Abstract
Humic lakes are systems often characterized by irregular high input of dissolved organic carbon (DOC) from the catchment. We hypothesized that specific bacterial groups which rapidly respond to changes in DOC availability might form large populations in such habitats. Seasonal changes of microbial community composition were studied in two compartments of an artificially divided bog lake with contrasting DOC inputs. These changes were compared to community shifts induced during short-term enrichment experiments. Inocula from the two compartments were diluted 1:10 into water from the more DOC-rich compartment, and inorganic nutrients were added to avoid microbial N and P limitation. The dilutions were incubated for a period of 2 weeks. The microbial assemblages were analyzed by cloning and sequencing of 16S rRNA genes and by fluorescence in situ hybridization with specific oligonucleotide probes. β-Proteobacteria from a cosmopolitan freshwater lineage related to Polynucleobacter necessarius (beta II) were rapidly enriched in all treatments. In contrast, members of the class Actinobacteria did not respond to the enhanced availability of DOC by an immediate increase in growth rate, and their relative abundances declined during the incubations. In lake water members of the beta II clade seasonally constituted up to 50% of all microbes in the water column. Bacteria from this lineage annually formed a significantly higher fraction of the microbial community in the lake compartment with a higher allochthonous influx than in the other compartment. Actinobacteria represented a second numerically important bacterioplankton group, but without clear differences between the compartments. We suggest that the pelagic microbial community of the studied system harbors two major components with fundamentally different growth strategies.
Humic lakes are naturally acidic freshwater systems with a large pool of dissolved organic carbon (DOC) that originates mainly from the catchment area (18, 35). Due to the bad-light climate and other factors, the biomass of primary producers in humic lakes is often lower than what might be predicted by nutrient levels (19), and mixotrophy is a common strategy of many phytoplankton species in such systems (21). The ratio of bacteria to phytoplankton is typically high in this type of lake (54). Bacterial secondary production is often nutrient (P and N) rather than carbon limited (22), and it may significantly exceed primary production (46, 54). Little is known about the bacterial community structure of humic lakes. High abundances of uncultured members of the class Actinobacteria have been found in the plankton of the acidic bog lake Groβe Fuchskuhle by 16S rRNA gene sequence analysis and subsequent fluorescence in situ hybridization (FISH) with oligonucleotide probes (13, 47). With samples from several Scandinavian bog lakes, an analysis of bands from denaturing gradient gel electrophoresis revealed sequence types that were also related to this lineage (30).
Organic substrates are doubtlessly an important factor determining the success of particular bacterial species in the environment. The main fraction of the DOC in bog lakes is recalcitrant high-molecular-weight humic acids (53). These substances may be partially transformed into more bioavailable forms by photochemical processes prior to microbial degradation (55). An exact determination of the composition of such DOC usually is beyond analytical capacities. It is thus difficult to relate a specific DOC component to the growth of individual bacterial groups in situ. In addition, heterotrophic aquatic bacteria are probably rather versatile in their choice of carbon source, and there might be a great overlap in the types of substrates consumed by many planktonic bacteria. For example, radiolabeled amino acids are readily consumed by the majority of pelagic marine bacteria (23).
In contrast, another aspect of bottom-up interactions between water column microbes can be studied more readily. Individual phylogenetic lineages within the bacterioplankton may differ in their abilities to succeed in habitats with steep or flat substrate gradients (39), and some bacterioplankton genera exhibit a more rapid growth response under batch culture “feast-and-famine” conditions (8, 50). There are indications that this growth strategy might be widespread among bacterial groups that successfully utilize small-scale nutrient patches or colonize microaggregates (14). In contrast, the growth of some typical free-living freshwater bacteria may even be negatively affected by enhanced substrate levels (15). A different growth response to changing substrate availability (39) thus might determine the relative abundances of individual bacterial groups in situations of high resource variability.
We investigated the identities of bacteria that rapidly respond to enhanced DOC availability in a naturally acidified bog lake, the Groβe Fuchskuhle. This lake is divided into artificial compartments with contrasting limnological properties and plankton community structure (17, 24, 49). Enrichment experiments with inocula from two of these compartments were performed with dilutions of ambient water from the more acidic, humic-rich compartment. The experiments were carried out during the winter season when the growth of lake bacteria was most likely controlled by factors other than grazing mortality (temperature and resources). Because of the high allochthonous DOC input into humic lakes, we hypothesized that some bacterial groups which rapidly respond to changes in available substrate levels might also be of importance to the plankton of the lake. This hypothesis was tested with samples collected from the studied compartments over a period of 1 year.
MATERIALS AND METHODS
Study site.
Lake Groβe Fuchskuhle is situated in the Mecklenburg-Brandenburg Lake District in northeastern Germany, 59 m above mean sea level. In 1990 the naturally acidic bog lake was divided into four compartments: southwest (SW), northwest (NW), northeast (NE), and southeast (SE) (24, 27) (Fig. 1). Since the division, the compartments have diverged in their physical and chemical parameters, in microbial activity (2, 4), and in the structure of the microbial food web (17, 49). For this study two of the compartments were selected: the SW compartment, which has an acidotrophic humic character and a higher influx of allochthonous DOC, and the NE compartment, which has a more mesotrophic character. Between September 2000 and September 2001, lake water from the two compartments was collected from a 0.5-m depth at approximately monthly intervals for total microbial counts and community analysis.
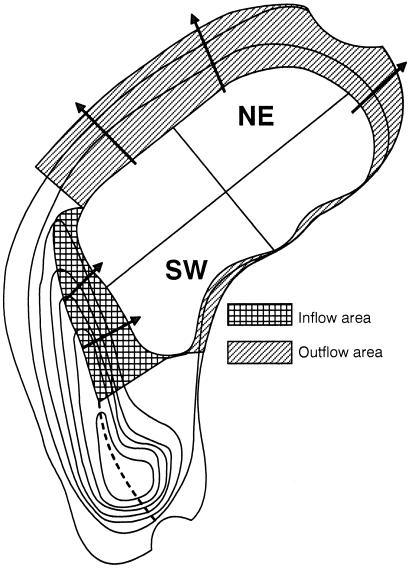
FIG. 1.
Schematic depiction of the compartments of Lake Groβe Fuchskuhle in relationship to the area of water in- and outflow of the catchment. The inflow area is largely formed by a Sphagnum bog. Contour lines indicate a 0.1-m change in altitude.
Experimental design, sampling, and fixation.
An enrichment experiment was conducted as a competition batch culture in 1:10 dilutions. Lake water from the SW compartment was collected in January 2002 from a 0.5-m depth and filtered twice through 0.1-μm-pore-size polycarbonate filters (diameter, 50 mm; Sartorius, Göttingen, Germany). The filtrate was supplemented with inorganic nutrients (final concentrations, 0.3 μM KH2PO4, 4.55 μM (NH4)2SO4, and 1.23 mM NaHCO3) to avoid bacterial N and P limitation (22) and used as incubation medium. Acid-prewashed and sterilized round flasks of the Duran type were filled with 1,800 ml of the medium and inoculated with 200 ml of prefiltered water (filter diameter, 47 mm; pore size, 0.8 μm; Osmonics) taken at a 0.5-m depth from either the SW or the NE compartment (termed SW and NE treatments). The experiment was performed in triplicate for inocula from each compartment, and changes were monitored for 14 days. For controls, 200-ml aliquots of inocula from the two compartments were diluted into 1,800 ml of an 1.23 mM NaHCO3 solution (final concentration) that was supplemented with the same inorganic nutrients as were in the lake water medium. The pH conditions in the treatments were set to reflect the pHs of the respective compartments (SW compartment, pH 5.0; NE compartment, pH 6.0). The flasks were placed in a dark room on magnetic stirrers. The treatment flasks were moderately aerated with filtered (pore size, 0.22 μm) air in order to maintain oxygenated conditions observed at the sampling location and time point (in situ oxygen saturation, 89%). During the experiment the temperature ranged between 11 and 12°C. Subsamples of ca. 150 ml were taken at day 0 and every second day thereafter for a period of 2 weeks.
Samples for total bacterial numbers were fixed with formaldehyde (1% [wt/vol] final concentration) and stored at 4°C until processed further. Samples for community analysis (10 to 20 ml) were fixed with formaldehyde (1% [wt/vol] final concentration) and stored for 24 h at 6°C. Then the samples were filtered onto a 0.2-μm-pore-size filter (type GTTC, 47 mm; Whatman), rinsed two times with 5 ml of sterile water, dried at room temperature, and stored at −20°C.
Total cell counts and bacterial production.
Total bacterial numbers were determined after filtration of subsamples (2 ml) onto black membrane filters (pore size, 0.2 μm; diameter, 25 mm; Osmonics) and staining with 4′,6′- diamidino-2′-phenylindole (DAPI; final concentration, 0.1 μg ml−1) (42). The enumeration was performed by epifluorescence microscopy (model DM RB microscope; Leica, Bensheim, Germany) coupled with a camera (model CF 8/1 RCC Air; KAPPA Technologies, Gleichen, Germany) connected to an image analysis system (IMAGE P2; resolution, 736 by 556 pixels; 256 grey levels; H&K, Berlin, Germany).
For the enumeration of protists, 50-ml subsamples were fixed with 20 to 25 μl of alkaline Lugol's solution, followed by immediate treatment with 1 to 1.25 ml of formaldehyde (1% [wt/vol] final concentration) and 40 to 50 μl of 3% sodium thiosulfate (48). The samples were filtered on black membrane filters (pore size, 0.8 μm; diameter, 25 mm; Osmonics Corp.) and stained with DAPI (final concentration, 0.1 μg ml−1), and cells were counted by epifluorescence microscopy (Axioplan microscope; Carl Zeiss, Jena, Germany).
Bacterial bulk growth activity during the enrichments was estimated from the incorporation rate of tritiated leucine modified as described by Kirchman et al. (25). Subsamples of 10 ml were incubated with 50 μl of [3H]leucine (final concentration, 0.1 μM; Amersham) and an equal amount of nonlabeled leucine. After 1 to 2 h of incubation at in situ temperature, the samples were preserved with formaldehyde (1% [wt/vol] final concentration) and filtered through polycarbonate filters (pore size, 0.2 μm; diameter, 25 mm; Corning Costar, Cambridge, Mass.). The filters were then treated four times with 1 ml of ice-cold 5% trichloroacetic acid. In the controls, formaldehyde was added before the addition of [3H]leucine. The dried filters were covered with 9 ml of scintillation cocktail (emulsifier safe; Canberra-Packard, Rodgau-Juegesheim, Germany), and cells were quantified in a liquid scintillation counter (TRI-CARD 1600; Canberra-Packard). A control parameter that indicated changes in quenching was obtained for each sample. The total incorporation of [3H]leucine into bacterial biomass was corrected by subtraction of counts of cells from those of prefixed controls.
Carbon analyses.
At the beginning of the experiment and at three subsequent time points (days 6, 8, and 14), the quality and quantity of the DOC pool (chromatographied DOC portion [cDOC]) were analyzed with a custom-designed automated size exclusion chromatograph connected to UV and organic carbon detection systems (45). Four different cDOC fractions were distinguished and quantified: polysaccharides, humic substances, low-molecular-weight acids, and other fractions (proteins and amino acids).
16S rRNA gene clone libraries.
For construction of 16S rRNA gene clone libraries, samples (500 to 600 ml) were taken from both compartments in February 2001 at a depth of 0.5 m. After prefiltration through polycarbonate filters (pore size, 1.2 μm; Schleicher & Schuell, Dueren, Germany) the samples were filtered on membrane filters (pore size, 0.2 μm; diameter, 47 mm; Durapore; Millipore Corp., Bedford, Mass.) and stored at −80°C until they were processed further. Unfixed samples from the beginning (1,000 ml) and from the end (620 to 720 ml) of the enrichment experiments were filtered directly onto membrane filters (pore size, 0.2 μm; diameter, 47 mm; Durapore; Millipore) and stored at −80°C. For the amplification of 16S rRNA genes by PCR, 5 μl of bovine serum albumin (stock concentration, 3 mg ml−1), 5 μl of 10× PCR buffer, 2 μl of the deoxynucleoside triphosphates (stock concentration, 2.5 mM), 0.5 μl of the general bacterial primers GM3F and GM4R (37) (stock concentration, 15 μM), and 0.25 μl of TaKaRa-Taq DNA polymerase (TaKaRa BIO Inc., Shiga, Japan) (stock concentration, 5 U μl−1) were adjusted to a final volume of 50 μl with sterile water. Small pieces (1 to 2 mm2) of the above-described filters were directly added to the PCR tubes as templates as described previously (26). The PCRs were run on a Mastercycler (Eppendorf, Harmurg, Germany) with the following cycling conditions: 1 cycle at 94°C for 5 min and 35 cycles at 94°C for 30 s, 47°C for 45 s, 74°C for 1 min 30 s, and 74°C for 10 min. The amplified 16S rRNA gene fragments were purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany), inserted into the TOPO vector (TOPO TA cloning kit; Invitrogen, Karlsruhe, Germany), and cloned into competent cells of Escherichia coli as described by the manufacturer. The transformed cells were plated on Luria-Bertani agar plates containing 50 μg of ampicillin ml−1 and incubated overnight at 37°C. The clones were screened for right-sized inserts, and plasmid preparations were made with a QIAprep Spin Miniprep kit (QIAGEN). For a first screening, the plasmid DNAs were sequenced with the primer M13F (5′-GTA AAA CGA CGG CCA G-3′), located on the vector pCR4-TOPO. Nearly full-length 16S rRNA sequences were obtained from selected inserts by additional sequencing with the primers GM1 (36) and M13R (5′-CAG GAA ACA GCT ATG AC-3′).
Phylogenetic analysis and probe design.
Partial sequences were assembled manually using the software Sequencher (Gene Codes Corp., Ann Arbor, Mich.) and tested for chimeras through the Ribosomal Database Project CHIMERA_CHECK program. All sequences were subsequently analyzed by the BLAST queuing system (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) to identify their closest relatives and their tentative phylogenetic positions. Those sequences that were related to the β-proteobacteria were then analyzed in more detail. Phylogenetic analyses were performed using the ARB software package (http:www.arb-home.de). The ARB database (release of June 2002) was complemented with other β-proteobacterial sequences from GenBank that were related to freshwater lineages (13, 56). For the reconstruction of a phylogenetic tree, only nearly complete (i.e., longer than 1,400 nucleotides) 16S rRNA sequences affiliated with this subphylum were considered. A 50% base frequency filter was used on these sequences to exclude highly variable positions. The respective ARB tools were used to perform maximum parsimony, neighbor joining, and maximum likelihood analyses. The resulting phylogenetic trees were compared manually. A consensus tree that shows bifurcations only if branchings were stable in the majority of analyses was constructed. Multifurcations were introduced if tree topologies could not be unambiguously resolved. Partial sequences were subsequently added to the consensus tree according to maximum parsimony criteria, without changes being allowed in the overall tree topology.
Specific oligonucleotide probes for the different β-proteobacterial clusters that contained sequence types from Lake Groβe Fuchskuhle were designed by using the ARB software package. Stringent conditions for FISH were established by analysis of the fluorescence intensities of the target cells from hybridizations at increasing concentrations of formamide in the hybridization buffer (40).
FISH.
Whole-cell in situ hybridizations of sections from the polycarbonate filters were performed with the oligonucleotide probes BET42a (β-proteobacteria, 23S rRNA targeted), HGC69a (Actinobacteria, 23S rRNA targeted) (1), and the newly designed specific probes. All probes were obtained from ThermoHybaid (Interactiva Division, Ulm, Germany). Hybridizations with directly Cy3-monolabeled probes BET42a and BET2-870 (length, 18 nucleotides; E. coli positions 870 to 888 [5], 5′-CCC AGG CGG CTG ACT TCA-3′) were performed as described previously (40) with 35% of formamide in the hybridization buffer. For FISH with probe HGC69a, whose 5′ end was labeled the horseradish peroxidase (HRP), and subsequent signal amplification by catalyzed reporter deposition (CARD-FISH), filters were first embedded in low-gelling-point agarose (0.2% concentration of MetaPhor; FMC Bioproducts, Rockland, Maine). Next, the embedded cells were permeabilized with lysozyme and achromopeptidase (47). Hybridizations and signal amplification with Cy3-labeled tyramides were performed as described previously (38). After FISH or CARD-FISH, the filters were air dried and mounted on glass slides with a previously described mounting mix amended with DAPI (4′,6′diamidino-2-phenylindole; final concentration, 1 μg ml−1) (38). The stained filter sections were evaluated on an Axioplan II microscope equipped with a 100× Plan Apochromat oil objective lens (Carl Zeiss, Jena, Germany) and an HBO 100-W Hg vapor lamp. We used a model HQ 41007 filter set (Chroma Technology Corp., Brattleboro, Vt.) for probe fluorescence and the Zeiss 01 filter set for DAPI. The counts were carried out by semiautomated image analysis.
To test the accuracy of FISH counts with the monolabeled probes, triplicate parallel hybridizations of samples from the SW treatment at day 0 with a monolabeled probe and HRP-labeled probe BET42a were performed. Statistical comparisons between the seasonal development of microbial communities in the two compartments were performed by the nonparametric Wilcoxon matched-pair test.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of the actinobacterial clones obtained during this study were deposited in GenBank with the following accession numbers: AJ575497 through AJ575556.
RESULTS
Phylogenetic analysis.
All together 62 unique partial sequences (approximately 700 bp) were obtained from the screening of 179 clones from four libraries (SW and NE compartments, day 0; SW and NE enrichments, day 12). Of these, 33 sequences were affiliated with the α-, β-, and γ-proteobacteria, 11 with the Bacteroidetes, 6 with the Actinobacteria, and 12 with the Verrucomicrobiae. Of the subsequently produced almost complete rRNA gene sequences, 18 were affiliated with the β-proteobacteria, 5 with the Bacteroidetes, 6 with the Actinobacteria, and 3 with the Verrucomicrobiae. Table 1 lists the sizes and phylogenetic affiliations of all newly produced sequences except for those affiliated with the β-proteobacteria. The phylogenetic positions of the obtained β-proteobacterial sequence types are instead depicted in Fig. 2.
TABLE 1.
16S rRNA gene sequences of clones from Lake Groβe Fuchskuhle and from the enrichments that were not affiliated with the β-proteobacteria
| Clone | Sample | nta | Affiliation | Closest hit in GenBank (accession no.) | Similarity (%) |
|---|---|---|---|---|---|
| 32 | Lake (SW) | 1,402 | α-Proteobacteria | Uncultured bacterium FukuS56 (AJ290014) | 99 |
| 22 | Lake (NE) | 1,420 | α-Proteobacteria | Uncultured bacterium FukuS56 (AJ290014) | 99 |
| 65 | Enrichment (SW) | 747 | α-Proteobacteria | Uncultured bacterium FukuS56 (AJ290014) | 99 |
| 41 | Enrichment (NE) | 1,450 | α-Proteobacteria | Uncultured bacterium FukuS56 (AJ290014) | 99 |
| 46 | Enrichment (NE) | 1,465 | α-Proteobacteria | Uncultured bacterium FukuN22 (AJ289994) | 99 |
| 69 | Enrichment (NE) | 1,441 | α-Proteobacteria | Uncultured bacterium FukuS56 (AJ290014) | 97 |
| 73 | Enrichment (NE) | 1,430 | α-Proteobacteria | α-Proteobacterium F06021 (AF235996) | 99 |
| 77 | Lake (SW) | 1,479 | γ-Proteobacteria | Unidentified γ-proteobacterium (AB015575) | 99 |
| 17 | Lake (NE) | 753 | γ-Proteobacteria | Pseudomonas chlororaphis DSM 50083T (Z76673) | 99 |
| 23 | Lake (NE) | 654 | γ-Proteobacteria | Pseudomonas aureofaciens (Z76656) | 99 |
| 26 | Enrichment (SW) | 717 | γ-Proteobacteria | Pseudomonas chlororaphis DSM 50083T (Z76673) | 99 |
| 28 | Enrichment (SW) | 1,519 | γ-Proteobacteria | Pseudomonas graminis (Y11150) | 98 |
| 67 | Enrichment (SW) | 1,463 | γ-Proteobacteria | Pseudomonas putida ATCC 17527 (AF094743) | 98 |
| 4 | Enrichment (NE) | 711 | γ-Proteobacteria | Pseudomonas sp. strain NZ017 (AY014805) | 99 |
| 79 | Enrichment (NE) | 950 | γ-Proteobacteria | Pseudomonas chlororaphis (AJ550465) | 99 |
| 11 | Lake (SW) | 1,505 | Bacteroidetes | Uncultured bacterium FukuS59 (AJ290042) | 98 |
| 30 | Lake (SW) | 1,523 | Bacteroidetes | Cytophagales strain MBIC4147 (AB022889) | 95 |
| 35 | Lake (SW) | 721 | Bacteroidetes | Cytophagales strain MBIC4147 (AB022889) | 96 |
| 36 | Lake (SW) | 1,482 | Bacteroidetes | Uncultured bacterium GKS2-106 (AJ290025) | 94 |
| 56 | Lake (SW) | 1,471 | Bacteroidetes | Uncultured bacterium FukuN23 (AJ290011) | 99 |
| 63 | Lake (SW) | 699 | Bacteroidetes | Cytophagales strain MBIC4147 (AB022889) | 96 |
| 66 | Lake (SW) | 699 | Bacteroidetes | Cytophagales strain MBIC4147 (AB022889) | 97 |
| 2 | Enrichment (NE) | 1,454 | Bacteroidetes | Uncultured bacterium FukuN24 (AJ289995) | 98 |
| 3 | Enrichment (NE) | 702 | Bacteroidetes | Uncultured bacterium FukuN24 (AJ289995) | 97 |
| 60 | Enrichment (NE) | 894 | Bacteroidetes | Uncultured Cytophagales clone PRD01a001B (AF289149) | 99 |
| 61 | Enrichment (NE) | 578 | Bacteroidetes | Uncultured bacterium FukuN24 (AJ289995) | 97 |
| 31 | Lake (SW) | 1,492 | Actinobacteria | Uncultured actinobacterium ML316M-7 (AF447767) | 95 |
| 33 | Lake (SW) | 1,489 | Actinobacteria | Uncultured bacterium FukuS5 (AJ290022) | 99 |
| 94 | Lake (SW) | 1,473 | Actinobacteria | Uncultured bacterium clone HT2E3 (AF418967) | 96 |
| 7 | Lake (NE) | 1,483 | Actinobacteria | Uncultured bacterium FukuN30 (AJ289996) | 98 |
| 51 | Lake (NE) | 1,453 | Actinobacteria | Uncultured bacterium FukuN30 (AJ289996) | 99 |
| 84 | Enrichment (NE) | 1,472 | Actinobacteria | Uncultured Crater Lake bacterium CL500-29 (AF316678) | 98 |
| 59 | Lake (SW) | 926 | Verrucomicrobiae | Uncultured bacterium FukuN111 (AJ289985) | 98 |
| 6 | Lake (NE) | 1,520 | Verrucomicrobiae | Uncultured bacterium FukuN18 (AJ289992) | 98 |
| 9 | Lake (NE) | 1,520 | Verrucomicrobiae | Uncultured bacterium FukuN18 (AJ289992) | 98 |
| 10 | Lake (NE) | 662 | Verrucomicrobiae | Uncultured bacterium FukuN18 (AJ289992) | 98 |
| 12 | Lake (NE) | 698 | Verrucomicrobiae | Uncultured bacterium FukuN18 (AJ289992) | 98 |
| 37 | Lake (NE) | 720 | Verrucomicrobiae | Uncultured Crater Lake bacterium CL120-10 (AF316720) | 96 |
| 53 | Lake (NE) | 658 | Verrucomicrobiae | Uncultured Crater Lake bacterium CL120-10 (AF316720) | 97 |
| 54 | Lake (NE) | 700 | Verrucomicrobiae | Uncultured Crater Lake bacterium CL120-10 (AF316720) | 96 |
| 55 | Lake (NE) | 1,479 | Verrucomicrobiae | Uncultured Crater Lake bacterium CL120-10 (AF316720) | 96 |
| 80 | Lake (NE) | 749 | Verrucomicrobiae | Uncultured bacterium FukuN18 (AJ289992) | 98 |
| 82 | Lake (NE) | 729 | Verrucomicrobiae | Uncultured bacterium FukuN18 (AJ289992) | 99 |
| 87 | Lake (NE) | 650 | Verrucomicrobiae | Uncultured bacterium FukuN18 (AJ289992) | 98 |
nt, nucleotides.
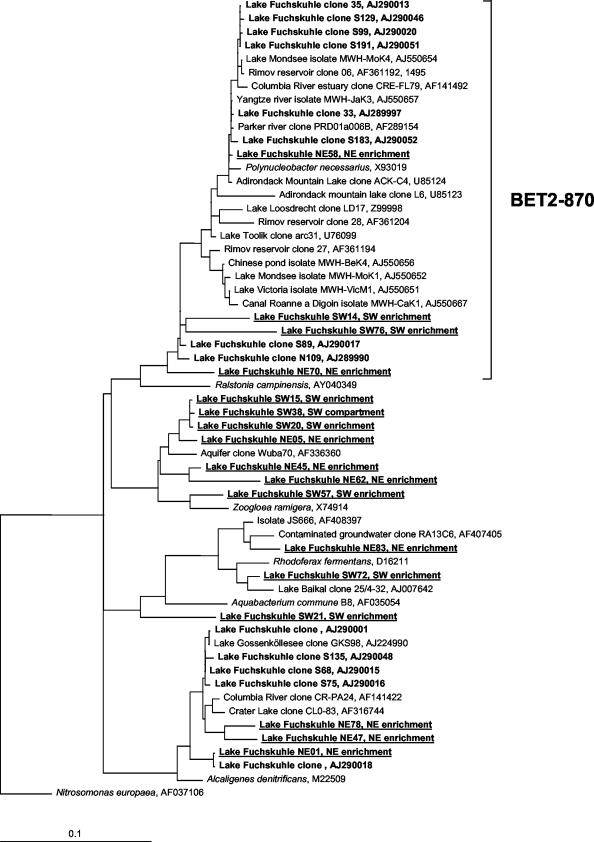
FIG. 2.
Phylogenetic relationship of β-proteobacterial 16S rRNA gene sequences from the study system and from enrichments of prefiltered (pore size, 0.1 μm) water from the SW compartment (1:10 dilutions). Sequences originating from Lake Groβe Fuchskuhle are depicted in boldface type, and sequences produced during this study are underlined. The scale bar indicates 10% estimated sequence divergence.
Two of the fully sequenced β-proteobacterial 16S rRNA sequence types were excluded as being chimeric. The remaining 16 sequences originated mainly from the enrichment libraries and fell into distinct clades of known freshwater bacteria (Fig. 2). Four sequences from the lake were affiliated with a cosmopolitan freshwater lineage (beta II) that includes the endosymbiont Polynucleobacter necessarius (13, 32, 56). Three sequences from the NE enrichment were related to the freshwater clone sequence GKS98, which was proposed to be a member of another freshwater clade (13, 56). Two sequences were affiliated with Rhodoferax sp. and thus with the so-called freshwater beta I/ACK2 lineage (13, 56), and one was affiliated with the drinking water biofilm isolate Aquabacterium commune. One set of sequences was affiliated with a phylotype obtained from a karstic aquifer in South Germany (GenBank accession number AF336360) within a broader lineage that also harbors Zoogloea ramigera.
Enrichment experiment.
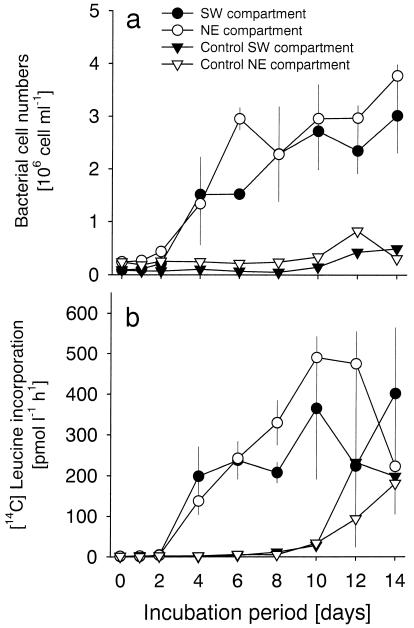
Bacterial total numbers in the enrichments increased from 0.1 × 106 to 3.0 ± 0.7 (mean ± standard deviation) × 106 cells ml−1 in the NE treatment, and from 0.24 × 106 to 3.8 ± 0.2 × 106 cells ml−1 in the SW treatment (Fig. 3a). A first doubling of bacterial abundances was observed after 48 h, and the most rapid growth occurred between days 2 and 4 in both sets of enrichments. This rapidity of growth was reflected in an approximately 40-fold increase in bacterial [3H]leucine incorporation between these sampling points (Fig. 3b). Bacterial leucine incorporation increased from 1 to 401 ± 162 and 491 ± 52 pmol liter−1 h−1 in the SW and NE compartments, respectively. The variability of leucine incorporation between the replicate incubations was high after day 8, which coincided with the appearance of low concentrations of heterotrophic flagellates in some of the replicates at this stage of the experiment (<50 flagellates ml−1) (data not shown). Bacterial abundances and productivity developed differently in the two treatments. In the SW treatment, a first plateau of total abundance on day 6 and of leucine incorporation on days 6 and 8 was discernible. In contrast, bacterial abundances and production continued to increase until days 8 and 10, respectively, in the NE treatment. No microbial growth was apparent in the control treatments until day 6 (Fig. 3a). Between days 8 and 14, bacterial abundances in these treatments increased to approximately 0.5 × 106 cells ml−1. This increase was paralleled by a steep rise in bacterial leucine incorporation (Fig. 3b).
FIG. 3.
Development of total bacterial abundances (a) and bacterial [3H]leucine incorporation (b) during the enrichment experiments. For the SW and NE treatments, inocula from the SW and NE compartments, respectively, were diluted 1:10 into prefiltered (pore size, 0.1 μm) water from the SW compartment.
The largest fraction of the cDOC in the lake water medium was formed by humic acids (66% ± 2%, mean ± range) (Table 2). This proportion did not change significantly during the experiment. All together, there were no significant decreases in the concentrations of the measured cDOC fractions in either treatment during the course of the incubations.
TABLE 2.
Fractions of DOC during incubations of microbial inocula from the two compartmentsa
| Inoculum | Day | Mean fraction (mg/liter of C) (SD)
|
||||
|---|---|---|---|---|---|---|
| cDOC | Humic acids | Polysaccharides | LMWA | Others | ||
| SW compartment | 0 | 13.1 (0.6) | 8.7 (0.1) | 0.5 (0.1) | ND (ND) | 3.9 (0.6) |
| 6 | 12.9 (0.5) | 9.0 (0.5) | 0.5 (0.1) | ND (ND) | 3.5 (0.0) | |
| 8 | 13.0 (0.2) | 9.0 (0.2) | 0.5 (0.0) | ND (ND) | 3.6 (0.1) | |
| 14 | 12.8 (0.1) | 8.9 (0.1) | 0.4 (0.1) | ND (ND) | 3.7 (0.0) | |
| Control | 1.5 (0.2) | 0.8 (0.1) | 0.1 (0.0) | ND (ND) | 0.5 (0.1) | |
| NE compartment | 0 | 12.5 (0.1) | 8.1 (0.1) | 0.6 (0.3) | 0.4 (0.4) | 3.3 (0.1) |
| 6 | 12.5 (0.4) | 8.4 (0.2) | 0.6 (0.3) | 0.1 (0.1) | 3.5 (0.2) | |
| 8 | 13.0 (0.7) | 8.8 (0.4) | 0.3 (0.1) | 0.1 (0.2) | 4.0 (0.7) | |
| 14 | 12.5 (0.1) | 8.6 (0.3) | 0.4 (0.2) | 0.1 (0.1) | 3.6 (0.1) | |
| Control | 1.3 (0.2) | 0.7 (0.1) | 0.1 (0.0) | ND (ND) | 0.5 (0.1) | |
Fractions are measured in prefiltered lake water from the SW compartment and in NaHCO3 solution (control) by size exclusion liquid chromatography and UV and organic carbon detection. LMWA, low-molecular-weight organic acids; ND, not detectable.
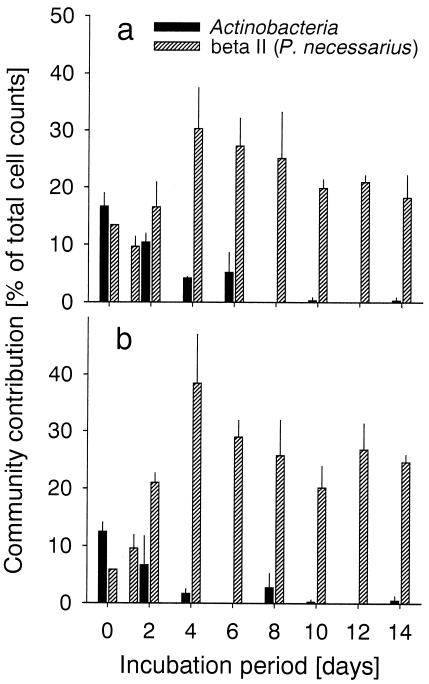
No significant differences were found between the total counts of β-proteobacteria in the inoculum from the SW compartment after FISH with a monolabeled probe and after CARD-FISH with an HRP-labeled probe (31% ± 3% and 33% ± 3% [means ± ranges], respectively). Several of the newly designed specific FISH probes detected bacterial populations that formed <1% of the total DAPI cell counts (data not shown). In contrast, high numbers of hybridized cells were found after FISH with a probe targeted to the beta II (P. necessarius) cluster of the β-proteobacteria (BET2-870; length, 18 nucleotides; E. coli positions 870 to 888 [5]; 5′-CCC AGG CGG CTG ACT TCA-3′; hybridized with 35% formamide in the hybridization buffer). Bacteria affiliated with the beta II clade initially constituted 13 and 6% of the microbial assemblages in the lake water inocula from the SW and NE compartments, respectively (Fig. 4). These rod-shaped bacteria, which are approximately 1 to 2 μm in length, overproportionally increased in numbers during the first days of incubation. On day 4 cells detected by probe BET2-870 formed 30% ± 7% and 38% ± 9% of all bacteria in the SW and NE treatments, respectively. Between days 6 and 14, the fraction of bacteria from the beta II clade gradually decreased in both treatments, to approximately 20% (SW) and 25% (NE).
FIG. 4.
Relative abundances of members of the class Actinobacteria and of the beta II clade (bacteria related to P. necessarius) during the enrichment on prefiltered (pore size, 0.1 μm) water from the SW compartment. (a and b) Inocula from the SW (a) and NE (b) compartments.
In samples from day 0, the Actinobacteria made up 17% ± 2% and 12% ± 2% of organisms in the SW and NE treatments, respectively (Fig. 4). Their relative abundances declined to <5 and 2%, respectively, within the first 4 days of incubation. During the second half of the experiment, cells from this lineage were frequently below our lower limit for reliable FISH counts (<1% of total counts) in individual samples.
Field data.
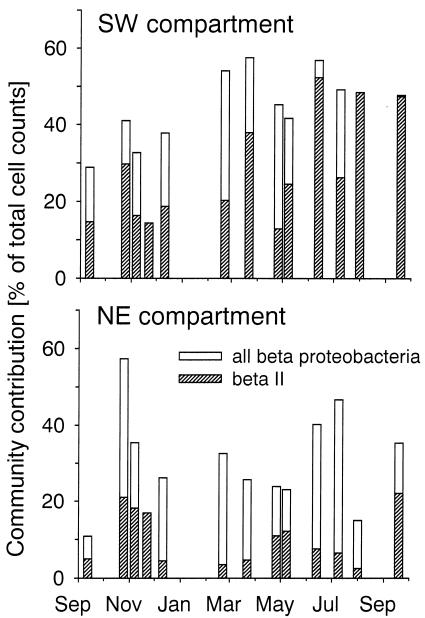
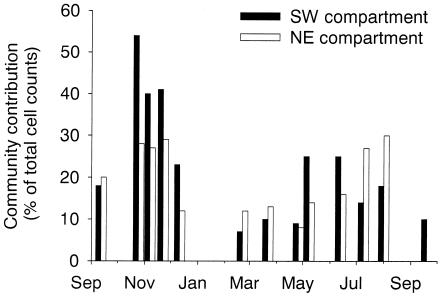
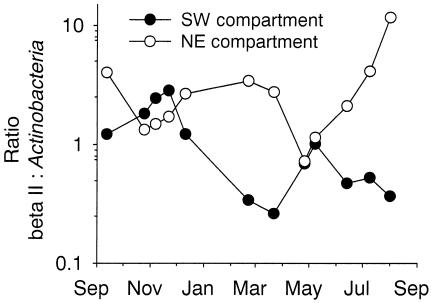
Between September 2000 and September 2001 bacterial total counts ranged between 0.8 × 106 and 3 × 106 cells ml−1 in the SW compartment of Lake Groβe Fuchskuhle, and from 1.5 × 106 to 4.3 ×106 cells ml−1 in the NE compartment (Table 3). Bacterial abundances were significantly higher in the NE compartment (P < 0.001), and distinct maxima were observed only in this compartment in September 2000 and 2001 and in February 2001. Annually, on average β-proteobacteria constituted 42% of all bacteria in the SW compartment and 29% in the NE compartment (Fig. 5). This difference was statistically significant (P < 0.01). Bacteria of the beta II lineage formed a significantly larger part of all β-proteobacteria (63%) and of the total microbial community (28%) in the SW compartment than in the NE compartment (33 and 10%, respectively) (Fig. 5). At three time points between July and September 2001, these bacteria made up >90% of the β-proteobacteria and approximately half of all bacteria in the SW compartment of Lake Groβe Fuchskuhle. Actinobacteria formed on average 23 and 20% of all cells in the SW and NE compartments, respectively, with no significant difference between the compartments (Fig. 6). Two distinct peaks in their relative community contributions could be distinguished in both compartments between November 2000 and January 2001 and from May to August 2001. The ratio between the community contributions of Actinobacteria and of members of the beta II lineage showed pronounced seasonal fluctuations (Fig. 7) in both compartments.
TABLE 3.
Cell numbers of bacteria in the SW and NE compartments of Lake Fuchskuhle between September 2000 and September 2001a
| Date of sampling | No. of cells ml−1 (106)
|
|||
|---|---|---|---|---|
| SW compartment
|
NE compartment
|
|||
| Mean | Range | Mean | Range | |
| 12 September 2000 | 3.0 | 0.2 | 4.1 | 0.8 |
| 25 October 2000 | 1.2 | <0.1 | 2.0 | 0.4 |
| 7 November 2000 | 1.5 | 0.1 | 2.5 | 0.1 |
| 22 November 2000 | 1.4 | 0.1 | 2.2 | 0.3 |
| 11 December 2000 | 1.3 | 0.3 | 2.0 | 0.4 |
| 21 February 2001 | 0.9 | 0.3 | 4.3 | 0.6 |
| 22 March 2001 | 0.9 | <0.1 | 2.3 | 0.3 |
| 26 April 2001 | 1.5 | 0.3 | 1.5 | 0.3 |
| 8 May 2001 | 1.6 | 0.3 | 1.8 | 0.7 |
| 13 June 2001 | 1.4 | <0.1 | 2.2 | 0.1 |
| 9 July 2001 | 0.8 | <0.1 | 2.4 | 0.4 |
| 1 August 2001 | 1.3 | 0.1 | 1.3 | <0.1 |
| 19 September 2001 | 2.1 | 0.2 | 3.3 | 0.3 |
Values are means and ranges of duplicate determinations.
FIG. 5.
Relative abundances of β-proteobacteria and of members of the beta II clade in surface waters of the SW and NE compartments of Lake Groβe Fuchskuhle between September 2000 and September 2001.
FIG. 6.
Relative abundances of Actinobacteria in surface waters of the SW and NE compartments of Lake Groβe Fuchskuhle between September 2000 and September 2001.
FIG. 7.
Ratios of members of the beta II clade to Actinobacteria in surface waters of the SW and NE compartments of Lake Groβe Fuchskuhle between September 2000 and September 2001.
DISCUSSION
Members of the beta II clade are rapidly enriched.
In incubations of water from Lake Groβe Fuchskuhle, microbes related to the freshwater beta II lineage were among the first bacteria that responded to the changed growth conditions (Fig. 4). In the dilution experiments on water from the SW basin, a first doubling of these bacteria was observed within 24 to 48 h, irrespective of the inoculum. Within 4 days of enrichment these bacteria increased in abundance by approximately 50-fold. P. necessarius (formerly known as omikron) is an obligatory symbiont of the hypotrichous freshwater ciliate Euplotes aediculatus (16). Its phylogenetic position within the β-proteobacteria has been demonstrated by 16S rRNA gene sequencing and subsequent FISH detection inside the host cells (52). Sequences affiliated with P. necessarius have been obtained from various freshwater systems, e.g., the acidified Adirondack mountain lakes, the oligotrophic Crater Lake, the Columbia River estuary, Antarctic Toolic Lake, and eutrophic Lake Loosdrecht (Fig. 2). Currently, no other bacteria from this lineage except for P. necessarius are validly described, but that group harbors culturable representatives from freshwater plankton (6, 15a).
Role of opportunistic water column bacteria.
The beta II clade represents a lineage of readily enriched pelagic bacteria that are important in the water column of Lake Groβe Fuchskuhle (Fig. 5). Members of this group seasonally constituted more than half of all bacterioplankton cells in the SW compartment. This suggests that the adaptation to rapid changes in substrate availability might be an important ecological feature of water column bacteria in this acidified bog lake. Bacteria related to beta II might, e.g., be specialized to respond to the sporadic high influx of allochthonous DOC from the catchment into the SW compartment of Lake Groβe Fuchskuhle (Fig. 1). Allochthonous carbon is a major substrate source for bacteria in humic lakes (54), and water influx has been proposed as an agent of bacterial community change in a boreal forest lake (28). Bottom-up factors have been shown to play a key role in controlling the composition of resource-limited microbial assemblages in other freshwater systems too (12, 50).
However, even though our data appear to support our hypothesis, it is too early for generalizations. First, it is currently unknown whether the high numbers of bacteria from the beta II clade are a typical feature of the studied humic lake or if these bacteria also form larger populations in other freshwaters with different limnological and hydrological characteristics. The answer cannot be deduced from isolates and the occurrence of 16S rRNA gene sequences from this clade in various other systems (13, 56) (Fig. 2), because there may be little relationship between the abundances of phylotypes or isolates in situ and their presence in clone libraries and culture collections (7, 8). Second, it is unclear whether the phylotypes of the beta II clade that were enriched during the incubation experiments were indeed identical with the dominant sequence types from this lineage that occur in the water column of Lake Groβe Fuchskuhle. The clade might harbor a high diversity of strains with contrasting ecological and ecophysiological features. For example, some members of the marine Roseobacter lineage from North Sea waters can be readily enriched and isolated, whereas the abundant Roseobacter phylotypes still resist cultivation (10).
Our results nevertheless point at a potential difference between the functioning of the bacterial assemblage in the studied lake and that of the microbial communities in other aquatic habitats, e.g., coastal North Sea waters. In the latter system, a 1:10 dilution with prefiltered ambient water and subsequent incubation typically leads to the rapid enrichment of culturable γ-proteobacteria related to the genera Vibrio and Pseudoalteromonas (8). These bacteria are often associated with biofilms, e.g., animal or plant surfaces (20, 34). They are apparently adapted to quickly respond to changes in environmental conditions (8, 11), and they are superior competitors when they are exposed to steep substrate gradients (39). Bacteria from such opportunistic genera are usually extremely rare in coastal North Sea plankton (9), likely because they are overproportionally reduced by size-selective protistan grazing (3). This rarity is in striking contrast to our present findings about the important role of the readily enriched beta II lineage in the surface waters of Lake Groβe Fuchskuhle (Fig. 5).
The relative abundances of bacteria related to the Actinobacteria clearly declined during the incubations (Fig. 4). This class of gram-positive bacteria with a high genomic G+C content are a ubiquitous component of the water column of many freshwater systems (13, 32, 56). High numbers of these bacteria were detected in lakes with different limnological characteristics only after substantial improvements of the original FISH technique (13, 47). Recently, members from one actinobacterial lineage have been isolated from various sites by Hahn and coworkers (15). In that study the actinobacterial cells had to be physically separated from other bacteria by filtration for successful cultivation. In addition, the inorganic media were gradually enriched with organic substrates over a period of several days in order to avoid substrate shock. These observations by Hahn et al. agree with our findings that Actinobacteria were outcompeted by more rapidly growing bacterial groups after sudden changes in substrate availability (Fig. 4).
DOC and bacterial production during the incubations.
Interestingly, no change in the total cDOC pool or in the fractions determined by size exclusion chromatography with UV and organic carbon detection could be observed during the incubation experiment (Table 2). If we apply the frequently used factor of Simon and Azam (51) for a conversion of [3H]leucine incorporation into bacterial carbon production, a total production of approximately 1 to 1.5 mg of carbon can be estimated for the duration of the incubations. This value represents 10% of the measured carbon pool. Such consumption should be reflected in changes of total cDOC (Table 2). Apparently, size exclusion chromatography was not sensitive enough for analyzing the potential changes in the cDOC fractions. Moreover, the above-noted conversion factor most likely under- rather than overestimated bacterial carbon production in the studied assemblage. During the first 6 days of incubation (i.e., before losses by heterotrophic flagellates need to be considered), the use of this factor results in estimates of per cell carbon bacterial content of only 5 to 10 fg. This is probably too low by a factor of 2 to 4 (43). Thus, either the enriched bacterial species exhibited unusually high growth efficiencies or these bacteria preferably utilized substrate sources other than the offered amino acid.
Field observations.
Due to the artificial division, Lake Groβe Fuchskuhle is a rather unusual system. During the summer months the lake forms a steep oxycline, and the two studied compartments differ in several fundamental parameters, e.g., water chemistry, primary productivity, the structure of the phytoplankton community, and the structure of the microbial food web (4, 17, 45, 49). In a previous study, the more acidified, humic-acid-richer SW compartment was inhabited by large cladocerans, which effectively suppressed protistan bacterial predators (49). Bacterial mortality in the more productive NE compartment was caused mainly by a small oligotrich ciliate, Cyclidium sp., which is rather untypical for freshwater systems. These differences between the compartments were again reflected in the contrasting annual developments of the microbial communities in the SW and NE compartments (Fig. 5 and 6). On an annual scale, total bacterial abundances were lower in surface waters of the SW compartment, yet the proportions of bacteria from the beta II clade were significantly higher (Fig. 5). The latter finding is consistent with the rapid enrichment on prefiltered water of beta II clade organisms that originated from this compartment (Fig. 4).
Actinobacteria clearly formed a second important component of the microbial assemblages in both compartments of Lake Groβe Fuchskuhle (Fig. 6). Sequence types related to this lineage have also been reported from several Scandinavian bog lakes (30). The ecological role of these bacteria in freshwaters is unknown. Some aquatic Actinobacteria are apparently better protected from mortality by protistan grazing than the community average. The enrichment of such bacteria was repeatedly observed in continuous culture after the addition of a mixotrophic flagellate, Ochromonas sp. strain DS (41). This flagellate was completely unable to consume a freshwater actinobacterial isolate in batch culture experiments (15). The phytoplankton communities of humic lakes often harbor a higher share of such mixotrophic flagellate species than other lakes, including Ochromonadaceae (22). On the other hand, actinobacterial strains from terrestrial habitats have been shown to produce peroxidases that are able to cleave complex organic compounds such as humic substances (31, 44). Actinobacteria in Lake Groβe Fuchskuhle might thus be involved in the turnover of the more recalcitrant DOC fraction.
At this stage little is known about the functional role of Actinobacteria or members of the beta II lineage in Lake Groβe Fuchskuhle apart from their contrasting behavior during enrichment. Pronounced phytoplankton blooms occurred in the two compartments during the study period, but there was no significant relationship of chlorophyll a and the changes in absolute and relative numbers of the studied groups (data not shown). Together, the two lineages represented half of the bacteria in the surface waters of the SW compartment and a third of the bacteria in the more mesotrophic NE compartment. In October 2000 and June 2001 they constituted approximately 80% of all cells in the SW compartment. The high fluctuations in the ratio between bacteria of the beta II clade and the Actinobacteria hint at remarkable differences in their respective roles during the seasonal development (Fig. 7). The fluctuations furthermore indicate that there were pronounced differences also between the SW and NE compartments. Interestingly, this parameter appeared to be almost mirrored in the two compartments. Primary producers, microzooplankton, DOC content, and pH have all been suggested to affect microbial community composition in acidified or humic-acid-rich lakes (29, 33). However, more-detailed experimental and comparative investigations and high-resolution time course studies are required to understand the factors that determine the occurrence and the population sizes of the two lineages in humic lakes.
Acknowledgments
We thank R. Amann and H.-P. Grossart for inspiring discussions and M. Degebrodt for measurements of leucine incorporation.
This work was supported by the Deutsche Forschungsgemeinschaft (Ba 1288/5-1-2), by the German Ministry of Education and Research (BIOLOG, BMBF 01 LC0021/TP4), and by the Max Planck Society.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babenzien, D., and C. Babenzien. 1990. Microbial activities in a naturally acidotrophic lake. Arch. Hydrobiol. Suppl. 34:175-181. [Google Scholar]
- 3.Beardsley, C., J. Pernthaler, W. Wosniok, and R. Amann. 2003. Are readily cultured bacteria in coastal North Sea waters suppressed by selective grazing mortality? Appl. Environ. Microbiol. 69:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bittl, T., and H.-D. Babenzien. 1996. Microbial activities in an artificially divided acidic lake. Arch. Hydrobiol. Adv. Limnol. 48:113-121. [Google Scholar]
- 5.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 6.Bruns, A., U. Nübel, H. Cypionka, and J. Overmann. 2003. Effect of signal compounds and incubation conditions on the culturability of freshwater bacterioplankton. Appl. Environ. Microbiol. 69:1980-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eilers, H., J. Pernthaler, and R. Amann. 2000. Succession of pelagic marine bacteria during enrichment: a close look on cultivation-induced shifts. Appl. Environ. Microbiol. 66:4634-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eilers, H., J. Pernthaler, J. Peplies, F. O. Glöckner, G. Gerdts, and R. Amann. 2001. Isolation of novel pelagic bacteria from the German Bight and their seasonal contribution to surface picoplankton. Appl. Environ. Microbiol. 67:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flärdh, K., P. S. Cohen, and S. Kjelleberg. 1992. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J. Bacteriol. 174:6780-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasol, J. M., M. Comerma, J. C. Garcia, J. Armengol, E. O. Casamayor, P. Kojecka, and K. Simek. 2002. A transplant experiment to identify the factors controlling bacterial abundance, activity, production, and community composition in a eutrophic canyon-shaped reservoir. Limnol. Oceanogr. 47:62-77. [Google Scholar]
- 13.Glöckner, F.-O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossart, H. P., and H. Ploug. 2001. Microbial degradation of organic carbon and nitrogen on diatom aggregates. Limnol. Oceanogr. 46:267-277. [Google Scholar]
- 15.Hahn, M. W., H. Lünsdorf, Q. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Hahn, M. W. 2003. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climate zones. Appl. Environ. Microbiol. 69:5248-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heckmann, K. 1975. Omikron, essential endosymbiont of Euplotes aediculatus. J. Protozool. 22:97-104. [Google Scholar]
- 17.Hehmann, A., L. Krienitz, and R. Koschel. 2001. Long-term phytoplankton changes in an artificially divided, top-down manipulated humic lake. Hydrobiologia 448:83-96. [Google Scholar]
- 18.Hemond, H. 1990. Wetlands as the source of dissolved organic carbon to surface waters, p. 301-313. In E. Perdue and E. Gjessing (ed.), Organic acids in aquatic ecosystems: report of the Dahlem workshop on organic acids in aquatic ecosystems. John Wiley & Sons, Chichester, United Kingdom.
- 19.Heyman, U. 1983. Relationship between production and biomass of phytoplankton in four Swedish lakes of different trophic status and humic content. Hybrobiologia 101:71-88. [Google Scholar]
- 20.Holmström, C., and S. Kjelleberg. 1999. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 21.Ilmavirta, V. 1988. Phytoflagellates and their ecology in Finnish brown-water lakes. Hydrobiologia 161:255-270. [Google Scholar]
- 22.Jansson, M., P. Blomqvist, A. Jonsson, and A.-K. Bergström. 1996. Nutrient limitation of bacterioplankton, autotrophic and mixotrophic phytoplankton, and heterotrophic nanoflagellates in Lake Örtäsket. Limnol. Oceanogr. 41:1552-1559. [Google Scholar]
- 23.Karner, M., and J. A. Fuhrman. 1997. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl. Environ. Microbiol. 63:1208-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasprzak, P. 1993. The use of an artificially divided bog lake in food web studies. Verh. Int. Verein. Limnol. 25:652-656. [Google Scholar]
- 25.Kirchman, D., E. Knees, and R. Hodson. 1985. Leucine incorporation and its potential as a measure of protein-synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirchman, D. L., L. Y. Yu, B. M. Fuchs, and R. Amann. 2001. Structure of bacterial communities in aquatic systems as revealed by filter PCR. Aquat. Microb. Ecol. 26: 13-22. [Google Scholar]
- 27.Koschel, R. 1995. Manipulation of whole lake ecosystems and long term limnological observations of the Brandenburg-Mecklenburg lake district. Int. Rev. Gesamten. Hydrobiol. 80:1-12. [Google Scholar]
- 28.Lindström, E. S. 1998. Bacterioplankton community composition in a boreal forest lake. FEMS Microbiol. Ecol. 27:163-174. [Google Scholar]
- 29.Lindström, E. S. 2000. Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb. Ecol. 40:104-113. [DOI] [PubMed] [Google Scholar]
- 30.Lindström, E. S., and E. Leskinen. 2002. Do neighboring lakes share common taxa of bacterioplankton? Comparison of 16S rDNA fingerprints and sequences from three geographic regions. Microb. Ecol. 44:1-9. [DOI] [PubMed] [Google Scholar]
- 31.Mercer, D., M. Iqbal, P. Miller, and A. McCarthy. 1996. Screening actinomycetes for extracellular peroxidase activity. Appl. Environ. Microbiol. 62:2186-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Methé, B. A., W. D. Hiorns, and J. P. Zehr. 1998. Contrasts between marine and freshwater bacterial community composition: analyses of communities in Lake George and six other Adirondack lakes. Limnol. Oceanogr. 43:368-374. [Google Scholar]
- 33.Methé, B. A., and J. P. Zehr. 1999. Diversity of bacterial communities in Adirondack lakes: do species assemblages reflect lake water chemistry? Hydrobiologia 401:77-96. [Google Scholar]
- 34.Montanari, M. P., C. Pruzzo, L. Pane, and R. R. Colwell. 1999. Vibrios associated with plankton in a coastal zone of the Adriatic Sea (Italy). FEMS Microbiol. Ecol. 29:241-247. [Google Scholar]
- 35.Mulholland, P., C. Dahm, M. David, D. Di Toro, T. Fisher, H. Hemond, I. Kögel-Knabner, M. Meybeck, J. Meyer, and J. Sedell. 1990. Group report: what are the temporal and spatial variations of organic acids at the ecosystem level?, p. 315-329. In E. Perdue and E. Gjessing (ed.), Organic acids in aquatic ecosystems: report of the Dahlem workshop on organic acids in aquatic ecosystems. John Wiley & Sons, Chichester, United Kingdom.
- 36.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationship of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 38.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pernthaler, A., J. Pernthaler, H. Eilers, and R. Amann. 2001. Growth patterns of two marine isolates: adaptations to substrate patchiness? Appl. Environ. Microbiol. 67:4077-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 41.Pernthaler, J., T. Posch, K. Simek, J. Vrba, A. Pernthaler, F. O. Glöckner, U. Nübel, R. Psenner, and R. Amann. 2001. Predator-specific enrichment of actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl. Environ. Microbiol. 67:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 43.Posch, T., M. Loferer-Krossbacher, G. Gao, A. Alfreider, J. Pernthaler, and R. Psenner. 2001. Precision of bacterioplankton biomass determination: a comparison of two fluorescent dyes, and of allometric and linear volume-to-carbon conversion factors. Aquat. Microb. Ecol. 25: 55-63. [Google Scholar]
- 44.Ramachandra, M., D. Crawford, and G. Hertel. 1988. Characterization of an extracellular lignin peroxidase of the lignocellulolytic actinomycete Streptomyces viridosporus. Appl. Environ. Microbiol. 54:3057-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sachse, A., D. Babenzien, G. Ginzel, J. Gelbrecht, and C. E. W. Steinberg. 2001. Characterization of dissolved organic carbon (DOC) in a dystrophic lake and an adjacent fen. Biogeochemistry 54:279-296. [Google Scholar]
- 46.Salonen, K., P. Kankaala, T. Tulonen, T. Hammar, M. James, T. R. Metsala, and L. Arvola. 1992. Planktonic food-chains of a highly humic lake. 2. A mesocosm experiment in summer during dominance of heterotrophic processes. Hydrobiologia 229:143-157. [Google Scholar]
- 47.Sekar, R., A. Pernthaler, J. Pernthaler, F. Warnecke, T. Posch, and R. Amann. 2003. An improved protocol for the quantification of freshwater actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherr, E. B., and B. F. Sherr. 1993. Protistan grazing rates via uptake of fluorescently labelled prey, p. 695-701. In P. Kemp, B. F. Sherr, E. B. Sherr, and J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis, Boca Raton, Fla.
- 49.Simek, K., D. Babenzien, T. Bittl, R. Koschel, M. Macek, J. Nedoma, and J. Vrba. 1998. Microbial food webs in an artificially divided acidic bog lake. Int. Rev. Hydrobiol. 83:3-18. [Google Scholar]
- 50.Simek, K., K. Hornak, M. Masin, U. Christaki, J. Nedoma, M. G. Weinbauer, and J. R. Dolan. 2003. Comparing the effects of resource enrichment and grazing on a bacterioplankton community of a meso-eutrophic reservoir. Aquat. Microb. Ecol. 31: 123-135. [Google Scholar]
- 51.Simon, M., and F. Azam. 1989. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51:201-213. [Google Scholar]
- 52.Springer, N., R. Amann, W. Ludwig, K. H. Schleifer, and H. Schmidt. 1996. Polynucleobacter necessarius, an obligate bacterial endosymbiont of the hypotrichous ciliate Euplotes aediculatus, is a member of the beta-subclass of Proteobacteria. FEMS Microbiol. Lett. 135:333-336. [DOI] [PubMed] [Google Scholar]
- 53.Thurman, E. 1985. Organic geochemistry of natural waters. Martinus Nijhoff Publishers, Dordrecht, The Netherlands.
- 54.Travnik, L. J. 1989. Bacterioplankton growth, grazing mortality, and quantitative relationship to primary production. J. Plankton Res. 11:985-1000. [Google Scholar]
- 55.Vähätalo, A. V., K. Salonen, U. Münster, M. Jarvinen, and R. G. Wetzel. 2003. Photochemical transformation of allochthonous organic matter provides bioavailable nutrients in a humic lake. Arch. Hydrobiol. 156:287-314. [Google Scholar]
- 56.Zwart, G., B. C. Crump, M. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28: 141-155. [Google Scholar]