Abstract
In this study, we examined the long-term development of the overall structural morphology and community composition of a biofilm formed in a model drinking water distribution system with biofilms from 1 day to 3 years old. Visualization and subsequent quantification showed how the biofilm developed from an initial attachment of single cells through the formation of independent microcolonies reaching 30 μm in thickness to a final looser structure with an average thickness of 14.1 μm and covering 76% of the surface. An analysis of the community composition by use of terminal restriction fragment length polymorphisms showed a correlation between the population profile and the age of the sample, separating the samples into young (1 to 94 days) and old (571 to 1,093 days) biofilms, whereas a limited spatial variation in the biofilm was observed. A more detailed analysis with cloning and sequencing of 16S rRNA fragments illustrated how a wide variety of cells recruited from the bulk water initially attached and resulted in a species richness comparable to that in the water phase. This step was followed by the growth of a bacterium which was related to Nitrospira, which constituted 78% of the community by day 256, and which resulted in a reduction in the overall richness. After 500 days, the biofilm entered a stable population state, which was characterized by a greater richness of bacteria, including Nitrospira, Planctomyces, Acidobacterium, and Pseudomonas. The combination of different techniques illustrated the successional formation of a biofilm during a 3-year period in this model drinking water distribution system.
Ecologists have long studied the development of communities of plants and animals on newly exposed landforms. They have described how species successively invade and colonize a site, inducing alterations in biomass, productivity, diversity, and so forth and culminating in a steady-state community or the climax community (6, 31). The stepwise change in a population is an effect of individual members interacting and results in a self-induced modification of the local environment, thereby creating new niches for colonization or excluding existing members (31). Alternatively, changing environmental factors, localized gradients, topography, stochastic processes, spatial separation, and other factors result in random or loosely coordinated populations, as in many bacterial soil communities (12, 30, 45).
The structural development of biofilm formation has been described as a sequence of events including attachment, microcolony development, and finally establishment of distinct structures, e.g., “mushrooms” (reviewed in references 7 and 27). However, these studies generally have examined changes in biofilm morphology (e.g., thickness and roughness), and few have combined such examinations with investigations of alterations in community composition (15). In addition, most biofilm studies have focused on short-term changes, assuming that they reflected long-term effects of various factors.
Can a successional model be applied to microbial communities? In terms of colonization of a surface, microbial biofilm formation could be viewed as a successional process in structure and composition. The process begins with the stochastic primary attachment of a number of species recruited from the bulk water population forming a monolayer on the surface. Since the bacteria initially are spatially separated and have a low selection pressure for attachment, most likely a community with a high level of diversity occurs. This initial attachment (facilitation) can be followed by a secondary colonization of bacteria that benefit from a protective environment in the biofilm and/or feed on the remnants of other bacteria. In this secondary community, better resource or space competitors may exclude less competitive organisms. Further biofilm formation then may cause a reduction in diversity, as a single or a few superior competitors start to dominate the community. As the bacterial biofilm matures, more niches are created due to the formation of gradients and the internal recycling of resources. At this seral stage, richness and evenness increase again, establishing a unimodal relationship between age and diversity. The final increase in species richness reflects a complex spatial structure with many functional groups of bacteria (16).
Biofilms formed in water distribution networks are known to cause public health problems, such as protecting and supporting pathogenic microorganisms (5), bacterial regrowth (20, 42), and depletion of disinfection agents (34). Several groups of investigators have examined the diversity of these biofilms to identify bacteria involved in these problems (18, 29, 34), and such knowledge may provide new tools for improving water quality for the consumer (21).
In this study, we examined the overall morphology and community composition of a biofilm in a model drinking water distribution system by using techniques independent of cultivation. In a previous investigation of this model system, colonization and the development of a microbial biofilm were analyzed by using direct and viable counts. The direct enumeration of cells illustrated how the biomass increased during the first 200 to 300 days, reaching a stationary state with regard to number of cells. Concurrently, the ratio of viable to total cells decreased during the initial colonization, from about 1/10 to 1/10,000 CFU/cell in the stationary biofilm (2), indicating a shift in either the diversity or the physiology of the bacteria.
The purposes of the present study included testing whether the structure of the biofilm developed through serial stages, as in other model biofilm communities, and examining the extent of spatial variation in community composition in the model system. This information is important for verifying the suitability and reproducibility of our model system for analyzing factors influencing microbial diversity in drinking water distribution systems. Finally, we wanted to test whether the development of a physical structure was accompanied by the succession of community composition and diversity and then further identify the dominant bacteria at different stages of biofilm development.
The outcome of our investigation suggests whether a biofilm is formed by stochastic processes leading to patches of diversity or whether the formation is a more orderly process toward a defined community.
Moreover, the results of our study indicate a baseline for the spatial and temporal variations in a biofilm and provide a reference for future studies investigating factors influencing the microbial community in a water distribution system.
MATERIALS AND METHODS
Description of the system.
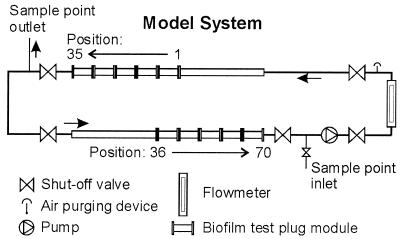
Biofilm and bulk water were sampled from a model water distribution system operated under turbulent-flow conditions (described in detail by Boe-Hansen et al. [3]). In brief, the model system was built as a completely mixed 12.2-m loop-shaped reactor with a retention time of 2 h and a flow velocity of 0.07 m/s. Aligned with the surface, 72 stainless steel plugs (grade 316; area, 7.1 cm2) were inserted (Fig. 1). Before insertion, the plugs were treated with 10% HNO3 for 24 h and heated to 220°C for 5 h. The model system was supplied with groundwater from a water works facility (pipe distance, 1 km; no branching points). The physicochemical properties of the inlet water are listed in Table 1; the water contained no disinfectant.
FIG. 1.
Model distribution system.
TABLE 1.
List of physicochemical properties for inlet watera
| Property | Value |
|---|---|
| Turbidity | 0.2 FTU |
| pH | 7.4 |
| Conductivity | 86.3 mS/m |
| NVOC | 2.2 |
| COD | 7 |
| Solids | 480 |
| Hardness (CaO) | 164 |
| CO2 | <2 |
| HCO3− | 344 |
| Methane | <0.01 |
| O2 | 8.1 |
| F− | 0.5 |
| Cl− | 93 |
| NH4+ | <0.05 |
| NO2− | <0.01 |
| NO3− | 3.1 |
| P (total) | <0.02 |
| SO42− | 11 |
| Na+ | 59.5 |
| Mg2+ | 18 |
| K+ | 3.5 |
| Ca2+ | 87.5 |
| Mn (total) | <0.005 |
| Fe (total) | 0.05 |
| Ni2+ | <3 μg/liter |
Measured according to Danish standards. Values are given in milligrams per liter unless otherwise indicated and are averages of two measurements. FTU, formazine turbidity unit; NVOC, nonvolatile organic compounds; COD, chemical oxygen demand.
Sampling.
Two sample series were analyzed. In the first series, 15 plugs from different positions in the model system but of the same age (818 days old) were analyzed to examine spatial variations in the microbial community. In the second series, 22 biofilm samples of different ages were analyzed to investigate temporal variations in biofilm morphology and composition. The plugs had been inserted at different times but were sampled simultaneously to limit the effects of seasonal variations on the microbial community. The two series were sampled 3 months apart. The plugs were kept at 5°C until analysis (less than 5 h after sampling). Eight 2-liter samples of water from the inlet and the outlet were filtered with 0.22-μm-pore-size Durapore filters (hydrophilic polyvinylidene difluoride; Millipore, Bedford, Mass.) as bulk water samples and stored at −20°C until analysis. The bulk water was sampled before and after each of the previously mentioned biofilm series.
Microscopy and image analysis of biofilm.
Bacteria in the biofilm were stained with 1 ml of 5 μM SYTO 62 (Molecular Probes, Eugene, Oreg.) for 15 min, washed, and counterstained with 0.9% NaCl for 15 min. Cells were visualized with an LSM 510 scanning confocal laser microscope (Zeiss, Jena, Germany). Several images from each plug were acquired at a random position, based on averaging of four vertical scans to increase the signal-to-noise ratio. Extended-focus images and vertical cross sections through the biofilm were generated by using the IMARIS software package (Bitplane AG, Zurich, Switzerland). An extended version of the Comstat software package (14) was developed for quantitative analysis of the heterogeneous structure of the biofilm. Biomass was quantified by performing a threshold operation and labeling all three-dimensionally connected pixel regions containing 30 pixels to remove background noise. The threshold levels were set by using the algorithm of Otsu (33). Biomass was estimated as the total volume of the pixel regions extracted from the background relative to the surface area of the sample. The biofilm coverage fraction was calculated as the relative percentage of lateral grid points in the three-dimensional image with biomass at one or more depth positions. The thickness at a given lateral grid position was determined as the distance from the lowest depth coordinate containing biomass at the given position to the highest depth coordinate. From this definition, the average thickness was estimated as the average thickness of the surface region covered with biofilm (according to the above definition of coverage).
DNA extraction.
DNA was extracted by using a modification of the protocol of Fuhrman et al. (11). In brief, cells from the plugs were removed from the surface with a wet cotton swab, leaving no visible biofilm behind; control staining of remaining cells showed only a few isolated cells on the surface. Filters with cells from the bulk water were cut into pieces. Biofilm and bulk water samples were then added to 1% sodium dodecyl sulfate in a buffer containing 50 mM Tris (pH 8), 5 mM EDTA, and 50 mM NaCl and boiled for 2 min, and DNA was precipitated with 96% ethanol at −20°C. The DNA was further purified by using phenol extraction and a QIAprep spin column (Qiagen, Hilden, Germany) to remove any inhibitory agents (including ferric oxides) that interfered with PCR amplification. Finally, the DNA was quantified by measuring the absorbance at 260 nm.
T-RFLP.
DNA from bulk water and biofilm samples was analyzed for the microbial population profile by using a 16S ribosomal DNA (rDNA)-targeted terminal restriction fragment length polymorphism (T-RFLP) protocol (23). Amplification of DNA was performed by PCR with Bacteria-specific primers 9F (5′-GAG TTT GAT CCT GGC TCA G-3′; labeled with 6-FAM, a 6-carboxyfluorescein-derived phosphoramidite fluorochrome) and 1512R (5′-ACG GCT ACC TTG TTA CGA CTT-3′). A total of 16.5 ng of genomic DNA per 50-μl reaction volume was used as a template in all PCRs. The following thermal cycling program was applied: a hot start at 94°C for 5 min; 30 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 2 min; and a final extension at 72°C for 10 min. The generated fragments were precipitated with ethanol for 2 h at −20°C, dissolved in 50 μl of water, and quantified by gel electrophoresis. The PCR product (300 ng) was digested with 20 U of HhaI (New England BioLabs, Beverly, Mass.) and purified by using YM-10 Microcon spin columns (Millipore). HhaI was used as the restriction enzyme because in silico analysis had predicted good separation of genetic diversity of 16S rRNA with this enzyme (9, 23). To limit PCR amplification bias, the amount of DNA used as the template and in the subsequent restriction analysis was quantified and standardized for all samples (32). Moreover, a surface area of the biofilm plugs of 7.1 cm2 ensured sufficient biomass for DNA extraction to avoid nested or high-cycle PCR.
Separation of the digestion products and the added base-pair marker and detection of labeled fragments were achieved by capillary electrophoresis with an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.) at the Genomic Technology Support Facility (Michigan State University, East Lansing). Electropherograms were analyzed by using STRand (http://www.vgl.ucdavis.edu/STRand) (41) to determine fragment lengths and transferred to a binary data format (presence or absence of peak). T-RFLP profiles were aligned and clustered with the neighbor-joining algorithm of Saitou and Nei (35), which is included in ClustalX (17). The Mantel test (25) for correlations among sample position, sample age, and the resulting population profile was performed by using the CADM program (test for congruence among several distance matrices) (22). In this test, the hypothesis was the incongruence of all matrices.
Cloning and sequencing.
Unlabeled 16S rDNA fragments were generated by the same PCR protocol as that used in the T-RFLP analysis and cloned by using a TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.). In addition, amplified DNA was gel purified to ensure that the cloned fragments were the correct sizes, since this cloning kit has a strong bias toward smaller fragments. Clones were sequenced with an ABI Prism 3700 DNA analyzer (Applied Biosystems) at the Genomic Technology Support Facility (Michigan State University); M13F was used as the sequence primer.
Phylogeny.
To identify bacteria in the samples, 16S rDNA sequences were aligned by using the fast-alignment tool implemented in the ARB software package (39), with manual correction for alignment errors. The new sequence was analyzed against the phylogenetic tree containing all sequences in the ARB database 6spring2001 by using the maximum-parsimony quick-addition tool to obtain an initial estimate of the affiliation of a new bacterial specimen. The family of each strain was identified, and a new tree was reconstructed with sequences from all species within the corresponding family, as described in Bergey's Manual (1), by using the neighbor-joining, maximum-parsimony, and FastDNA maximum-likelihood algorithms included in the ARB software package. A base frequency filter was generated on the basis of the selected sequences and excluding all positions different in more than 70% of the sequences to enable a comparison of homologous positions.
RESULTS AND DISCUSSION
The model drinking water distribution system used for this investigation has been operating for more than 3 years under constant hydraulic and temperature conditions that are similar to those of supply systems in Denmark. The system is designed as a completely mixed loop-shaped reactor allowing control of the water retention time and the flow velocity. Samples from 1 to 94 days old are termed young biofilm, whereas samples from 571 to 1,093 days old are defined as old biofilm, a distinction that will become clear in the following text.
Quantification of biofilm structure.
Biofilm samples from 1 day to approximately 3 years old were stained and visualized with a scanning confocal laser microscope to identify temporal patterns of the establishment of the biofilm. Our results showed that on average, a minimum of four vertical scans was necessary to distinguish biomass from background noise, due to the weak signal from and small size of the cells (data not shown).
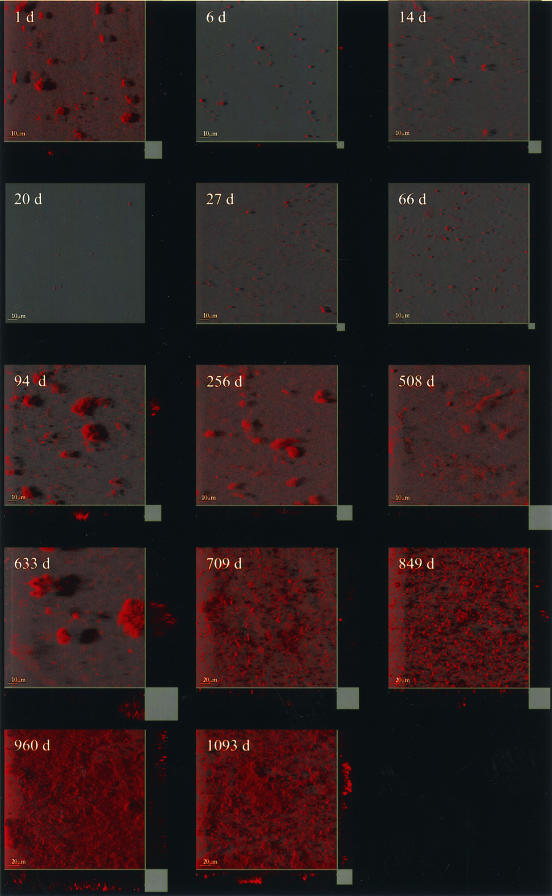
The micrographs (Fig. 2; e.g., the cross section at day 960) revealed how the bacteria inhabited crevices and depressions in the metal surface, creating an uneven base of the biofilm. Accordingly, the substratum of the biofilm could not be considered to be a plane surface. In addition, the visible biofilm consisted of individual cells separated by empty areas. Therefore, the biofilm could not be defined as single pixels connected to a base layer, as is normally assumed when the Comstat package is used. Instead, the biomass was extracted as all connected regions above a defined volume.
FIG.2.
Scanning confocal laser microscope micrographs showing the structural development of the biofilm aging from 1 day (d) to 3 years. Samples from 1 to 633 days were acquired at a magnification of ×630, whereas samples from 709 to 1,093 days were acquired at a magnification of ×400. The two different magnifications were applied to enable concurrent visualization of single cells and larger morphological structures.
Development of biofilm structure.
From the micrographs (Fig. 2), it was clear that the morphological structure of the biofilm developed in stages. Several clumps were observed at day 1. It was unclear whether these clumps were of biological origin or constituted nonspecific staining, since the signal here was uniform and bright in contrast to that of the older samples. In general, single cells scattered on the surface mainly constituted the young biofilm. A change in biofilm morphology was observed at about day 94, when individual larger microcolonies started to develop on the substratum, reaching a maximum thickness of 30 μm after 600 days. The structure slowly progressed from individual microcolonies to a looser organization covering most of the substratum from day 700 onward.
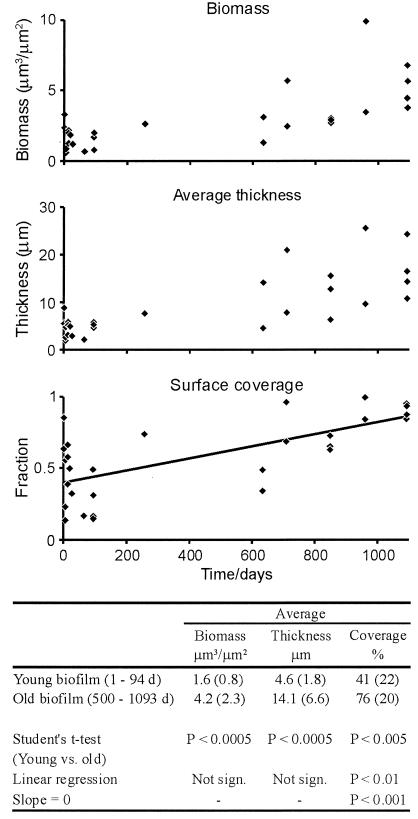
The biofilm increased significantly in average thickness, biomass, and surface coverage (Student's t test), but only the coverage fraction could be fitted by linear regression (Fig. 3). This finding illustrated the heterogeneity of the structure of this community.
FIG. 3.
Quantification of biofilm development over time. The table below the panels shows the average biomass, thickness, and coverage and a statistical comparison of young and old biofilms by using Student's t test and linear regression. Values in parentheses are standard deviations. d, days; sign., significant.
The increase in biomass was approximately threefold, in comparison to the fivefold increase in cell numbers observed earlier with this system (2). Previous studies revealed a coupling between increased metabolic activity and cell size in both laboratory strains and natural communities (36, 37), and this relationship could explain why the ratio of biomass to cell numbers decreased.
Like total biomass, the average thickness of the biofilm progressed from 2 to 26 μm during its development. Remarkably, the thickness of the biofilm for a few samples that were 600 to 800 days old was no greater than that observed for the young biofilm and could have been an outcome of sloughing events.
The coverage of the substratum by the bacterial cells progressed from 48.5 to 95.2% in the oldest samples (day 1093). The coverage fraction reflected how samples 633 days old were composed of large microcolonies that still covered only 50% of the substratum, as shown on the micrographs (Fig. 2).
The observed pattern of biofilm formation agrees with the conceptual model which has been proposed by Costerton and colleagues for the establishment of microbial communities on surfaces (7) and which has been seen in many other systems (19, 28). A net doubling time for cell numbers of 23 days in this system (2) may explain why it takes 3 months before microcolonies appear.
From day 709, the biofilm developed into a looser structure defined by nearly complete surface coverage and increased biomass and biofilm thickness. No increase in cell numbers was seen when microcolonies developed into a looser structure with high surface coverage. The general model for biofilm formation does not predict this final change in structure, which may be explained by the production of extracellular polymeric substances. Extracellular polymeric substances, including polysaccharides, DNA, and peptides, influence the morphology of the biofilm, allowing more elaborate structures to form (13, 44). Another factor is the presence of a substantial amount of iron oxides on the substratum in the old biofilm samples (observed as rust on the test plugs), due to precipitation from the water phase. Iron oxides may act as a matrix supporting the observed looser biofilm structure (4). Finally, sloughing and erosion could influence the physical structure of the biofilm by removing large microcolonies, resulting in a less rough biomass with a smaller average thickness.
Spatial variation of biofilm composition in the reactor.
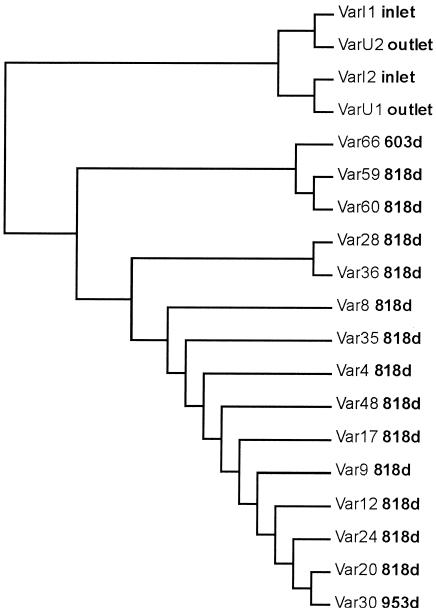
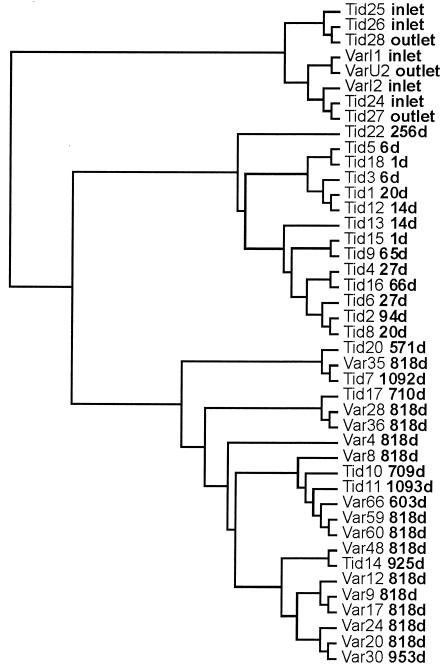
The model system was operated as a completely mixed reactor with a retention time of 2 h, which ensured that all biofilm samples were exposed to the same water phase (3). Therefore, the position of the plug should not influence the population profile of the attached microbial community. T-RFLP confirmed that the bacterial composition of the old biofilm formed was relatively homogeneous, as shown by the random clustering of the communities rather than clustering by actual distance (Fig. 4). Only one area sampled, covering 51 cm in the model system, appeared to cluster according to distance (positions 59 to 66). To test for statistical significance, the dissimilarity matrix of population profiles was compared to the physical distance between the samples by using the Mantel test for incongruence between matrices. A correlation between position and composition was observed (P < 0.0011) when samples from positions 59 to 66 were included in the analysis. Without these samples, the remaining plugs were distributed independently of position (P > 0.159). On the basis of these results, it can be concluded that the position in the model system had only a minor influence on the population of the biofilm. These results demonstrate the suitability of the model system for examining the influence of the age of a biofilm on the microbial community, even though one minor area behaved differently.
FIG. 4.
Comparison of 15 samples of similar ages to analyze for spatial variation. The number after “Var” for each sample indicates the position in the model system shown in Fig. 1. The dendrogram was calculated by using a dissimilarity matrix from the T-RFLP profiles and a subsequent cluster analysis with the neighbor-joining algorithm. d, days.
Temporal variation of biofilm composition.
To investigate the temporal variation of biofilm community composition in the model system, we used a strategy in which plugs were inserted at different times but sampled simultaneously (2). This approach controlled for variation due to time of sampling and thereby enabled an investigation of temporal variation in the attached microbial population over a long time span. Biofilm samples that were 1 to 1,093 days old were analyzed in duplicate by T-RFLP. Two samples did not yield enough DNA for comparison and were excluded from the analysis.
The analysis clustered the young biofilm (1 to 94 days) and the old biofilm (500 to 1,093 days) in distinct clades (Fig. 5). It is not completely clear whether the biofilm gradually changed during the first 94 days. There seemed to be some separation between samples that were 1 to 20 days old and samples that were 27 to 94 days old, but these differences were not as distinct as those between young biofilm and old biofilm. The 256-day-old sample appeared to group apart from the young biofilm, but more samples from this time point are needed for the results to be conclusive. Among the old biofilm samples, no temporal separation was observed. To ensure that the dendrogram was not an artifact of the clustering algorithm, a second tree was calculated by using the Jaccard index and clustering with the unweighted pair-group method with arithmetic averages (24, 43). The separation of young and old biofilm samples was again observed (data not shown), confirming the initial grouping. Since cluster analysis is purely a classification tool and does not provide a level of significance, a statistical assessment of the correlation between age and population profile was performed by using the Mantel test. When the diversity matrix used for clustering and a matrix for the difference in age were compared, a significant correlation was seen (P < 0.0002), confirming the clustering pattern.
FIG. 5.
Comparison of all samples to analyze for temporal variation. The dendrogram was calculated by using a dissimilarity matrix from the T-RFLP profiles and a subsequent cluster analysis with the neighbor-joining algorithm. See the legend to Fig. 4 for details.
With the use of cluster analysis as a sorting tool, it is possible to statistically compare the peak numbers of the different clades. The number of peaks can be regarded as an indirect measurement of species richness in a sample but should be interpreted with care, since peak numbers can be dependent on the strength of the T-RFLP signal (8). Nevertheless, the analysis showed a significantly larger number of peaks in the old biofilm (29) than in the young biofilm (17) (P < 0.008; Student's t test). Likewise, a larger number of peaks were observed in bulk water (29) than in the young biofilm (P < 0.0005; Student's t test), whereas no significant difference in the number of peaks was observed between the old biofilm and bulk water. Ten specific peaks were present in all of the old biofilm samples but could not be detected in the young biofilm samples, whereas five of those peaks were seen in bulk water samples. Two peaks were detected in the young biofilm but not in the old biofilm.
The combination of a clear clustering pattern, differences in numbers of peaks, the presence of distinct peaks, and the relative distances between the different subpopulations analyzed (young, old, and bulk) revealed a clear influence of age on the species composition of the biofilm.
Recently, it was demonstrated how cells are slowly transported along the substratum due to shear under turbulent flow conditions, resulting in the net movement of a biofilm (38). This phenomenon would have had a strong impact on biofilm formation on an introduced virgin surface in between older biofilm areas, as with the approach applied in this study. The observed correlation between age and community composition indicates that the primary mechanism for biofilm formation involves the deposition and growth of bacteria in this system and no or limited transport of cells along the surface.
Even though a stationary biofilm in terms of cell density was observed after 200 to 300 days (2), the physical structure continued to develop. If the result is applicable to other systems, it illustrates the importance of examining temporal variation, since a biofilm may continue to evolve even at constant cell densities. This finding may have some implications for the interpretation of model studies of the survival of pathogens, which are often carried out in young biofilms, since biofilms in real-world systems distribution are usually much older, with quite different properties.
Identification of community members.
In order to identify the dominant bacteria in the biofilm at various stages of development as well as to confirm the clustering pattern based on T-RFLP analysis, 228 16S rRNA fragments were cloned, sequenced, and subjected to a phylogenetic analysis (A. C. Martiny, H.-J. Albrechtsen, E. Arvin, and S. Molin, unpublished data). By performing in silico digestion of the generated sequences, one can predict the organism(s) responsible for individual peaks in the T-RFLP profile (26). A few peaks could be predicted by this analysis, but for the majority of the peaks, no match with the sequences was found. The combination of the dominance of a few bacteria and a high diversity at a low abundance of the target population demands that a considerably larger number of clones be sequenced in order to make this comparison of T-RFLP profiles and sequences. This goal is beyond our capabilities at present and shows one advantage obtaining a full profile of a community by using T-RFLP compared to cloning and sequencing.
Fluorescent in situ hybridization was used to try to examine the relationship between biofilm structure and community composition. Several approaches, including multilabeled polyribonucleotides, were applied to visualize the cells, but all were unsuccessful. The outcome may have been due to the low activity and therefore the low ribosome level of the bacteria in the biofilm.
The phylogenetic analysis showed that a strain affiliated with Nitrospira moscoviensis dominated the bulk water (30 to 40%) and was present in all biofilm samples at a lower abundance (a detailed analysis of the phylogenetic relationships of the bacteria seen in the system was carried out by Martiny et al. [submitted]). This genus is known to use nitrite as an energy source (10). Another strain included in the Nitrospirae phylum but phylogenetically distinct from N. moscoviensis appeared in a high abundance (44%) in the 94-day-old sample, reaching 78% in the 256-day-old sample, and dropping to 17.6% after 571 days; it was not detected after this time point or in the bulk water.
Compared to the young biofilm, the old biofilm was inhabitated by a large number of different strains, including those of Planctomyces, Acidobacterium, and Pseudomonas (Martiny et al., submitted). These strains are mostly known to proliferate through heterotrophic growth, even though some planctomycetes can perform anaerobic ammonium oxidation. These findings suggest that as the biofilm develops, new niches are created, allowing for alternative food chains to be introduced.
Species richness.
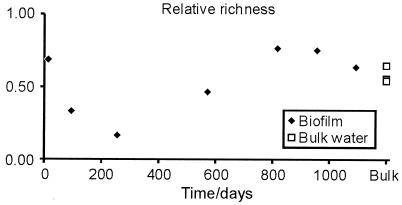
Based on the phylogenetic analysis, each group of sequences that formed a monophyletic clade was defined as an operational taxonomic unit (OTU), and the relative richness was calculated as the number of OTU divided by the number of sequences. Relative richness was used because many sequences were detected only once, indicating a much higher absolute diversity. Figure 6 illustrates the unimodal progression in richness as the biofilm developed together with richness in bulk water. An initial diversity at the level of that in bulk water was observed in the 14-day-old biofilm and dropped to three species by day 256. Following this period, the diversity increased, reaching a level in the old biofilm slightly higher than that in the water phase. This progression in species richness confirms the observed increase in peak numbers generated from the T-RFLP profiles. Nevertheless, since the richness estimate is relative, it is impossible to demonstrate whether the decrease in richness seen during the first 256 days is caused by species exclusion or by an altered relative abundance of species, thereby “hiding” less abundant bacteria.
FIG. 6.
Changes in OTU richness during biofilm development. To adjust for a slightly uneven sample size, the richness was normalized against the total number of sample sequences.
Successional development in a biofilm?
Does succession happen in this biofilm? The first step in the formation of a biofilm in the model drinking water distribution system is the attachment of single cells scattered on the surface. This attachment results in a high level of initial diversity of microbes recruited from the planktonic population, including known culturable organisms such as Pseudomonas, Sphingomonas, and Aquabacterium as well as Nitrospira.
The second step in biofilm formation is characterized by an increase in cell numbers, potentially from the growth of a population affiliated with the Nitrospirae, and the first microcolonies appear. These organisms dominate the microbial community by day 256 and may be responsible for the microcolony formation by clonal growth, which is a known mechanism for microcolony formation in biofilms of mixed communities (40). Concurrently, the richness of the community decreases, a finding which may suggest that an efficient competitor is selected for in the biofilm. The “rise and fall” of this population affiliated with the Nitrospirae indicates how the biofilm may create new ecological niches that are important for its composition.
The third step happens between 256 and 700 days, when a looser biofilm structure covering most of the substratum is formed. This period is correlated with a population change into a distinct community, as observed by T-RFLP and sequencing, and as a result, the mature biofilm population does not consist of patches of independent populations. The old biofilm is defined by a flat community structure of high diversity including both heterotrophic and autotrophic organisms. These findings suggest that a reduction in the activities of a few dominating organisms allows a variety of organisms to settle and survive in the biofilm, potentially proliferating from the organic carbon generated by autotrophic organisms. This diversity pattern fits a recently proposed conceptual model for the succession of bacteria in a biofilm (16).
The mature biofilm has overridden differences in the microenvironment, topography, etc., at the scale of sample size and of giving rise to a distinct community, although biofilm visualization reveals a heterogeneous structure of the biofilm at a fine scale, with large compact areas of cells separated by patches of a monolayer of bacteria on the substratum. Therefore, the homogeneous composition of the population in the mature biofilm could mask a dynamic situation at a smaller scale. Nevertheless, the analysis reported here shows how the formation of the biofilm at the structural and community levels in this system is a result of successional development into a mature, defined community.
Acknowledgments
We thank Rasmus Boe-Hansen for help in sampling the model drinking water distribution system. We also express our gratitude to Terence L. Marsh at Michigan State University for many helpful comments on T-RFLP and sequencing, Jennifer B. Hughes at Brown University for helpful suggestions on the manuscript, and Birte Brejl for technical assistance on designing the figures.
This work was supported by a grant from the Technical University of Denmark.
REFERENCES
- 1.Bergey's Manual Trust. April 2001, posting date. Taxonomic outline of the prokaryotes. Bergey's manual for systematic bacteriology, version 1.0. [Online.]. http://www.cme. msu.edu/bergeys/.
- 2.Boe-Hansen, R., H.-J. Albrechtsen, E. Arvin, and C. Jørgensen. 2002. Dynamics of biofilm formation in a model drinking water distribution system. J. Water SRT-AQUA 51:399-406. [Google Scholar]
- 3.Boe-Hansen, R., A. C. Martiny, E. Arvin, and H.-J. Albrechtsen. 2003. Monitoring biofilm formation and activity in drinking water distribution networks under oligotrophic conditions. Wat. Sci. Technol. 47:91-97. [PubMed] [Google Scholar]
- 4.Brown, D. A., T. J. Beveridge, C. W. Keevil, and B. L. Sherriff. 1998. Evaluation of microscopic techniques to observe iron precipitation in a natural microbial biofilm. FEMS Microbiol. Ecol. 26:297-310. [Google Scholar]
- 5.Buswell, C. M., Y. M. Herlihy, L. M. Lawrence, J. T. M. McGuiggan, P. D. Marsh, C. W. Keevil, and S. A. Leach. 1998. Extended survival and persistence of Campylobacter spp. in water and aquatic biofilms and their detection by immunofluorescent-antibody and -rRNA staining. Appl. Environ. Microbiol. 64:733-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clements, F. E. 1916. Plant succession. Carnegie Institution, Washington, D.C.
- 7.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 8.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrich, S., D. Behrens, E. Lebedeva, W. Ludwig, and E. Bock. 1995. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch. Microbiol. 164:16-23. [DOI] [PubMed] [Google Scholar]
- 11.Fuhrman, J. A., D. E. Comeau, Å. Hagström, and A. M. Chan. 1988. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl. Environ. Microbiol. 54:1426-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundmann, G. L., and D. Debouzie. 2000. Geostatistical analysis of the distribution of NH4+ and NO2-oxidizing bacteria and serotypes at the millimeter scale along a soil transect. FEMS Microbiol. Ecol. 34:57-62. [DOI] [PubMed] [Google Scholar]
- 13.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., S. Okabe, H. Satoh, and Y. Watanabe. 2002. Successional development of sulfate-reducing bacterial populations and their activities in a wastewater biofilm growing under microaerophilic conditions. Appl. Environ. Microbiol. 68:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson, C. R., P. F. Churchill, and E. E. Roden. 2001. Successional changes in bacterial assemblage structure during epilithic biofilm development. Ecology 82:555-566. [Google Scholar]
- 17.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 18.Kalmbach, S., W. Manz, and U. Szewzyk. 1997. Isolation of new bacterial species from drinking water biofilms and proof of their in situ dominance with highly specific 16S rRNA probes. Appl. Environ. Microbiol. 63:4164-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence, J. R., D. R. Korber, B. D. Hoyle, J. W. Costerton, and D. E. Caldwell. 1991. Optical sectioning of microbial biofilms. J. Bacteriol. 173:6558-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeChevallier, M. W., W. Schulz, and R. G. Lee. 1991. Bacterial nutrients in drinking water. Appl. Environ. Microbiol. 57:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeChevallier, M. W., N. J. Welch, and D. B. Smith. 1996. Full-scale studies of factors related to coliform regrowth in drinking water. Appl. Environ. Microbiol. 62:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legendre, P. April 2001, posting date. Congruence among distance matrices: program CADM user's guide. [Online.] Département de Sciences Biologiques, Université de Montréal, Montréal, Québec, Canada. http://www.fas.umontreal.ca/biol/casgrain/en/labo/cadm.html.
- 23.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magurran, A. E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, N.J.
- 25.Mantel, N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209-220. [PubMed] [Google Scholar]
- 26.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 27.Marshall, K. C. 1992. Biofilms: an overview of bacterial adhesion, activity, and control at surfaces. ASM News 58:202-207. [Google Scholar]
- 28.Massol-Deya, A. A., J. Whallon, R. F. Hickey, and J. M. Tiedje. 1995. Channel structures in aerobic biofilms of fixed-film reactors treating contaminated groundwater. Appl. Environ. Microbiol. 61:769-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norton, C. D., and M. W. LeChevallier. 2000. A pilot study of bacteriological population changes through potable water treatment and distribution. Appl. Environ. Microbiol. 66:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunan, N., K. Wu, I. M. Young, J. W. Crawford, and K. Ritz. 2002. In situ spatial patterns of soil bacterial populations, mapped at multiple scales, in an arable soil. Microb. Ecol. 44:296-305. [DOI] [PubMed] [Google Scholar]
- 31.Odum, E. P. 1969. The strategy of ecosystem development. Science 164:262-270. [DOI] [PubMed] [Google Scholar]
- 32.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 33.Otsu, N. 1979. A threshold selection method from gray-level histograms. IEEE Trans. Sys. Man. Cyb. 9:62-66. [Google Scholar]
- 34.Regan, J. M., G. W. Harrington, and D. R. Noguera. 2002. Ammonia- and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system. Appl. Environ. Microbiol. 68:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Schaechter, M., O. Maaløe, and N. O. Kjeldgaard. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19:592-606. [DOI] [PubMed]
- 37.Servais, P., C. Courties, P. LeBaron, and M. Troussellier. 1999. Coupling bacterial activity measurements with cell sorting by flow cytometry. Microb. Ecol. 38:180-189. [DOI] [PubMed] [Google Scholar]
- 38.Stoodley, P., L. Hall-Stoodley, and H. M. Lappin-Scott. 2001. Detachment, surface migration, and other dynamic behavior in bacterial biofilms revealed by digital time-lapse imaging. Methods Enzymol. 337:306-319. [DOI] [PubMed] [Google Scholar]
- 39.Strunk, O., O. Gross, M. Reichel, M. May, S. Hermann, N. Stuckman, B. Nonhoff, M. Lenke, A. Ginhart, A. Vilbig, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig. April 2001, posting date. ARB: a software environment for sequence data. [Online.] Department of Microbiology, Technische Universität München, Munich, Germany. http://www.arb-home.de.
- 40.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and S. Molin. 2000. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182:6482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toonen, R. J., and S. Hughes. 2001. Increased throughput for fragment analysis on an ABI PRISM 377 automated sequencer using a membrane comb and STRand software. BioTechniques 31:1320-1324. [PubMed] [Google Scholar]
- 42.van der Kooij, D., A. Visser, and J. P. Oranje. 1982. Multiplication of fluorescent pseudomonads at low substrate concentrations in tap water. Antonie Leeuwenhoek 48:229-243. [DOI] [PubMed] [Google Scholar]
- 43.van Ooyen, A. 2001. Theoretical aspects of pattern analysis, p. 31-45. In L. Dijkshoorn, K. J. Towner, and M. Struelens (ed.), New approaches for the generation and analysis of microbial typing data. Elsevier, Amsterdam, The Netherlands.
- 44.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, J., B. Xia, D. S. Treves, L. Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]