Abstract
A new microarray method, the isotope array approach, for identifying microorganisms which consume a 14C-labeled substrate within complex microbial communities was developed. Experiments were performed with a small microarray consisting of oligonucleotide probes targeting the 16S rRNA of ammonia-oxidizing bacteria (AOB). Total RNA was extracted from a pure culture of Nitrosomonas eutropha grown in the presence of [14C]bicarbonate. After fluorescence labeling of the RNA and microarray hybridization, scanning of all probe spots for fluorescence and radioactivity revealed that specific signals were obtained and that the incorporation of 14C into rRNA could be detected unambiguously. Subsequently, we were able to demonstrate the suitability of the isotope array approach for monitoring community composition and CO2 fixation activity of AOB in two nitrifying activated-sludge samples which were incubated with [14C]bicarbonate for up to 26 h. AOB community structure in the activated-sludge samples, as predicted by the microarray hybridization pattern, was confirmed by quantitative fluorescence in situ hybridization (FISH) and comparative amoA sequence analyses. CO2 fixation activities of the AOB populations within the complex activated-sludge communities were detectable on the microarray by 14C incorporation and were confirmed independently by combining FISH and microautoradiography. AOB rRNA from activated sludge incubated with radioactive bicarbonate in the presence of allylthiourea as an inhibitor of AOB activity showed no incorporation of 14C and thus was not detectable on the radioactivity scans of the microarray. These results suggest that the isotope array can be used in a PCR-independent manner to exploit the high parallelism and discriminatory power of microarrays for the direct identification of microorganisms which consume a specific substrate in the environment.
Deciphering the structure and function of complex microbial communities is a central theme in microbial ecology. However, traditional cultivation-dependent methods are inadequate to fulfill this task because most members of microbial communities in natural and engineered systems cannot be cultured (2, 69). In contrast, the cultivation-independent 16S rRNA approach allows one to obtain more complete inventories of such microorganisms present in a particular system. If this approach is combined with quantitative gene probe techniques, such as dot blot (66) or fluorescence in situ hybridization (FISH) (14), the compositions of microbial communities can be determined accurately (27, 37, 56, 57, 61, 62). However, these techniques are very time-consuming; therefore, the number of samples for which quantitative data can be obtained is relatively limited. This limitation can be (at least partially) overcome by the inclusion of DNA microarray technology for the detection of bacterial 16S rRNA or its genes (3, 7, 23, 29, 32, 48, 64, 77). Although precise quantitative data are difficult to obtain with this format, it allows one to apply many rRNA-targeted oligonucleotide probes in parallel and is therefore ideally suited to the selection of probes for subsequent quantitative dot blot or FISH analyses. However, only a few studies have actually applied DNA microarrays to the detection of 16S rRNA (genes) of indigenous microorganisms in environmental samples (19, 29, 32, 75, 77).
Unfortunately, 16S rRNA-based identification generally does not provide information on the physiology of the detected microorganisms. For cultured bacteria, the physiological potential can be determined in the laboratory, but the data obtained do not necessarily provide information on the actual functions of these bacteria in an ecosystem. For uncultured bacteria, for which only 16S rRNA sequence information is available, the inference of physiological traits is impossible if these microorganisms are not closely related to cultured microorganisms. Therefore, in recent years there have been considerable efforts to develop methods which allow study of the functions of microorganisms without cultivation (for a review, see reference 21). Several studies have used the cellular rRNA concentration, which can be quantified by FISH and image analyses, as a measure of the rate of growth of a given organism in the environment (28, 50). However, at least for some microorganisms, the cellular rRNA content is influenced largely not only by the actual growth rate but also by the physiological history of the microorganisms (46). Furthermore, some slowly growing chemolithoautotrophs retain a high cellular rRNA content even after complete physiological inhibition (63, 71) or after starvation (39). Consequently, it is almost impossible to deduce the physiological status of an organism in its environment by measuring its cellular rRNA content.
The addition of substrates labeled with stable or radioactive isotopes to microbial communities has been applied to the identification of microorganisms which consume specific compounds in the environment. Identification can be performed by using molecular tools after the separation of labeled DNA (40, 54, 55, 76) or RNA (35, 36) from unlabeled nucleic acids. Alternatively, the incorporation of labeled substrates into polar lipid ester-linked fatty acids (6, 25, 59) or biphytanyl membrane lipids (79) has been used to monitor bacteria with specific activities in the environment, but the identification of as-yet-uncultured microorganisms generally is not possible with membrane lipids.
The recently developed combination of FISH and microautoradiography (MAR) (9, 20, 31, 47) allows one to record the substrate uptake profiles of probe-identified prokaryotes at the single-cell level under different environmental conditions. This method has found widespread application in environmental microbiology (10, 13, 42-44) and is ideally suited to monitoring the functions of selected cultured and uncultured microorganisms; however, the numbers of experiments which can be performed in parallel are limited due to the tediousness of the method.
The goal of the present study was to develop a method which allows simultaneous monitoring of the diversity and substrate incorporation of complex microbial communities by DNA microarray technology. Nitrifying activated sludge amended with radioactive bicarbonate served as a model system. After extraction and fluorescence labeling of community rRNA, the diversity and bicarbonate incorporation of ammonia-oxidizing bacteria (AOB) were measured by using a prototype DNA microarray for AOB detection.
MATERIALS AND METHODS
Isotope labeling of pure cultures.
A pure culture of Nitrosomonas eutropha Nm57 (provided by H. P. Koops, University of Hamburg) was grown in liquid mineral medium (LM medium) with the following composition (per liter): (NH4)2SO4, 0.5 g; KH2PO4, 0.2 g; CaCl2 · 2H2O, 20 mg; MgSO4 · 7H2O, 40 mg; FeNaEDTA, 3.8 mg; phenol red, 1 μg; HEPES buffer, 4.8 g; and trace element solution, 1 ml (17). The medium was adjusted to pH 7.5 with 10 M NaOH and autoclaved. NaHCO3 was subsequently added to a final concentration of 1.5 mM. Growing cultures were stirred continuously at 30°C in the dark, until phenol red turned yellow and all ammonia was oxidized to nitrite. The culture was routinely screened for contaminants by drop plating on tryptic soy agar incubated at 30°C (Becton Dickinson, Bedford, Mass.). The culture was assumed to be axenic if no growth on agar occurred after 4 days.
N. eutropha was harvested from a late-log-phase culture (250 ml) by centrifugation (10,000 × g, 20 min) (Centrikon T-42K; Kontron Instruments, Milan, Italy) and resuspended in 15 ml of fresh LM medium devoid of added NaHCO3, (NH4)2SO4, and phenol red. H14CO3− (58 mCi of C mmol−1; Amersham Life Science, Little Chalfont, United Kingdom) was added to the concentrated culture (60 μM H14CO3−; 3.6 μCi ml−1). Immediately after this step, 2.5 mM (NH4)2SO4 was added, and the culture was incubated in a shaker (150 rpm) at 30°C in the dark for 48 h. Incorporation of radioactive substrate into the biomass was monitored after 48 h by filtration with cellulose filters (see below). For RNA extraction, labeled cells were harvested by centrifugation (10,000 × g, 10 min) and resuspended in 90% RNAlater (Ambion, Austin, Tex.)-10% LM medium. According to the instructions of the manufacturer, samples in RNAlater were kept on ice for approximately 2 h, followed by incubation for 24 h at 4°C, and then were stored at −20°C until RNA extraction.
Isotope labeling of activated sludge.
Activated sludge was collected from the nitrification tanks of industrial (Oberding) and municipal (Aalborg West) wastewater treatment plants. Activated-sludge samples were kept at 5°C in the dark for no more than 3 days before isotope labeling. Before the addition of labeled bicarbonate, the concentration of bicarbonate in the sludge was minimized by centrifugation of the activated sludge (10,000 × g, 15 min), disposal of the supernatant, and subsequent resuspension of the pellet in HEPES-buffered LM medium devoid of added NaHCO3, (NH4)2SO4, and phenol red. In addition, the resuspended sludge was flushed with CO2-free air for 1 h to remove the remaining bicarbonate, as previously suggested by Whitby et al. (76). The concentrations of bicarbonate (alkalinity) in the sludge before and after this treatment were determined by gas chromatography after acidification, as described previously by Chai et al. (8). H14CO3− was added to a final concentration of approximately 1.85 mM H14CO3− (107 μCi ml−1). Immediately after this step, 2.5 mM (NH4)2SO4 was added, and the amended sludge was incubated on a shaker (150 rpm) at 30°C in the dark for up to 26 h. Parallel activated-sludge samples were incubated with H14CO3−, (NH4)2SO4, and 5 mg of allylthiourea liter−1 as a specific inhibitor of ammonia monooxygenase (5). Allylthiourea is a reversible inhibitor of ammonia monooxygenase which prevents ammonia oxidation by chelating Cu ions. The incorporation of radioactive material into activated-sludge biomass was continuously monitored after filtration of the activated sludge with cellulose filters (see below). Parallel activated-sludge samples were incubated with nonradioactive HCO3− and used to monitor the concentrations of NO2−, NO3−, and NH4+ with color-reaction strips (Merck AG, Darmstadt, Germany) at different times. NH4+ was continuously added to radioactive and nonradioactive samples when the concentrations decreased to values below 1 mM. After radioactive labeling, all samples were stored in RNAlater as described for pure-culture samples.
Quantification of radioactivity incorporation.
Subsamples (10 μl) from radioactive incubations (pure culture or activated sludge) were harvested with a needle and syringes. The subsamples were added to approximately 10 ml of LM medium and filtered through 0.2-μm-pore-size mixed-cellulose filters (Advantec MFS Inc., Pleasanton, Calif.). Subsequently, 5 ml of 0.1 N HCl was added to the filtration unit in order to remove inorganic carbonates. After 3 min of acidification, the HCl was removed by filtration, and the filters were transferred to scintillation vials and dissolved in 10 ml of scintillation fluid (Filter-Count; Packard, Groningen, The Netherlands). Dissolved filters were analyzed for 3 min by liquid scintillation counting (Packard 1600 TR). All samples were corrected for quenching by using external standards.
RNA extraction.
To eliminate RNase activity, all reagents and plastic ware were treated with 0.1% (vol/vol) diethylpyrocarbonate (DEPC; Sigma, St. Louis, Mo.) for at least 3 h at 37°C and subsequently autoclaved. Reagents containing primary amino groups were not directly exposed to DEPC but were prepared with DEPC-pretreated double-distilled H2O (ddH2O). Total RNA was extracted from biomass by using a modified RNAwiz (Ambion) protocol. Briefly, RNAlater was removed from the samples before extraction by centrifugation (23,645 × g, 15 min, 4°C) (Rotina 35R; Hettich, Tuttlingen, Germany). The pelleted samples were resuspended in 1 ml of RNAwiz reagent in a 2-ml screw-lid tube (Sarstedt, Nürnbrecht, Germany), and the contents of one tube of lysing matrix E (containing a mixture of ceramic and silica particles) (FastDNA kit for soil; Bio 101, La Jolla, Calif.) were added. Homogenization of N. eutropha pure-culture cells was carried out by bead beating (FastPrep120; Bio 101) for 40 s at a speed of 4.5. For activated-sludge samples, bead beating was repeated four times. After 5 min of incubation at room temperature, 0.2 ml of chloroform (Sigma) was added; after vigorous shaking, the samples were incubated for 10 min at room temperature. In the next step, phase separation was achieved by centrifugation (15 min, 23,645 × g, 4°C). The aqueous phase was transferred to a new tube and stored on ice. To increase the RNA extraction efficiency for activated-sludge samples, RNA was extracted again from the organic phase. For this purpose, 0.4 ml of RNAwiz was added to the organic sample fraction (which still contained the lysing matrix), vigorously vortexed for 1 min, heated at 65° for 10 min, vortexed again for 30 s, and chilled on ice for 5 min. Then, 0.2 ml of chloroform was added, followed by thorough shaking and incubation for 10 min at room temperature. After centrifugation (15 min, 23,645 × g, 4°C), the aqueous phase was pooled with that from the first extraction step. RNA was precipitated after the addition of 0.5 ml of water and 1 ml of isopropanol (J. T. Baker, Deventer, The Netherlands) by incubating the tubes in a dry ice-ethanol mixture for 5 min. Subsequently, RNA was pelleted by centrifugation (20 min, 23,645 × g, 4°C), washed with 75% ethanol, air dried for 3 min at 90°C, resuspended in a buffer containing 10 mM Tris and 1 mM EDTA (pH 8), and fully dissolved by heating at 90°C for 3 min. RNA from activated sludge was further purified by using an RNA purification kit according to the recommendations of the manufacturer (Qiagen, Hilden, Germany). Extracted RNA was stored in aliquots at −20°C after precipitation with 0.1 volume of 5 M NaCl and 3 volumes of 96% ethanol (J. T. Baker).
The amount and purity of extracted RNA were determined spectrophotometrically (SmartSpec 3000; Bio-Rad, Hercules, Calif.) at 260 and 280 nm and by gel electrophoresis (0.9% SeaPlaque GTG agarose; BioWhittaker Molecular Applications, Rockland, Maine) in a buffer containing 40 mM Tris, 10 mM sodium acetate, and 1 mM EDTA (pH 8.0).
Quantification of radioactivity in RNA.
The incorporated radioactivity in rRNA was determined by excising the 16S and 23S rRNA bands from the agarose gel. The RNA-containing gel pieces were melted at 95°C in 1 ml of ddH2O, mixed with scintillation fluid (InstaGel-Plus; Packard), and measured for 11 min in a liquid scintillation counter (LS-1800; Beckman Instruments, Inc., Pasadena, Calif.)
Probes and microarray processing.
A prototype array with eight probes was applied for the detection of AOB (Table 1). All oligonucleotide probes were purchased from MWG Biotech (Ebersberg, Germany). Probes were resuspended in spotting buffer (Exiqon AS, Vedbaek, Denmark) to a final concentration of 50 pmol μl−1 and subsequently spotted onto Immobilizer MicroArray slides (Exiqon AS). Slides with two different spot diameters (125 and 500 μm) were manufactured by using a GMS 417 contact array spotter (Affymetrix, Santa Clara, Calif.). Additionally, manually spotted arrays (approximate diameter, 1,000 μm) were prepared by applying probe droplets with a pipette tip to the slide surface. Coupling of the aminated oligonucleotides to the slides was performed as proposed by the supplier (Exiqon AS).
TABLE 1.
Oligonucleotide probes used for FISH and microarray hybridization experimentsa
| Probe name | Probe sequence (5′-3′) | Target site (positions) | Target organism(s) | Reference(s) |
|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | 338-355 | Most but not all Bacteria | 1, 12 |
| EUB338-II | GCAGCCACCCGTAGGTGT | 338-355 | Planctomycetales | 12 |
| EUB338-III | GCTGCCACCCGTAGGTGT | 338-355 | Verrucomicrobiales | 12 |
| Nso1225 | CGCCATTGTATTACGTGTGA | 1224-1243 | Most but not all beta-proteobacterial AOB (30) | 38 |
| Nso190 | CGATCCCCTGCTTTTCTCC | 189-207 | Many but not all beta-proteobacterial AOB (30) | 38 |
| Nsv443 | CCGTGACCGTTTCGTTCCG | 443-461 | Nitrosospira cluster 1-3 (30) | 38 |
| Ncmob | TCCTCAGAGACTACGCGG | 174-191 | Nitrosococcus mobilis (30) | 26 |
| NEU | CCCCTCTGCTGCACTCTA | 651-668 | Most halophilic and halotolerant nitrosomonads (30) | 71 |
| Cte | TTCCATCCCCCTCTGCCG | 659-676 | Competitor probe for NEU; Comamonas spp., Acidovorax spp., Hydrogenophaga spp., Aquaspirillum spp. (30) | 71 |
| 6a192 | CTTTCGATCCCCTACTTTCC | 192-212 | Nitrosomonas oligotropha lineage | 52 |
| c6a192 | CTTTCGATCCCCGACTTTCC | 192-212 | Competitor probe for 6a192; Nitrosomonas eutropha | 52 |
| NONEUB | ACTCCTACGGGAGGCAGC | 74 | ||
| NONSENSE | AGAGAGAGAGAGAGAGAG | 32 | ||
| CONT | AGGAAGGAAGGAAGGAAG | Control oligonucleotide | 32 | |
| CONT-COMP | CTTCCTTCCTTCCTTCCT | Complementary to CONT | 32 |
More detailed information for each probe can be obtained at the oligonucleotide online resource probeBase (33). All but CONT and CONT-COMP were used for FISH. All but EUB338-II, EUB338-III, and Cte were used for microarray analysis.
In vitro transcription of rRNA genes.
To obtain sufficient amounts of rRNA from several AOB which were used as reference organisms for DNA microarray evaluation, their cloned 16S rRNA genes (51) were transcribed in vitro. For this purpose, pCR2.1-TOPO (Invitrogen, Frederick, Md.) plasmids containing the respective 16S rRNA gene insert were linearized by digestion with restriction enzyme BamHI (MBI Fermentas, St. Leon-Rot, Germany). The RNA was synthesized by using a commercially available T7 transcription kit (MBI Fermentas) containing the T7 RNA polymerase in accordance with the recommendations of the manufacturer. The DNA template was removed by DNase I digestion (Promega, Madison, Wis.) in accordance with the manufacturer's instructions. For purification of the RNA, the phenol-chloroform extraction method was used as described by Griffiths et al. (22). The correct size of the in vitro-transcribed RNA was verified by gel electrophoresis, and the RNA concentration was determined spectrophotometrically.
Fluorescence labeling of RNA and microarray hybridization.
For RNA labeling with fluorescent dyes (Cy3 and Cy5), a CyScribe direct mRNA labeling kit (Amersham Biosciences, Uppsala, Sweden) was used. This kit covalently labels guanine residues at the N7 position and thus does not interfere with subsequent base pairing. For labeling, 0.5 μg of precipitated RNA was pelleted by centrifugation (15 min, 23,645 × g, 4°C), washed with 70% ethanol, air dried, and resuspended in an appropriate amount of ddH2O. Labeling was carried out according to the manufacturer's recommendations. To remove the excess dye, the labeled RNA was precipitated with 0.1 volume of 5 M NaCl and 3 volumes of 96% ethanol at −80°C on a dry ice-ethanol mixture, centrifuged (30 min, 23,645 × g, 4°C), washed with 70% ethanol, and air dried. The RNA pellet was resuspended in 30 μl of freshly prepared hybridization buffer (6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5×Denhardt's solution, 5 to 30% formamide, 0.5% sodium dodecyl sulfate, 50 mM Na2HPO4 [pH 8.0]), denatured by heating to 90°C for 5 min, and subsequently chilled on ice. The RNA-containing hybridization solution was transferred to the slide surface, covered with a coverslip, and inserted into a hybridization chamber (32) containing 120 μl of hybridization buffer. Hybridization was performed overnight (18 h) at 42°C with a water bath and terminated by washing the slide for 5 min in 2× SSC at room temperature, in ice-chilled ddH2O for 1 min, in ice-chilled 96% ethanol for 10 s, and in ice-chilled ddH2O for 10 s. It should be noted that the hybridization and wash buffers used in this study differed significantly in composition from those used by Loy et al. (32) for the detection of 16S rRNA gene fragments. The buffer composition was changed to improve signal-to-noise ratios for rRNA detection on the microarrays (data not shown).
Subsequently, the slide was air dried at room temperature in the dark. For qualitative and quantitative evaluation of fluorescence signals, a GMS 418 array scanner (Affymetrix) equipped with ImaGene 4.0 software (BioDiscovery, Inc., Los Angeles, Calif.) was used. For each spot, signal-to-noise ratios were calculated as outlined by Loy et al. (32), and hybridizations were scored positive for ratios above 2. Radioactivity signals were recorded with a β-Imager (BioSpace Mesure, Paris, France), and the radioactivity for each spot was quantified with β-Vision software (BioSpace Mesure). Cumulative fluorescence intensity values for manually applied spots were determined by using the public domain software package Image J (http://rsb.info.nih.gov/ij/).
FISH and MAR.
The abundance of AOB in activated-sludge samples was determined by quantitative FISH with fluorescence-labeled derivatives of the respective rRNA-targeted probes by following the approach outlined by Daims et al. (14) and Schmid et al. (62). Probe NEU and probe 6a192 were used with the respective competitor probes to enhance specificity (52, 71). Probes were used as Cy3- and Cy5-labeled derivatives (Thermo Hybaid, Ulm, Germany). All probes were applied under the optimal hybridization conditions determined in the original publications. Simultaneous hybridizations with probes requiring different stringencies were performed with subsequent hybridization procedures (70).
In parallel to the isotope array approach, the incorporation of a radioactive substrate into activated-sludge microorganisms was also monitored by using the combination of FISH and MAR (31). For this purpose, activated-sludge samples were incubated with H14CO3− (10 μCi ml−1). Nonradioactive bicarbonate was added to obtain the same total concentration of carbonate as in the samples used for the isotope array experiment (1.85 mM). The FISH-MAR procedure was carried out as described previously (31).
amoA gene fragment-based diversity analysis of AOB.
Total genomic DNA was extracted from 0.5-ml sludge subsamples with a FastDNA spin kit for soil (Bio 101) according to the manufacturer's instructions. A 453-bp fragment (excluding primers) of the amoA gene was amplified from 50 ng of DNA by using the optimized (67) primers amoA-1F and amoA-2R (60) for PCR with a Primus cycler (MWG Biotech). Reaction mixtures containing 50 pmol of each primer (1 μM) were prepared in a total volume of 50 μl by using 2 mM MgCl2 reaction buffer and 1.5 U of Taq polymerase (Promega). Thermal cycling was carried out with an initial denaturation step at 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 20 s, annealing at 52°C for 30 s, and elongation at 72°C for 40 s. Cycling was completed by a final elongation step at 72°C for 10 min. The amplified amoA gene fragments were separated by gel retardation (62), visualized with SYBR Gold (Molecular Probes, Inc., Eugene, Oreg.), excised from the gel, and cloned into pCR2.1-TOPO TA vectors (Invitrogen, San Diego, Calif.) according to the manufacturer's instructions. After plasmid purification (Qiagen), sequences were determined by using a Thermo Sequenase cycle sequencing kit (Amersham Life Science), infrared-labeled primers (IRD800 and IRD700), and an automated DNA sequencer (Li-Cor, Inc., Lincoln, Nebr.). The sequences obtained were imported into the amoA database by using the ARB program package (http://www.arb-home.de) and aligned manually. Phylogenetic analyses were performed based on nucleic acid (amoA) and amino acid (AmoA) sequences by applying distance-matrix (Phylip and FITCH), maximum-parsimony, and maximum-likelihood methods with the respective tools in the ARB program package.
Nucleotide sequence accession numbers.
For each wastewater treatment plant, representative amoA sequences were deposited at GenBank under accession numbers AY367275 to AY367276 (Aalborg West) and AY367277 (Oberding).
RESULTS
DNA microarray prototype for AOB.
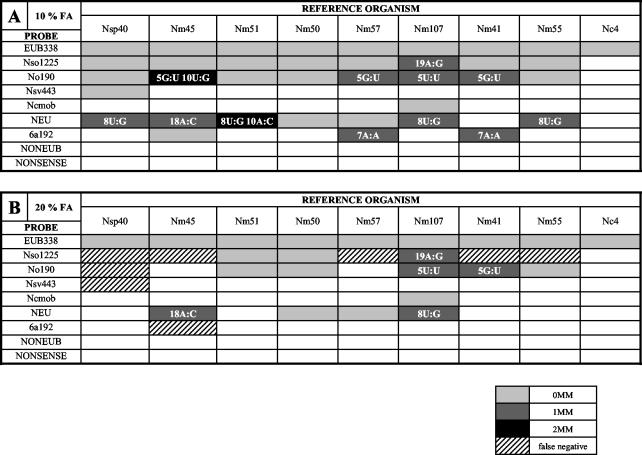
Initially, a prototype DNA microarray for the detection of beta-proteobacterial AOB in the environment was spotted by using six previously published 16S rRNA-targeted oligonucleotide probes. The microarray was complemented by a bacterial probe and several control probes. For evaluation, in vitro-transcribed 16S rRNA of nine different AOB (eight affiliated with the Betaproteobacteria and one affiliated with the Gammaproteobacteria) was used under conditions of increasing stringency by varying the formamide concentration in the hybridization buffer (Fig. 1). With 10% formamide, all probes hybridized to the 16S rRNA of AOB which possessed a fully matched target site. However, several probes also bound to target sites containing one or (in two cases) two mismatches. Increasing the formamide concentration to 20% increased the specificity of the probes, but for probes Nso1225, Nsv443, Nso190, and 6a192, false-negative signals were observed (Fig. 1). At 30% formamide, signal-to-noise ratios above the threshold level were observed only for probe EUB338 (data not shown). Based on these results, we selected 10% formamide in the hybridization buffer for all subsequent experiments.
FIG. 1.
DNA microarray evaluation. Hybridizations were performed with 10% (A) and 20% (B) formamide (FA) in the hybridization buffer. Individual hybridization results are shown for probes with the following AOB reference organisms (phylogenetic affiliations are as in references 30 and 53): Nitrosospira sp. strain Nsp40 (belonging to Nitrosospira cluster 3); N. oligotropha Nm45 (affiliated with the N. oligotropha lineage); Nitrosomonas sp. strain Nm51 (affiliated with the N. marina lineage); N. europaea Nm50, N. eutropha Nm57, and N. mobilis Nm107 (all belonging to the N. europaea-N. mobilis lineage); Nitrosomonas sp. strain Nm41 (affiliated with the N. communis lineage); N. cryotolerans Nm55; and N. oceani Nc4 (a gamma-proteobacterial AOB). Shaded boxes indicate hybridization signals with a signal-to-noise ratio above the threshold value of 2, as indicated by the key below the panels. 0MM indicates hybridization to an AOB with a target sequence fully complementary to the probe. 1MM and 2MM indicate hybridizations to target sequences having one and two mismatches, respectively. False-negative signals represented the absence of duplex formation despite the presence of a fully matched target sequence. For each of the observed mismatch hybridizations, the position and type of the mismatch(es) within the duplex are shown.
In addition to using nonfragmented 16S rRNA, we also evaluated fragmented 16S rRNA for microarray hybridization. Fragmented rRNA was produced by application of the Affymetrix protocol (15) or by alkaline hydrolysis. However, in our experiments, fragmentation of rRNA did impair the signal-to-noise ratio of microarray hybridization (data not shown). Consequently, a fragmentation protocol was excluded from all further experiments.
Isotope array experiments with N. eutropha Nm57.
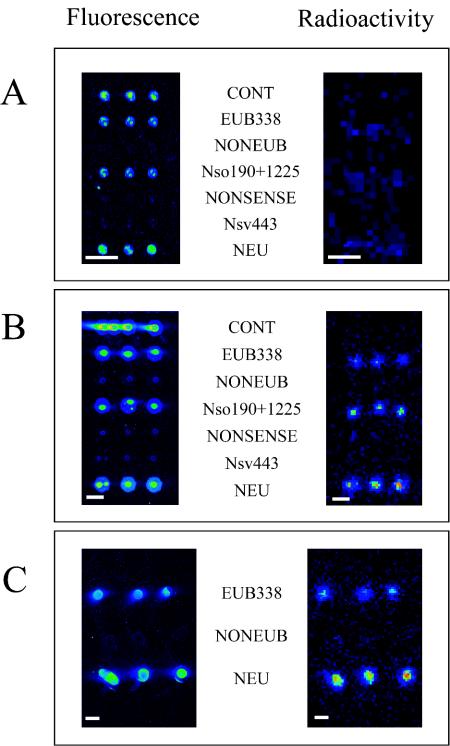
A pure culture of N. eutropha was incubated with radioactively labeled bicarbonate. After 48 h, 39% of the added 14C was detected in the biomass, and 19,000 cpm of 14C was found per μg of RNA (corresponding to 9 nCi of 14C per μg of RNA) (Table 2). Extracted rRNA was fluorescently labeled, and 0.5 μg of rRNA was hybridized to the DNA microarray with 125-μm-diameter spots, which were subsequently scanned for fluorescence and radioactivity. While the expected fluorescence signals were observed, no radioactivity could be detected with the β-Imager for the individual spots (Fig. 2A). The same result was obtained when 1 μg of rRNA was applied per microarray hybridization (data not shown). The use of higher rRNA concentrations led to significantly enhanced background fluorescence and did not allow the detection of radioactivity (data not shown).
TABLE 2.
Incorporation of 14C into biomass and rRNA of N. eutropha and activated sludge from two wastewater treatment plants after incubation with radioactive bicarbonate
| Sample | Incubation time (h) | Addition of:
|
Incorporation of 14C into:
|
|||
|---|---|---|---|---|---|---|
| ATU (mg liter−1) | 14C (μCi ml−1) | Total biomass (%) | Purified rRNA
|
|||
| % | cpm of RNA μg−1 | |||||
| N. eutropha Nm57 | 48 | 0 | 3.6 | 38.8 | 0.19 | 19,000 |
| Wastewater treatment plant | ||||||
| Aalborg West | 3 | 0 | 107 | 4.4 | 0.05 | 9,000 |
| 13 | 0 | 107 | 10.2 | 0.06 | 16,000 | |
| 26 | 0 | 107 | 12.4 | 0.13 | 37,000 | |
| 3 | 5 | 107 | 0.7 | 0.02 | 3,000 | |
| 13 | 5 | 107 | 2.6 | 0.03 | 6,000 | |
| 26 | 5 | 107 | 2.7 | 0.04 | 10,000 | |
| Oberding | 2.5 | 0 | 107 | 7.5 | 0.10 | 10,000 |
| 11 | 0 | 107 | 20.7 | 0.33 | 31,000 | |
| 25 | 0 | 107 | 22.8 | 0.29 | 29,000 | |
| 2.5 | 5 | 107 | 0.6 | 0.01 | 2,000 | |
| 11 | 5 | 107 | 2.1 | 0.03 | 2,000 | |
| 25 | 5 | 107 | 3.2 | 0.02 | 4,000 | |
FIG. 2.
Isotope array experiment with N. eutropha Nm57. rRNA was extracted from N. eutropha cultures after incubation with [14C]bicarbonate. After labeling of the rRNA with Cy5, a single slide carrying three prototype arrays with different spot sizes was hybridized, washed, and scanned for fluorescence and radioactivity. (A) Spot diameter, 125 μm. (B) Spot diameter, 500 μm. (C) Approximate spot diameter, 1 mm. Bars, 500 μm.
Since the 125-μm-diameter spot was close to the resolution limit of the β-Imager, we tested larger spot sizes to improve the detection of 14C incorporation into the rRNA of N. eutropha. For this purpose, microarrays with 500-μm-diameter spots were produced by automated spotting. Furthermore, even larger spots with an approximate diameter of 1 mm were produced by manually spotting approximately 0.1 μl of the probe solutions onto the microarray slides with a 1-μl pipette. After hybridization with 0.5 μg of rRNA of N. eutropha, the expected fluorescence and radioactivity signals were detected for the 500-μm and 1-mm spots (Fig. 2B and C). However, after initial successful experiments, the pins used for automated spotting of the 500-μm-diameter spots became bent and started to damage the surface of the plastic slides. Therefore, for practical reasons, we used manually spotted arrays (with spot diameters of about 1 mm) for all subsequent experiments, despite the obvious disadvantage that the amount of probe applied per spot varies more significantly than it does with automatically generated spots.
Isotope array experiments with activated sludge.
The suitability of the developed isotope array approach for analysis of complex microbial communities was tested by using two different nitrifying activated-sludge sources (Aalborg West and Oberding). Initially, however, the AOB community structure in both sludge sources was analyzed by quantitative FISH and the amoA approach in order to generate microarray-independent reference data for comparison.
In the Aalborg West sludge, 4.7% ± 0.5% (mean and standard deviation) of the bacterial population detectable with the EUB338 probe set (12) hybridized with probe Nso1225. All cells which showed a signal with this probe also bound probe 6a192 in the presence of the competitor probe c6a192. No hybridization signals were detectable with probes NEU (in the presence of unlabeled competitor probe Cte), Nsv443, and Ncmob, and only a few cells (<1%) hybridized with probe Nso190. This hybridization pattern indicates that members of the Nitrosomonas oligotropha lineage almost exclusively represented the AOB community in the Aalborg West activated sludge. In the Oberding sludge, 9.3% ± 1.1% of the cells detectable with the bacterial probe mix hybridized with probe Nso1225. All cells hybridizing with this probe also bound probes Nso190 and NEU (in the presence of unlabeled competitor probe Cte). No hybridization signals could be detected after the application of probes Ncmob, Nsv443, and 6a192. This hybridization pattern suggests that in the Oberding activated sludge, members of the Nitrosomonas europaea and N. eutropha lineages were dominant. After the addition of allylthiourea to the activated-sludge samples from Aalborg West and Oberding, AOB remained detectable by FISH for the entire experimental period. Nonspecific binding of probes could be excluded for all samples by the application of probe NONEUB338 (complementary to probe EUB338) labeled with the respective fluorophores (data not shown).
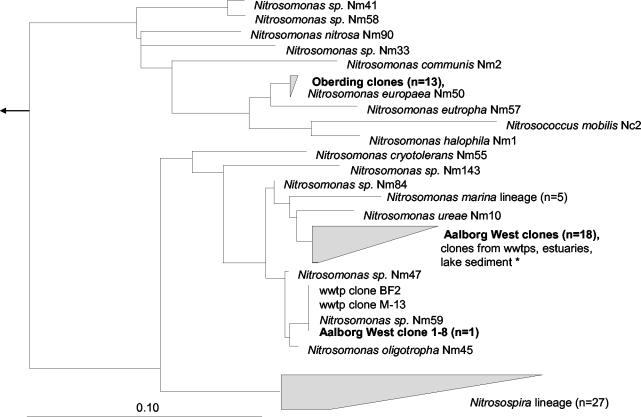
In addition to FISH analysis, comparative amoA gene sequence analysis was used to determine the species richness of AOB from both sludge sources. Nineteen randomly chosen amoA clones were sequenced for the Aalborg West plant. Eighteen of these sequences were highly similar to each other and most closely related to amoA clones from other environments, including other wastewater treatment plants. These clones formed a monophyletic grouping within the N. oligotropha and Nitrosomonas marina lineages and could not be separated from each other at the amoA level (30). One amoA clone from Aalborg West had an amoA sequence which was identical at the amino acid level to that from Nitrosomonas sp. strain Nm59, which is a close relative of N. oligotropha (Fig. 3). For the Oberding plant, the 13 amoA clones sequenced were identical to each other and very closely related to N. europaea.
FIG. 3.
Phylogenetic tree showing the affiliations of the AmoA sequences obtained from the Aalborg West and Oberding wastewater treatment plants (wwtps). The tree was calculated based on the AmoA amino acid sequences deduced from the FITCH algorithm with global rearrangements and a randomized input order. The bar shows 10% estimated sequence divergence. AmoA sequences of gammaproteobacterial AOB were used as outgroups (not shown). The accession numbers for the clones from wwtps, estuaries, and lake sediment were as follows: estuary clone—AF367463 (Duc-27); freshwater clones—AJ388566 (pGtA.2), AJ388570 (pG5B.1), and AJ388571 (pG5B.2); wwtps clones—AF420298 (M-13), AF272442 (IA-32), AF272482 (BF1-1), AF489661 (S_3), and AF489660 (S_2); and lake sediment clone—AF489645 (T_2). amoA sequences of reference organisms were published elsewhere (30, 41, 51, 53, 65).
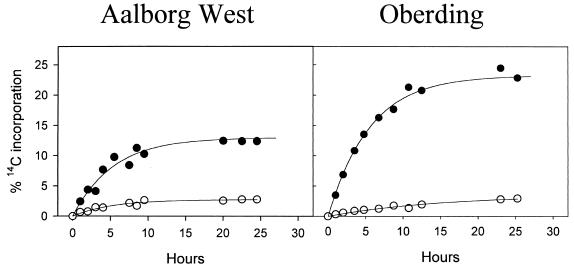
Samples from both sludge sources were incubated with [14C]bicarbonate (at a concentration typically found in activated sludge) after the natural bicarbonate content of the sludge samples was minimized by washing and degassing. The incorporation of radioactivity in the activated sludge after different time periods was monitored by scintillation counting of filtered biomass. As controls, parallel experiments were performed for both plants with activated sludge amended with allylthiourea, a specific inhibitor of autotrophic ammonia oxidation (Fig. 4 and Table 2). During the first 10 h, a rapid increase in 14C incorporation was observed for samples from both noninhibited nitrifying activated-sludge sources. Incubation beyond 10 h did not significantly increase the amount of incorporated radioactivity. In the presence of allylthiourea, significantly less incorporation of radioactively labeled bicarbonate into the sludge biomass was detected. This incorporation in the presence of allylthiourea probably was caused by heterotrophic CO2 fixation.
FIG. 4.
Incorporation of radioactive 14C into nitrifying activated-sludge biomass after incubation with [14C]bicarbonate. Filled symbols indicate incorporation in the absence of the inhibitor allylthiourea. Open symbols indicate incorporation in the presence of allylthiourea.
Intact rRNA was extracted from the activated-sludge samples (for an agarose gel image, please see www.microbial-ecology.net/supplements.html), and after agarose gel separation and purification, the amount of incorporated radioactivity within the 16S and 23S rRNA molecules (5S rRNA is selectively excluded by the purification columns) was determined by scintillation counting (Table 2). The sum of incorporated 14C in 16S and 23S rRNA molecules extracted from noninhibited activated sludge was 0.05 to 0.33% the amount of 14C added to the activated sludge and corresponded to 1 to 2% the isotope assimilated in the biomass. As expected, lower percentages of rRNA incorporation (0.01 to 0.04%) were found for the inhibited sludge samples (Table 2).
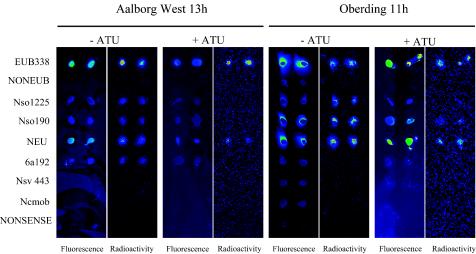
After fluorescence labeling of the rRNA, manually spotted microarrays were hybridized and then scanned for fluorescence and radioactivity (Fig. 5). In total, each sludge sample (with and without allylthiourea) was analyzed after three different incubation times with [14C]bicarbonate. The fluorescence hybridization patterns observed were generally congruent with the ammonia oxidizer diversity analyses performed by FISH and amoA sequencing. For the Aalborg West plant, probes Nso1225, 6a192, Nso190, and NEU had signal-to-noise ratios above the threshold value of 2 and were thus scored positive. Consistent with the results of the FISH analysis, probes Nsv443 and Ncmob had signal-to-noise ratios below the threshold value in the microarray analysis. For the Oberding plant, probes Nso1225, Nso190, and NEU had signal-to-noise ratios above the threshold value of 2, while probes 6a192, Nsv443, and Ncmob had values below this threshold. For the Oberding plant, the fluorescence microarray hybridization patterns of the activated-sludge samples did not differ significantly between uninhibited and inhibited samples. However, for the Aalborg West plant, probe NEU was the only AOB-specific probe which gave a positive signal after inhibition with allylthiourea.
FIG. 5.
Isotope array experiment with activated sludge. rRNA was extracted from inhibited and noninhibited activated sludge from the Aalborg West and Oberding wastewater treatment plants after incubation with [14C]bicarbonate for 13 and 11 h, respectively. After labeling of the rRNA with Cy3, the manually spotted microarrays were hybridized and scanned for fluorescence and radioactivity. In order to enhance clarity, recorded spot signals were aligned by using Adobe Photoshop. Within each slide, blue indicates low signals and green, yellow, and red indicates increased signals. It should be noted that signal intensities in the radioactivity scans cannot be compared directly between slides. ATU, allylthiourea.
After scanning of the microarrays for radioactivity, we found that the rRNA immobilized by the bacterial and ammonia oxidizer probes had incorporated sufficient amounts of 14C to be detectable (Fig. 5). As expected, the level of incorporation of 14C into ammonia oxidizer rRNA was very low or even absent in activated-sludge samples inhibited by allylthiourea. The radioactive labeling of rRNA in the presence of allylthiourea, as detected with probe EUB338, probably reflects heterotrophic CO2 fixation.
In the next step, digital image analysis was used to extract quantitative information on 14C incorporation into rRNA from the fluorescence and radioactivity scans of the microarrays. For quantification, the ratio of the radioactive signal to the fluorescence signal for rRNA hybridized to a probe (A-value) was determined with the formula (XP − XPLB) × (IP − IPLB)−1, where XP and IP are the cumulative intensities of a manually applied probe spot in radioactivity and fluorescence scans, respectively, and XPLB and IPLB are the cumulative intensities of a local background area identical in size to the analyzed probe spot in radioactivity and fluorescence scans, respectively.
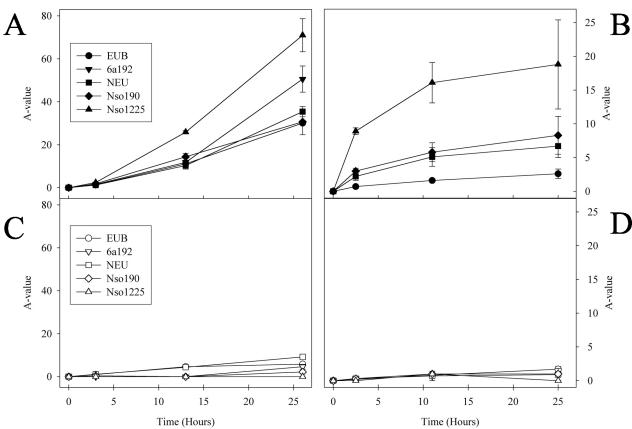
Figure 6A and B show that 14C incorporation into ammonia oxidizer rRNA steadily increased with incubation time. Interestingly, for both plants, the increases in A-values for the different ammonia oxidizer-specific probes varied at given incubation times. The A-value of probe Nso1225 was always higher than the A-values of probes Nso190, NEU, 6a192, and EUB338. For sludge samples inhibited with allylthiourea, very low A-values were determined for all probes and incubation times (Fig. 6C and D).
FIG. 6.
Incorporation of radioactivity into rRNA of phylogenetic subgroups, as measured by A-value determination for each probe (see the text for details). Isotope experiments (as displayed in Fig. 5) were performed after three different lengths of incubation of activated sludge with [14C]bicarbonate. A-values were determined for each probe and experiment. Error bars indicate standard deviations of the A-values from duplicate or triplicate experiments. (A) Aalborg West activated sludge. (B) Oberding activated sludge. (C) Aalborg West activated sludge inhibited by allylthiourea. (D) Oberding activated sludge inhibited by allylthiourea. In panels A and B, only probes which had a fluorescence signal-to-noise ratio above the threshold value of 2 are depicted.
The incorporation of [14C]bicarbonate into ammonia oxidizers was independently monitored for both wastewater treatment plants by using a combination of FISH and MAR. As expected, the ammonia oxidizers in noninhibited sludge incorporated significant amounts of labeled bicarbonate, while no incorporation could be observed after inhibition with allylthiourea (for a FISH-MAR image of this experiment, please see www.microbial-ecology.net/supplements.html).
DISCUSSION
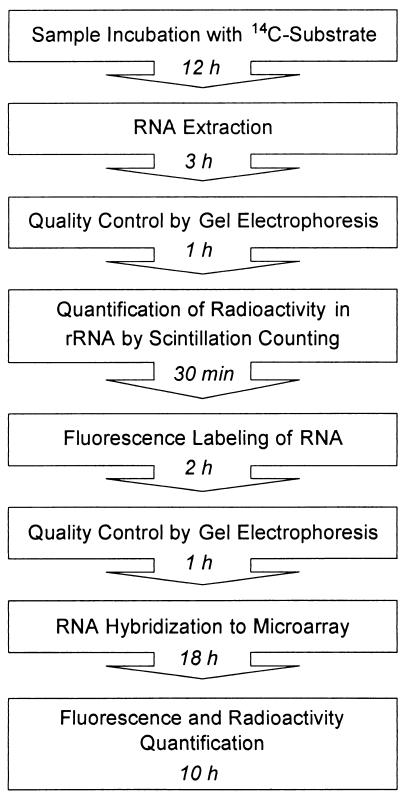
A major challenge in microbial ecology is to reveal the composition and function of complex microbial communities. While several methods for cultivation-independent analysis of community structure are available, the number of approaches which allow inference of the activity and specific function of a microorganism in its environment is still very limited. In this study, an approach was developed to allow simultaneous investigation of community structure and specific substrate consumption by community members via direct detection of environmentally retrieved 16S rRNA on a microarray (Fig. 7).
FIG. 7.
Flowchart of the isotope array approach.
AOB prototype microarray.
A prototype microarray for AOB consisting of probes which were developed previously for AOB detection via FISH or dot blot hybridization (26, 38, 52, 71) was used. Evaluation of the prototype microarray with 16S rRNAs of nine different AOB reference cultures under optimized conditions showed that all probes detected their target 16S rRNAs. However, duplex formation also occurred for some 16S rRNA molecules, with one (and, in two cases, even two) mismatches within the probe target sites (Fig. 1). No false-positive results were obtained with 16S rRNA molecules having more than two mismatches with the probes. For probes NEU and 6a192, the lack of perfect specificity on the microarray may be explained by the absence of the respective competitor probes in the hybridization solution. In addition, the length and GC content of the probes obviously influenced their specificity on the microarray. Although not perfect, the level of specificity achieved with the prototype array allowed the acquisition of important information about the AOB community structure of environmental samples. Under the conditions applied, the AOB prototype microarray is suited (i) to the detection of all recognized beta-proteobacterial AOB (positive signals with probe Nso190 and probe Nso1225) and (ii) to the specific identification of the occurrence of nitrosospiras (probe Nsv443) and N. mobilis (probe Ncmob) in a sample. Furthermore, if probe Nsv443 is negative and other AOB-specific probes on the array have a signal-to-noise ratio above the threshold, this finding is strongly indicative of the presence of nitrosomonads. We considered this level of resolution sufficient for the purpose of this study, because (i) the AOB microarray was primarily used to demonstrate the feasibility of the isotope array approach and (ii) the AOB community composition of the activated-sludge samples was analyzed in parallel by quantitative FISH and by the amoA approach.
Analysis of AOB in activated sludge by using the isotope array approach.
To demonstrate the feasibility of the isotope array approach, this method was used to investigate the community composition and CO2 fixation activity of AOB in samples from two nitrifying activated-sludges. Parallel analysis of both sludges with FISH and comparative amoA sequence analysis showed that they harbored different AOB. The Aalborg West sludge was dominated by AOB of the N. oligotropha lineage (30, 53), while N. europaea was detected as the only AOB in the Oberding sludge. These AOB are typical members of nitrifying activated-sludge communities (51, 72, 73). The same probes which were used for FISH analysis were also applied to the isotope array analysis. For both types of sludge, all probes which gave positive FISH signals were also positive in the fluorescence readouts of the isotope array experiments (Fig. 5). However, for the Aalborg West sludge, probe NEU was scored positive on the isotope array but did not yield detectable FISH signals. This discrepancy reflects the fact that the competitor probe (Cte), which is required to obtain specific FISH signals with probe NEU (71), could not be applied on the microarray format. Because probe NEU has a single mismatch with the 16S rRNAs of several heterotrophic bacteria, it detects these organisms in the absence of the competitor probe. After the inhibition of AOB in the Aalborg West sludge by the addition of allylthiourea, no signals could be obtained with the AOB-specific probes (with the exception of probe NEU) on the isotope array. In contrast, no significant changes in isotope array hybridization patterns were found between uninhibited and inhibited sludge samples from Oberding. These data suggest that the AOB of the N. oligotropha lineage in the Aalborg West sludge did reduce their cellular ribosome contents during inhibition (but still retained ribosome concentrations sufficient for FISH detection), while no indications of this effect were found for N. europaea in the Oberding sludge. Past research has shown that at least some AOB do maintain high cellular ribosome concentrations during inhibition with allylthiourea (71) and starvation (39). We are presently investigating whether species-specific differences in the control of cellular ribosome concentrations exist among AOB.
The isotope array successfully detected 14CO2 incorporation into the rRNA of AOB (Fig. 5). As expected, very low levels of radioactive rRNA labeling were observed in the control experiments with sludge samples containing an inhibitor for AOB. Radioactive labeling of rRNA in the presence of the inhibitor allylthiourea likely is caused by heterotrophic CO2 assimilation, as previously described (16, 58). For each probe and incubation time, the A-value was determined (Fig. 6). This value allows one to compare substrate incorporation between different time points in an incubation experiment and between different probe-defined populations. As expected, the A-values obtained with the AOB-specific probes and with the general bacterial probe increased during incubation with radioactive bicarbonate for both types of sludge, while very low A-values were determined for the samples amended with the inhibitor allylthiourea. The higher A-values obtained with the AOB-specific probes than with the general bacterial probe reflect the fact that the latter probe also targets nonradioactive rRNA from heterotrophic microorganisms. Interestingly, different A-values were observed for different AOB-specific probes after hybridization with rRNA from the same sludge and incubation time point. These differences could reflect differences in CO2 fixation activity between different AOB populations, illustrating the possibility of using A-value determinations for revealing important ecophysiological traits of analyzed microbial communities. However, the observation that, at least in the Oberding plant, only one AOB population was present is inconsistent with this assumption. If all AOB-specific probes were absolutely specific in this plant, then one would expect for a given incubation time point identical A-values for each of these probes Therefore, the most likely explanation for differences in A-values between the AOB-specific probes is that the probes with lower A-values bind (different amounts of) nonradioactive rRNAs of heterotrophic microorganisms. For all experiments, probe Nso1225 showed the highest A-values, indicating that this probe had the highest specificity for AOB and thus bound no rRNA or only very small amounts of rRNA from heterotrophic organisms on the microarray. This notion is in accordance with a recent specificity reevaluation of all AOB-specific probes (30), which showed that in present publically available 16S rRNA databases, there are only four 16S rRNA sequences of non-AOB with no mismatches or one mismatch with this probe. For probe Nso190, which had much lower A-values, database checks revealed the same small number of non-AOB with no mismatch or one mismatch. However, the dissociation temperature of this probe is much higher than that of probe Nso1225 (38); therefore, the hybridization conditions applied to the microarray were less stringent for probe Nso190 than for probe Nso1225, allowing for more nonspecific binding of nontarget rRNA by probe Nso190. The lower A-values of probes NEU and 6a192 than of probe Nso1225 also reflected their nonperfect specificity because their respective competitor probes, designed to increase their specificity, were not applied during microarray hybridization.
Strength and limitations of the isotope array approach.
The isotope array approach exploits environmentally retrieved rRNA as a marker for the phylogenetic affiliation and substrate consumption of target organisms. Therefore, this approach is not affected by the multiple biases associated with PCR amplification of rRNA genes (4, 11, 34, 49, 68, 78) but requires efficient extraction of rRNA from the environment under investigation. In contrast to the results reported in previous publications (3, 19, 23, 29, 64), fragmentation of rRNA prior to microarray hybridization did not enhance hybridization efficiency but increased background fluorescence after hybridization. This inconsistency might be explained by the fact that we applied a different microarray format and varied the composition of the hybridization buffer. The use of intact rRNA for microarray hybridization has the advantage that nonrandom destruction of probe target sites, which might be caused by fragmentation, can be excluded. Consequently, for our approach the quality of the extracted rRNA must be checked carefully by agarose or polyacrylamide electrophoresis prior to labeling and microarray hybridization.
The sensitivity of the isotope approach was sufficient to detect 14C incorporation into the 16S rRNA of community members which made up less than 10 and 5% of the bacterial communities in the analyzed systems, as demonstrated by quantitative FISH. However, in ecosystems with less active and dense prokaryotic communities, the applicability of the isotope array in its present format probably will be limited to numerically more abundant microorganisms. In general, the sensitivity of the isotope array depends on several factors. First, the number of bacterial target cells in the system and their mean cellular ribosome content determine the limit of detection of the organism by microarray hybridization. Second, the specific activity of the labeled substrate and the concentration of the nonlabeled substrate in the sample influence sensitivity. Third, the substrate consumption of a detectable organism can be measured only if sufficient 14C is incorporated into its rRNA molecules. For [14C]bicarbonate, we could demonstrate that 19,000 cpm of 14C per μg of RNA was sufficient to detect CO2 fixation by an ammonia-oxidizing pure culture via the isotope array approach (Table 2 and Fig. 2). The substrate incubation time required for successful analysis of AOB autotrophy was significantly lower for the isotope array approach than for DNA stable-isotope probing (76). This finding confirms that during incubation experiments, the rRNA of a particular organism is enriched for labeled carbon much faster than is its DNA (35), reflecting the more rapid turnover of the former molecules in the cell. However, it must be considered that rates of incorporation of labeled carbon into rRNA might vary significantly depending on the type of labeled substrate administered, on the stability of rRNA, and on the metabolism of the consuming organism (although the biosynthetic pathways for RNA production are quite similar among bacterial species). This limitation is not restricted to the isotope array approach and also influences DNA and RNA stable-isotope probing (35, 36, 54), whereas this bias does not apply to FISH-MAR experiments (31), which purely monitor the assimilation of labeled substrates.
The exploitation of labeled substrates also causes other potential limitations. In principle, the possibility that members of a microbial community discriminate between an isotope-labeled substrate and its natural counterpart cannot be excluded. Although bacteria discriminate against 14C-labeled compounds more than against 13C-labeled substrates, the extent of fractionation is often negligible (18, 24, 45) and unlikely to affect the assimilation of substrates in experiments (as ours) with large amounts of isotope enrichment.
Another important factor which must be considered is substrate cross-feeding, which is of particular importance at prolonged substrate incubation times. This effect could be exploited to investigate the transfer of carbon through microbial communities but can cause problems in differentiating between primary substrate consumers and microorganisms which live on the secretions or lysis products of those primary consumers. Again, these limitations are not inherent to the isotope array approach but affect stable isotope probing and the combination of FISH and MAR.
Unlike DNA stable-isotope probing, isotope array analysis does not allow one to harvest and exploit the genomic DNA of metabolically active microorganisms for subsequent analyses and thus does not provide insights into the genetic compositions of the target organisms. On the other hand, isotope array analysis has several unique advantages. In contrast to DNA or RNA stable-isotope probing, substrate incorporation into target nucleic acids is measured directly by using the isotope array approach; thus, organisms which do not possess labeled nucleic acids cannot be detected in error. Compared to FISH-MAR, the isotope array allows one to apply many probes in parallel, a feature which will be of major importance if the ecophysiology of complex microbial communities is of interest. Additionally, the isotope array may (if stringent hybridization conditions can be achieved for all probes) permit examination of the specific incorporation of isotopes into the rRNAs of members of different phylogenetic groups, expressed as A-values.
In conclusion, this study reports on the development and evaluation of a novel approach for structure-function analyses of complex microbial communities with oligonucleotide microarrays. Continuing advances in microarray technology (19, 32, 48, 64) suggest that it will soon become feasible to develop encompassing habitat-specific microarrays which will allow an investigator to monitor the entire microbial community of a system. Equipped with such tools, the isotope array will provide a new means to directly link the detected organisms with their specific activities and functions in an ecosystem.
Acknowledgments
J.A. and M. Hesselsoe contributed equally to this work.
The excellent technical assistance of Sibylle Schadhauser, Carmen Krammer, Kirsten Maagaard, and Marianne Stevenson is acknowledged.
This research was supported by the following grants: bmb+f 01 LC 0021 (Subproject 2 in the framework of the BIOLOG Program) (to M.W.), a Bayerischen Forschungsstiftung grant (for development of oligonucleotide DNA chips, in cooperation with MWG Biotech; project 368/99) (to M.W.), travel grant 21-02-0166 from the Danish Natural Science Research Council (to M. Hesselsoe), grant QLK3-2000-01528 from the European Commission (Framework Program 5) (to M.H., N.I., and P.R.), and a grant from the Danish Technical Research Council (Framework Program Activity and Diversity in Complex Microbial Systems) (to P.H.N., N.I., and P.R.).
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bavykin, S. G., J. P. Akowski, V. M. Zakhariev, V. E. Barsky, A. N. Perov, and A. D. Mirzabekov. 2001. Portable system for microbial sample preparation and oligonucleotide microarray analysis. Appl. Environ. Microbiol. 67:922-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, S., P. Boger, R. Oehlmann, and A. Ernst. 2000. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Appl. Environ. Microbiol. 66:4945-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedard, C., and R. Knowles. 1989. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev. 53:68-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boschker, H. T. S., S. C. Nold, P. Wellsburry, D. Bos, W. de Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 7.Busti, E., R. Bordoni, B. Castiglioni, P. Monciardini, M. Sosio, S. Donadio, C. Consolandi, L. Rossi Bernardi, C. Battaglia, and G. De Bellis. 2002. Bacterial discrimination by means of a universal array approach mediated by LDR (ligase detection reaction). BMC Microbiol. 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chai, X. S., Q. Luo, and J. Y. Zhu. 2001. Analysis of nonvolatile species in a complex matrix by headspace gas chromatography. J. Chromatogr. 909:249-257. [DOI] [PubMed] [Google Scholar]
- 9.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 11.Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques 34:790-794, 796, 798. [DOI] [PubMed] [Google Scholar]
- 12.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 13.Daims, H., J. L. Nielsen, P. H. Nielsen, K. H. Schleifer, and M. Wagner. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daims, H., N. B. Ramsing, K. H. Schleifer, and M. Wagner. 2001. Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization. Appl. Environ. Microbiol. 67:5810-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Saizieu, A., U. Certa, J. Warrington, C. Gray, W. Keck, and J. Mous. 1998. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat. Biotechnol. 16:45-48. [DOI] [PubMed] [Google Scholar]
- 16.Dijkhuizen, L., and W. Harder. 2000. Microbial metabolism of carbon dioxide, p. 409-423. In H. Dalton (ed.), Comprehensive bio/technology, vol. 1. Pergamon Press Ltd., Oxford, United Kingdom.
- 17.Donaldson, J. M., and G. S. Henderson. 1989. A dilute medium to determine population size of ammonium oxidizers in forest soils. Soil. Sci. Soc. Am. J. 53:1608-1611. [Google Scholar]
- 18.Ekblad, A., G. Nyberg, and P. Hogberg. 2000. C-13-discrimination during microbial respiration of added C-3-, C-4- and C-13-labelled sugars to a C-3-forest soil. Oecologia 131:245-249. [DOI] [PubMed] [Google Scholar]
- 19.El Fantroussi, S., H. Urakawa, A. E. Bernhard, J. J. Kelly, P. A. Noble, H. Smidt, G. M. Yershov, and D. A. Stahl. 2003. Direct profiling of environmental microbial populations by thermal dissociation analysis of native rRNAs hybridized to oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2377-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray, N. D., R. Howarth, R. W. Pickup, J. G. Jones, and I. M. Head. 2000. Use of combined microautoradiography and fluorescence in situ hybridization to determine carbon metabolism in mixed natural communities of uncultured bacteria from the genus Achromatium. Appl. Environ. Microbiol. 66:4518-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray, N. D., and I. M. Head. 2001. Linking genetic identity and function in communities of uncultured bacteria. Environ. Microbiol. 3:481-492. [DOI] [PubMed] [Google Scholar]
- 22.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, A., and J. Pardue. 1999. Quantifying the mineralization of contaminants using stable carbon isotope ratios. Org. Geochem. 30:787-792. [Google Scholar]
- 25.Johnsen, A. R., A. Winding, U. Karlson, and P. Roslev. 2002. Linking of microorganisms to phenanthrene metabolism in soil by analysis of 13C-labeled cell lipids. Appl. Environ. Microbiol. 68:6106-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Roser, H.-P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 28.Kerkhof, L., and P. Kemp. 1999. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 30:253-260. [DOI] [PubMed] [Google Scholar]
- 29.Koizumi, Y., J. J. Kelly, T. Nakagawa, H. Urakawa, S. El-Fantroussi, S. Al-Muzaini, M. Fukui, Y. Urushigawa, and D. A. Stahl. 2002. Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology. Appl. Environ. Microbiol. 68:3215-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koops, H.-P., U. Purkhold, A. Pommerening-Röser, G. Timmermann, and M. Wagner. March 2003, posting date. The lithoautotrophic ammonia-oxidizing bacteria. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. An evolving electronic resource for the microbiological community, 3rd ed., release 3.13. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 31.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescence in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manefield, M., A. S. Whiteley, N. Ostle, P. Ineson, and M. J. Bailey. 2002. Technical considerations for RNA-based stable isotope probing: an approach to associating microbial diversity with microbial community function. Rapid Commun. Mass Spectrom. 16:2179-2183. [DOI] [PubMed] [Google Scholar]
- 37.Minz, D., S. Fishbain, S. J. Green, G. Muyzer, Y. Cohen, B. E. Rittmann, and D. A. Stahl. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl. Environ. Microbiol. 65:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. (Erratum, 63:815, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgenroth, E., A. Obermayer, E. Arnold, A. Brühl, M. Wagner, and P. A. Wilderer. 2000. Effect of long-term idle periods on the performance of sequencing batch reactors. Water Sci. Technol. 41:105-113. [Google Scholar]
- 40.Morris, S. A., S. Radajewski, T. W. Willison, and J. C. Murrell. 2002. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen, J. L., S. Juretschko, M. Wagner, and P. H. Nielsen.2002. Abundance and phylogenetic affiliation of iron reducers in activated sludge as assessed by fluorescence in situ hybridization and microautoradiography. Appl. Environ. Microbiol. 8:629-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen, J. L., D. Christensen, M. Kloppenborg, and P. H. Nielsen. 2003. Quantification of cell-specific substrate uptake by probe-defined bacteria under in situ conditions by microautoradiography and fluorescence in situ hybridization. Environ. Microbiol. 5:202-211. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen, P. H., M. A. de Muro, and J. L. Nielsen. 2000. Studies on the in situ physiology of Thiothrix spp. present in activated sludge. Environ. Microbiol. 2:389-398. [DOI] [PubMed] [Google Scholar]
- 45.Northrop, D. B. 1981. The expression of isotope effects on enzyme-catalyzed reactions. Annu. Rev. Biochem. 50:103-131. [DOI] [PubMed] [Google Scholar]
- 46.Oda, Y., S. Slagman, W. G. Meijer, L. J. Forney, and J. C. Gottschal. 2000. Influence of growth rate and starvation on fluorescent in situ hybridization of Rhodopseudomonas palustris. FEMS Microbiol. Ecol. 32:205-213. [DOI] [PubMed] [Google Scholar]
- 47.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peplies, J., F. O. Glockner, and R. Amann. 2003. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 69:1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purkhold, U., A. Pommering-Röser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purkhold, U. 2003. Untersuchungen zur Phylogenie und Verbreitung Ammoniak oxidierender Bakterien. Ph.D. thesis. Technische Universität München, Munich, Germany.
- 53.Purkhold, U., M. Wagner, G. Timmermann, A. Pommerening-Röser, and H.-P. Koops. 2003. 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia oxidizing isolates: extension of the data set and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485-1494. [DOI] [PubMed]
- 54.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 55.Radajewski, S., G. Webster, D. S. Reay, S. A. Morris, P. Ineson, D. B. Nedwell, J. I. Prosser, and J. C. Murrell. 2002. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331-2342. [DOI] [PubMed] [Google Scholar]
- 56.Ravenschlag, K., K. Sahm, and R. Amann. 2001. Quantitative molecular analysis of the microbial community in marine arctic sediments (Svalbard). Appl. Environ. Microbiol. 67:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Risatti, J. B., W. C. Capman, and D. A. Stahl. 1994. Community structure of a microbial mat: the phylogenetic dimension. Proc. Natl. Acad. Sci. USA 91:10173-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romanenko, V. I. 1964. Heterotrophic assimilation of CO2 by bacterial flora of water. Microbiology 33:610-614.14238140 [Google Scholar]
- 59.Roslev, P., P. L. Madsen, J. B. Thyme, and K. Henriksen. 1998. Degradation of phthalate and di-(2-ethylhexyl)phthalate by indigenous and inoculated microorganisms in sludge-amended soil. Appl. Environ. Microbiol. 64:4711-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahm, K., B. J. MacGregor, B. B. Jorgensen, and D. A. Stahl. 1999. Sulphate reduction and vertical distribution of sulphate-reducing bacteria quantified by rRNA slot-blot hybridization in a coastal marine sediment. Environ. Microbiol. 1:65-74. [DOI] [PubMed] [Google Scholar]
- 62.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. Jetten, J. W. Metzger, K.-H. Schleifer, and M. Wagner. 2000. Molecular evidence for a genus-level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 63.Schmid, M., S. Schmitz-Esser, M. Jetten, and M. Wagner. 2001. 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium oxidizing bacteria: implications for phylogeny and in situ detection. Environ. Microbiol. 3:450-459. [DOI] [PubMed] [Google Scholar]
- 64.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Speksnijder, A. G., G. A. Kowalchuk, K. Roest, and H. J. Laanbroek. 1998. Recovery of a Nitrosomonas-like 16S rDNA sequence group from freshwater habitats. Syst. Appl. Microbiol. 21:321-330. [DOI] [PubMed] [Google Scholar]
- 66.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stephen, J. R., Y. J. Chang, S. J. Macnaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil beta-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wagner, M., R. Amann, H. Lemmer, and K.-H. Schleifer. 1993. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 59:1520-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagner, M., R. Amann, P. Kämpfer, B. Assmus, A. Hartmann, P. Hutzler, N. Springer, and K.-H. Schleifer. 1994. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. Syst. Appl. Microbiol. 17:405-417. [Google Scholar]
- 71.Wagner, M., G. Rath, R. Amann, H.-P. Koops, and K.-H. Schleifer. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 72.Wagner, M., and A. Loy. 2002. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 13:218-227. [DOI] [PubMed] [Google Scholar]
- 73.Wagner, M., A. Loy, R. Nogueira, U. Purkhold, N. Lee, and H. Daims. 2002. Microbial community composition and function in wastewater treatment plants. Antonie Leeuwenhoek 81:665-680. [DOI] [PubMed] [Google Scholar]
- 74.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 75.Wang, R. F., M. L. Beggs, L. H. Robertson, and C. E. Cerniglia. 2002. Design and evaluation of oligonucleotide-microarray method for the detection of human intestinal bacteria in fecal samples. FEMS Microbiol. Lett. 213:175-182. [DOI] [PubMed] [Google Scholar]
- 76.Whitby, C. B., G. Hall, R. Pickup, J. R. Saunders, P. Ineson, N. R. Parekh, and A. McCarthy. 2001. 13C incorporation into DNA as a means of identifying the active components of ammonia-oxidizer populations. Lett. Appl. Microbiol. 32:398-401. [DOI] [PubMed] [Google Scholar]
- 77.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wintzigerode, F., and U. B. Goebel. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 79.Wuchter, C., S. Schouten, H. T. Boschker, and J. S. Sinninghe Damste. 2003. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol. Lett. 219:203-207. [DOI] [PubMed] [Google Scholar]