considered the possibility of coisolation of two morphologically indistinguishable organisms far more likely than the occurrence of many different operons in a limited number of deviant strains. Also, all strains that were most likely unialgal (because they originated from culture collections or had been isolated by using plating techniques) gave rise to one band on DGGE. Following this line of reasoning, the explanation for the two sequences in culture collection strain SAG17.85 (Fig. 1C and D) must be that they acquired unnoticed contamination during years of cultivation in the lab. Support for the occurrence of only one operon (or several identical operons) in the genus Microcystis comes from the unambiguous sequences that Otsuka et al. (30) obtained from sequencing the rRNA-ITS of 47 Microcystis strains.
The occurrence of multiple operons in one organism resulting in rRNA-ITS amplicons of different sizes (Table 3) has been found before in the genera Anabaena, Aphanizomenon, Anabaenopsis, Nostoc, Cylindrospermopsis, and Nodularia (13, 14, 26). Sequencing of the rRNA-ITS from Nodularia, Nostoc, Calothrix, and Scytonema strains (3, 7, 13) revealed that not all rRNA operons of cyanobacteria contain tRNAIle genes. However, since other operons in these organisms do contain tRNAIle genes, they are detectable with ITSa amplification (as shown for Nodularia and Nostoc in Fig. 3A). The presence of tRNA genes on only one copy of the rRNA may explain for a number of genera the production of only one band after ITSa amplification (Fig. 3A and Table 3), despite the existence of different operons revealed by ITSb or ITSc amplification (Fig. 3B and Table 3). Alternatively, generation of a single ITSa DGGE band could be explained by identical sequences at the 5′ end of different rRNA-ITS operons.
For the generation of DGGE profiles from longer stretches of rRNA-ITS, primer set ITSc is preferable in most cases. Compared to ITSb analysis, the resolution was similar (compare Fig. 1C and D), yet more genera could be amplified and there is more sequence information contained in the amplicons (60 to 100 bp extra; Table 3). Nevertheless, for some genera, primer set ITSb was more suitable for DGGE profiling. For instance, ITSb amplification products of Nodularia yielded sharp DGGE bands (data not shown), whereas ITSc DGGE resulted in diffuse bands unsuitable for analysis (Fig. 3B). Also, DGGE profiles resulting from ITSb amplification of Planktothrix aghardii were relatively simple (one band) compared to the profile resulting from ITSc amplification (three bands). Due to the occurrence of multiple rRNA-ITS operons in one organism, identification of all dominant bands produced with ITSb and ITSc primers is necessary to come to a reliable estimate of the diversity of cyanobacteria in complex DGGE profiles.
It must be emphasized that DGGE diversity profiles do not necessarily reflect the true diversity in the field. DNA extraction efficiency and the number of rRNA operons may vary between genera, and the PCR step has several inherent pitfalls, which complicate the interpretation of DGGE profiles of communities. Artifacts introduced by the PCR amplification reaction of homologous sequences include chimera (17) and heteroduplex formation (15), which may lead to overestimation of the number of organisms, and template annealing (36) and preferential amplification of some DNA templates (33), which may result in a shift in the apparent ratio of the different organisms after PCR amplification. Heteroduplex bands were formed in some cultures suspected of containing more than one Microcystis isolate (Fig. 1) and in some organisms containing multiple operons (Fig. 3). In contrast, deliberate mixing of template DNA from several Microcystis strains showed no evidence of chimera or heteroduplex formation, since no additional bands were detected in the DGGE profiles (Fig. 2). Equal band intensities pointed to the absence of preferential amplification. However, this effect is more likely between genera, since primer binding may differ due to imperfect matching.
Conclusion.
By using the primers and protocols described in this paper, cyanobacterial communities and isolates can be studied at high resolution. The selectivity of the primers makes it possible to focus on cyanobacteria in the presence of other organisms. The rationale for using different rRNA-ITS primer sets has become clear from our results. Each set has its advantages and disadvantages, and the choice of which one to use depends on the study that is to be executed. For studying cyanobacteria, ITSa primers have the broadest applicability and produce the most straightforward DGGE profiles. However, bands from some organisms may end up at identical positions in the gel even though sequence differences do exist (either because sequence differences occur in the nonamplified 3′ part of the rRNA-ITS or because different sequences dictate identical melting behavior). The information obtained from DGGE of ITSa may be insufficient to describe an ecosystem in detail or to make sure that a culture contains one cyanobacterial isolate. The longer stretches of rRNA-ITS that are amplified by ITSb and ITSc primers make detection of additional sequence differences possible and thus provide supplementary resolution (Fig. 1). Also, these amplicons contain more sequence information, which is especially interesting when field profiles are analyzed. The disadvantages of ITSb primers are their inability to amplify DNA of some genera (Table 3) and the complex DGGE profiles they produce for some genera. Most sequence information can be retrieved from ITSc primer amplicons. Moreover, for most tested genera, they yielded sharp bands on DGGE gels.
Acknowledgments
This work was funded by the Technology Foundation STW (project no. ACH 4874).
Josje Snoek is gratefully acknowledged for isolation and maintenance of cyanobacterial cultures. Miguel Dionisio-Pires, Olav M. Skulberg, Sven Jansson, and Henk M. Jonkers are gratefully acknowledged for supplying cyanobacterial cultures.
REFERENCES
- 1.Abed, R. M. M., and F. Garcia-Pichel. 2001. Long-term compositional changes after transplant in a microbial mat cyanobacterial community revealed using a polyphasic approach. Environ. Microbiol. 3:53-62. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker, G. L. A., P. K. Hayes, S. L. O'Mahony, P. Vacharapiyasophon, and A. E. Walsby. 1999. A molecular and phenotypic analysis of Nodularia (cyanobacteria) from the Baltic Sea. J. Phycol. 35:931-937. [Google Scholar]
- 4.Becker, S., M. Fahrbach, P. Böger, and A. Ernst. 2002. Quantitative tracing, by Taq nuclease assays, of a Synechococcus ecotype in a highly diversified natural population. Appl. Environ. Microbiol. 68:4486-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltran, E. C., and B. A. Neilan. 2000. Geographical segregation of the neurotoxin-producing cyanobacterium Anabaena circinalis. Appl. Environ. Microbiol. 66:4468-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bittencourt-Oliveira, M. D., M. C. de Oliveira, and C. J. S. Bolch. 2001. Genetic variability of Brazilian strains of the Microcystis aeruginosa complex (Cyanobacteria/Cyanophyceae) using the phycocyanin intergenic spacer and flanking regions (cpcBA). J. Phycol. 37:810-818. [Google Scholar]
- 7.Boyer, S. L., V. R. Flechtner, and J. R. Johansen. 2001. Is the 16S-23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol. Biol. Evol. 18:1057-1069. [DOI] [PubMed] [Google Scholar]
- 8.Burger Wiersma, T., L. J. Stal, and L. R. Mur. 1989. Prochlorothrix hollandica gen-nov, sp-nov, a filamentous oxygenic photoautotrophic procaryote containing chlorophyll a and chlorophyll b—assignment to Prochlorotrichaceae fam-nov and order Prochlorales Florenzano, Balloni, and Materassi 1986, with emendation of the ordinal description. Int. J. Syst. Bacteriol. 39:250-257. [Google Scholar]
- 9.Chorus, I., and J. Bartram. 1999. Toxic cyanobacteria in water. E & FN Spon, London, England.
- 10.De Rijk, P., and R. De Wachter. 1993. DCSE v2.54, an interactive tool for sequence alignment and secondary structure research. Comput. Appl. Biosci. 9:735-740. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Pichel, F., M. Kühl, U. Nübel, and G. Muyzer. 1999. Salinity-dependent limitation of photosynthesis and oxygen exchange in microbial mats. J. Phycol. 35:227-238. [Google Scholar]
- 12.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 13.Iteman, I., R. Rippka, N. Tandeau de Marsac, and M. Herdman. 2000. Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiology 146:1275-1286. [DOI] [PubMed] [Google Scholar]
- 14.Iteman, I., R. Rippka, N. Tandeau de Marsac, and M. Herdman. 2002. rDNA analyses of planktonic heterocystous cyanobacteria, including members of the genera Anabaenopsis and Cyanospira. Microbiology 148:481-496. [DOI] [PubMed] [Google Scholar]
- 15.Jensen, M. A., and N. Straus. 1993. Effect of PCR conditions on the formation of heteroduplex and single-stranded DNA products in the amplification of bacterial ribosomal DNA spacer regions. PCR Methods Appl. 3:186-194. [DOI] [PubMed] [Google Scholar]
- 16.Jonkers, H. M., R. Ludwig, R. De Wit, O. Pringault, G. Muyzer, H. Niemann, N. Finke, and D. De Beer. 2003. Structural and functional analysis of a microbial mat ecosystem from a unique permanent hypersaline inland lake: ′La Salada de Chiprana' (NE Spain). FEMS Microbiol. Ecol. 1488:1-15. [DOI] [PubMed] [Google Scholar]
- 17.Kopczynski, E. D., M. M. Bateson, and D. M. Ward. 1994. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl. Environ. Microbiol. 60:746-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurmayer, R., E. Dittmann, J. Fastner, and I. Chorus. 2002. Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. in Lake Wannsee (Berlin, Germany). Microb. Ecol. 43:107-118. [DOI] [PubMed]
- 19.Kwok, S., D. E. Kellog, N. McKinney, D. Spasic, L. Goda, C. Levenson, and J. J. Sninsky. 1990. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus 1 model studies. Nucleic Acids Res. 18:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laamanen, M. J., L. Forsström, and K. Sivonen. 2002. Diversity of Aphanizomenon flos-aquae (cyanobacterium) populations along a Baltic Sea salinity gradient. Appl. Environ. Microbiol. 68:5296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laloui, W., K. A. Palinska, R. Rippka, F. Partensky, N. Tandeau de Marsac, M. Herdman, and I. Iteman. 2002. Genotyping of axenic and non-axenic isolates of the genus Prochlorococcus and the OMF-‘Synechococcus' clade by size, sequence analysis or RFLP of the internal transcribed spacer of the ribosomal operon. Microbiology 148:453-465. [DOI] [PubMed] [Google Scholar]
- 22.Lovell, C. R., M. J. Friez, J. W. Longshore, and C. E. Bagwell. 2001. Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora. Appl. Environ. Microbiol. 67:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, W. Q., E. H. Evans, S. M. McColl, and V. A. Saunders. 1997. Identification of cyanobacteria by polymorphisms of PCR-amplified ribosomal DNA spacer region. FEMS Microbiol. Lett. 153:141-149. [Google Scholar]
- 24.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neilan, B. A., J. L. Stuart, A. E. Goodman, P. T. Cox, and P. R. Hawkins. 1997. Specific amplification and restriction polymorphisms of the cyanobacterial rRNA operon spacer region. Syst. Appl. Microbiol. 20:612-621. [Google Scholar]
- 27.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orcutt, K. M., U. Rasmussen, E. A. Webb, J. B. Waterbury, K. Gundersen, and B. Bergman. 2002. Characterization of Trichodesmium spp. by genetic techniques. Appl. Environ. Microbiol. 68:2236-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otsuka, S., S. Suda, R. H. Li, M. Watanabe, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 1998. 16S rDNA sequences and phylogenetic analyses of Microcystis strains with and without phycoerythrin. FEMS Microbiol. Lett. 164:119-124.
- 30.Otsuka, S., S. Suda, R. H. Li, M. Watanabe, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 1999. Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence. FEMS Microbiol. Lett. 172:15-21. [DOI] [PubMed] [Google Scholar]
- 31.Palinska, K. A., W. Liesack, E. Rhiel, and W. E. Krumbein. 1996. Phenotype variability of identical genotypes: the need for a combined approach in cyanobacterial taxonomy demonstrated on Merismopedia-like isolates. Arch. Microbiol. 166:224-233. [DOI] [PubMed] [Google Scholar]
- 32.Rasmussen, U., and M. M. Svenning. 2001. Characterization by genotypic methods of symbiotic Nostoc strains isolated from five species of Gunnera. Arch. Microbiol. 176:204-210. [DOI] [PubMed] [Google Scholar]
- 33.Reysenbach, A.-L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rychlik, W. 1995. Selection of primers for polymerase chain reaction. Mol. Biotechnol. 3:129-134. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tillett, D., and B. A. Neilan. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36:251-258. [Google Scholar]
- 38.Whitton, B. A., and M. Potts. 2000. The ecology of cyanobacteria, their diversity in time and space, 1st ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 39.Wilmotte, A. 1994. Molecular evolution and taxonomy of the cyanobacteria, p. 1-25. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 40.Wilmotte, A., G. Van der Rauwera, and R. De Wachter. 1993. Structure of the 16-S ribosomal RNA of the thermophilic cyanobacterium chlorogloeopsis HTF (‘mastigocladus laminosus HTF') strain PCC7518, and phylogenetic analysis. FEBS Lett. 317:96-100. [DOI] [PubMed] [Google Scholar]
- 41.Young, J. P. W. 1992. Phylogenetic classification of nitrogen-fixing organisms, p. 43-86. In G. Stacey, H. J. Evans, and R. H. Burris (ed.), Biological nitrogen fixation. Chapman & Hall, New York, N.Y.
- 42.Zehr, J. P., M. T. Mellon, and W. D. Hiorns. 1997. Phylogeny of cyanobacterial nifH genes: evolutionary implications and potential applications to natural assemblages. Microbiology (Reading) 143:1443-1450. [DOI] [PubMed] [Google Scholar]
- 43.Zwart, G., W. D. Hiorns, B. A. Methe, M. P. Van Agterveld, R. Huismans, S. C. Nold, J. P. Zehr, and H. J. Laanbroek. 1998. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst. Appl. Microbiol. 21:546-556. [DOI] [PubMed] [Google Scholar]