Abstract
Pseudomonas stutzeri A15 is a nitrogen-fixing bacterium isolated from paddy rice. Strain A15 is able to colonize and infect rice roots. This strain may provide rice plants with fixed nitrogen and hence promote plant growth. In this article, we describe the use of dapB-based in vivo expression technology to identify P. stutzeri A15 genes that are specifically induced during colonization and infection (cii). We focused on the identification of P. stutzeri A15 genes that are switched on during rice root colonization and are switched off during free-living growth on synthetic medium. Several transcriptional fusions induced in the rice rhizosphere were isolated. Some of the corresponding genes are involved in the stress response, chemotaxis, metabolism, and global regulation, while others encode putative proteins with unknown functions or without significant homology to known proteins.
Rice (Oryza sativa L.) is the staple food of over 40% of the world's population. Considering the increase in the world's population and the limited possibility to expand the acreage under cultivation, increasing the yield of rice production is of great concern. Yields in systems with a low input of N fertilizer can be increased considerably by a higher level of fertilization. However, the environmental concerns raised against the extensive use of fertilizers necessitate the search for alternatives. One of the explored alternatives is biofertilization through the interaction between nitrogen-fixing plant-growth-promoting rhizobacteria and rice.
The diazotrophic strain A15 was isolated during surveys for nitrogen-fixing bacteria in the rhizosphere of paddy rice grown in China with suboptimal N fertilization (78). Yield increases of 3 to 7% have been reported for field-grown rice inoculated with strain A15 (77). Initially, this strain was phenotypically characterized as Alcaligenes faecalis, but the taxonomic position of strain A15 was reinvestigated, and it was reclassified as Pseudomonas stutzeri (69). It has been shown that the strain A15 nitrogen fixation genes (nif) are expressed in the rice rhizosphere (68). Since strain A15 is able to infect rice roots and survive within rice plants (24, 67, 79), it may provide rice plants with fixed nitrogen and hence promote plant growth. However, direct evidence for bacterial N transfer to the plants is still lacking.
At present, the mechanisms that enable strain A15 to colonize and infect rice roots and survive within rice plants are not known. The lack of a readily scored plant phenotype has hampered the identification and characterization of the P. stutzeri A15 genes that are required for interaction with the host plant. Because conditions during bacterium-host interactions are difficult to mimic in vitro, new techniques have been devised to study in vivo gene expression; these include differential fluorescence induction, signature-tagged mutagenesis, RNA arbitrarily primed PCR, and in vivo expression technology (IVET) (reviewed in references 8, 22, and 51). In this study, an IVET strategy was devised to enable the identification of genes specifically expressed during the interaction between P. stutzeri A15 and rice.
Briefly, IVET is a promoter-trapping technique that selects microbial promoters active in a specified niche, for instance, during the interaction of a microorganism with its host. Such promoters are identified by the ability to drive the expression of a promoterless selection marker gene in vivo (36), resulting in the complementation of a mutation in an essential gene and, consequently, in survival under the conditions encountered during the interaction. In contrast to traditional mutagenesis techniques, the advantage of the IVET strategy lies in the positive selection of genes that are specifically induced by environmental parameters. Furthermore, this technique does not disrupt genes that may be essential for survival in vivo.
IVET, originally developed to study Salmonella enterica serovar Typhimurium infection of animals (36), was adapted for use in the study of various other phylogenetically diverse pathogenic bacteria, namely, Vibrio cholerae (5), Pseudomonas aeruginosa (71), Yersinia enterocolitica (80), Staphylococcus aureus (35), Actinobacillus pleuropneumoniae (15), Listeria monocytogenes (16), Klebsiella pneumoniae (31), Porphyromonas gingivalis (74), and Shigella flexneri (1), as well as the pathogenic fungi Candida albicans (60) and Histoplasma capsulatum (54). IVET was also used to identify genes of Pseudomonas putida specifically expressed in vivo upon colonization by the plant-pathogenic fungus Phytophthora parasitica (33), plant-associated Pseudomonas fluorescens (49), legume symbiont Sinorhizobium meliloti (44), and the phytopathogen Pseudomonas syringae pv. tomato (2).
In this article, we describe the development and application of an IVET strategy based on a P. stutzeri A15 dapB mutant strain defective in the biosynthesis of peptidoglycan and lysine. Recently, a similar system was used to study sugar beet colonization by P. fluorescens (17). By applying this technique, we were able to identify several genes specifically induced during the colonization of rice by P. stutzeri A15.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. stutzeri was grown overnight at 37°C in Luria-Bertani (LB) medium under vigorous aeration or for 2 days at 30°C in minimal M9 medium (56) or MMAB medium, containing, per liter, 5 g of malic acid, 3 g of K2HPO4, 1 g of NaH2PO4, 1 g of NH4Cl, 0.3 g of MgSO4 · 2H2O, 0.15 g of KCl, 0.01 g of CaCl2, and 0.0025 g of FeSO4 · 7H2O at pH 7. Escherichia coli strains were grown at 37°C in LB medium. For mating experiments, the conjugation mixture was grown on D medium, containing, per liter, 8 g of nutrient broth and 15 g of agar. Antibiotics were added at the following concentrations when required: rifampin, 100 μg/ml; tetracycline, 10 μg/ml; kanamycin, 50 μg/ml; and ampicillin, 100 μg/ml. Diaminopimelate (DAP) and lysine were added at 100 μg/ml, unless indicated otherwise. Indicator plates for strains carrying gusA fusions contained 50 μg of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc)/ml.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristics or sequence | Reference or source |
|---|---|---|
| Strains | ||
| Pseudomonas stutzeri | ||
| A15 | Wild-type strain, isolated from rice rhizosphere soil | 78 |
| FAJ2050 | A15 ΔdapB::Kmr | This study |
| FAJ2062 | Spontaneous rifampin-resistant mutant of A15 | This study |
| Pseudomonas fluorescens SBW25ΔdapB | DAP- and lysine-requiring auxotroph (dapB gene deleted) | 17 |
| Escherichia coli | ||
| cc118λpir | Δ(lac-pro) argE(Am) recA56 nalA Rifr (λpir) | 23 |
| S17-1 | mob+tra+ | 57 |
| TOP10 | F−mcrA ΔlacX74 galK Δ(mrr-hsdRMS-mcrBC) galU deoR Δ(ara-leu)7697 endA1 φ80dlacZΔM15 recA1 araD139 rpsL (Strr) | Invitrogen |
| Plasmids | ||
| pCMPG6031 | Promoterless P. stutzeri A15 dapB gene in pGFPGUS | This study |
| pCR2.1-TOPO | Cloning vector; ColE1 replicon; Apr Kmr | Invitrogen |
| pFAJ2935 | 20-kb fragment carrying P. stutzeri A15 dapB inserted in pLAFR3 | This study |
| pFAJ2936 | 4.4-kb EcoRI fragment of pFAJ2935 in pUC18 | This study |
| pFAJ2937 | pFAJ2936 ΔdapB::Kmr | This study |
| pFAJ2938 | 5.7-kb EcoRI fragment of pFAJ2937 inserted in pSUP202; ΔdapB::Kmr | This study |
| pGFPGUS | pIVET vector with a promoterless gfp gene transcriptionally coupled to a promoterless uidA reporter gene; oriR6K origin of replication; Apr Tcr | Bertrand and Rainey (unpublished) |
| pSUP202 | Derived from pBR322; mob; Apr Cmr Tcr | 57 |
| pUC18 | Cloning vector; ColE1 replicon; Apr | 75 |
| pUC4K | pUC18 derivative containing a 1.2-kb Kmr gene | Amersham Biosciences |
| Primers | ||
| Pseu-419 | 5′-GGCGCNGCCGGSCGNATGGGS-3′ | |
| Pseu-420 | 5′-RTCGCCSAGSACGCGSGCSGC-3′ | |
| Pseu-458 | 5′-TCGGTCTGCGATTCCGACT-3′ | |
| Pseu-459 | 5′-TTGCCAATGATGTTACAGAT-3′ | |
| Pseu-518 | 5′-CACTAGTCGCCGTCGGTTTCCGGCTTGC-3′ | |
| Pseu-520 | 5′-GGAATTCCACACAGCCAGTGTGCGCAACG-3′ | |
| Pseu-526a | 5′-CAGGGTTATTGTCTCATGAGCG-3′ | |
| Pseu-647a | 5′-GCTCCAAGCTGCAAGCCGG-3′ |
Primer labeled with Cy5 (5-N,N′-diethyltetramethylindodicarbocyanine) for sequencing.
DNA manipulation and sequencing.
Standard techniques for subcloning and agarose gel electrophoresis were used as described by Sambrook and Russell (56). Plasmid DNA was isolated by using either a GFX microplasmid preparation kit (Amersham Biosciences) or a QIAprep spin minipreparation kit (Westburg). DNA restriction and modification enzymes were obtained from Roche and Westburg. DNA sequencing was done by the chain termination method with an ALFexpress2 automated sequencer (Amersham Biosciences).
A homology search and a sequence analysis were performed by using the Blast program at http://www.ncbi.nih.nlm.gov/blast.cgi and http://www.pseudomonas.com. The presence of possible stem-loop structures was investigated by using the mfold program (http://www.bioinfo.rpi.edu/applications/mfold/). The conservation of gene organization was analyzed by using the STRING database (70).
Northern and Southern hybridizations.
Hybridizations were performed by using a digoxigenin (DIG) detection kit (Roche) with the chemiluminescence substrate disodium 3-{4-methoxyspiro-[1,2-dioxetane-3,2′-(5′-chloro)tricyclo(3.3.1.13,7)decan]-4-yl}phenyl phosphate (Roche). For detection of the Kmr cassette, a PCR-generated DIG-labeled probe (0.6 kb) was obtained with primers Pseu-458 and Pseu-459 (Table 1). For detection of the dapB gene, a 1.3-kb BamHI/EclXI restriction fragment of pFAJ2936 was purified and DIG labeled with the Klenow polymerase. For Northern analysis, RNA of wild-type P. stutzeri A15 was isolated as described by Eggermont et al. (13). For detection of dapB mRNA, a DIG-labeled PCR fragment generated with primers Pseu-419 and Pseu-420 was used as a probe. Northern hybridizations were carried out at 50°C.
Construction of a P. stutzeri A15 dapB mutant.
Previously published dapB sequences were used to design a pair of degenerate primers (Pseu-419 and Pseu-420) for amplification of a 400-bp internal dapB fragment. These primers were used to isolate a dapB-containing cosmid clone (pFAJ2935) from an A15 genome library (H. Vermeiren, unpublished data). A dapB-hybridizing 4.4-kb EcoRI restriction fragment from pFAJ2935 was cloned into pUC18, generating pFAJ2936. Sequencing of both strands of the insert confirmed the presence of a dapB homologue. The predicted DapB protein displayed a high level of sequence identity with other Pseudomonas homologues, ranging from 77% for P. fluorescens to 87% for P. syringae.
The internal 0.5-kb EclXI fragment of pFAJ2936 was replaced with the kanamycin resistance cassette from pUC-4K (Kmr; 1.2 kb), cut with BamHI, and blunt ended with the Klenow DNA polymerase. Subsequently, the Kmr cassette was ligated in blunt-ended, EclXI-restricted pFAJ2936 to generate pFAJ2937. The resulting 5.7-kb EcoRI restriction fragment of pFAJ2937 was transferred to the unique EcoRI restriction site of pSUP202, yielding pFAJ2938. Finally, this construct was transferred to P. stutzeri A15 by triparental mating with pRK2013 as a helper plasmid. Clones in which the wild-type dapB gene was putatively replaced with the mutated dapB gene by double homologous recombination were selected on MMAB medium supplemented with kanamycin, DAP, and lysine. The correct insertion of the Kmr cassette was verified by colony PCR with primers Pseu-518 and Pseu-520, primers Pseu-518 and Pseu-459, and primers Pseu-520 and Pseu-458 and confirmed by Southern hybridizations.
Germination, inoculation, and cultivation of rice seedlings.
Rice seeds (O. sativa indica cv. IR42; International Rice Research Institute) were peeled and surface sterilized by consecutive immersions in 70% ethanol (1 min) and Domestos solution (45 min), consisting of 3% commercial bleach, 0.1% Na2CO3, 3% NaCl, and a few drops of 10% sodium dodecyl sulfate (SDS). The seeds were rinsed three times (15 min each time) with sterile distilled water and germinated on humid sterile filter paper at 30°C. Three-day-old seedlings with radicles of approximately 1 cm were planted in aseptic test tubes (20 by 200 mm) containing 50 ml of half-strength Yoshida nutrient solution (76). The seeds were supported by a layer of perlite floating just beneath the surface of the nutrient solution. The tubes were covered by placing other test tubes on top of them and then were placed in the growth chamber (constant temperature of 26°C, 12-h day, and 70% relative humidity). Bacterial inoculation was done on the next day. Overnight cultures of P. stutzeri A15 were harvested, washed with 10 mM MgSO4 solution, and resuspended to an optical density at 600 nm (OD600) of 0.2, corresponding to a cell density of approximately 108 CFU per ml. From this suspension, 1-ml portions were added to the nutrient solution of the rice seedlings.
Bacterial growth tests and competition experiments.
To monitor the growth characteristics of free-living cells, overnight precultures of the bacterial strains of interest were brought to equal cell densities (OD600) and subsequently diluted 6,000 times in the appropriate medium. The growth of these cultures at 30°C was monitored for 72 h by using a Bioscreen C apparatus (Labsystems) with a continuous shaking regimen. The OD600s of the liquid cultures were determined every 3 h and reported as the mean of five different measurements.
To investigate growth dynamics in the rice rhizosphere, rice seedlings were inoculated with a 1:1 ratio of strains FAJ2050 and A15. For selective plate counting, a spontaneous P. stutzeri A15 rifampin-resistant mutant (FAJ2062) was used instead of the wild type. This mutant was obtained by plating a stationary-phase culture on LB agar containing 150 μg of rifampin/ml. The growth of FAJ2062 was comparable to that of the wild type when grown in LB medium or in minimal MMAB medium or M9 medium without rifampin (data not shown). In media containing rifampin, the wild type did not grow, while the growth of FAJ2062 was not affected (data not shown). Cells were harvested from the rice rhizosphere of seven rice seedlings at 7, 10, 13, and 17 days after inoculation. Harvesting was done by placing the roots of inoculated rice plants in a 15-ml Falcon tube containing 5 ml of phosphate-buffered saline (PBS; 1.24 g of K2HPO4/liter, 0.39 g of KH2PO4/liter, 8.80 g of NaCl/liter [pH 7.2]) and glass beads. The tube was vortexed vigorously for 1 min, and CFU were determined by using a spiral plater (model D; Spiral Systems Inc.). Since bacterial populations approximate a log-normal distribution (34), values were log transformed before analysis.
Construction of a fusion library in pCMPG6031.
The IVET vector used in our study (pCMPG6031) was constructed by inserting a promoterless P. stutzeri A15 dapB gene into a unique SpeI site upstream of the gfp reporter gene in pGFPGUS (N. Bertrand and P. B. Rainey, unpublished data). The promoterless A15 dapB gene was amplified by PCR with the Pfx polymerase (Roche) and primers Pseu-518 and Pseu-520. Next, the PCR fragment was ligated in the blunt-ended SpeI restriction site of pGFPGUS.
Genomic DNA of P. stutzeri A15 was isolated by using a Puregene kit (Gentra Systems) and completely digested with BglII/BclI/BamHI or partially digested with Sau3AI. Restriction fragments were separated on an agarose gel, and fragments that were 1 to 5 kb long were purified by using a QIAquick gel extraction kit (Westburg). Subsequently, these fragments were inserted into the unique BglII-dephosphorylated restriction site of pCMPG6031. These ligation mixtures were used to transform E. coli cc118λpir with selection on LB medium with ampicillin and tetracycline. For each transformation, the randomness of the DNA inserts was verified by colony PCR with primers Pseu-518 and Pseu-520. Next, plasmids from approximately 100 E. coli clones pooled in LB medium were mobilized to FAJ2050 by triparental mating with helper plasmid pRK2013. Since pCMPG6031 is unable to replicate in FAJ2050, the pIVET fusion constructs must integrate into the genome by single homologous recombination at sites homologous to the cloned genomic fragment. Transconjugants were selected for growth on MMAB medium containing kanamycin, tetracycline, DAP, and lysine. From each E. coli pool, approximately 200 recombinant FAJ2050 clones were picked.
Isolation of genes induced in vivo.
Approximately 100 FAJ2050 clones, containing an integrated pCMPG6031 transcriptional fusion, were pooled and grown overnight in LB medium containing DAP, lysine, tetracycline, and kanamycin. Cells were washed once with PBS, after which three rice seedlings were inoculated. After 2 to 3 weeks, bacteria were reisolated from rice roots as described above and plated on MMAB medium with the addition of kanamycin, tetracycline, DAP, lysine, and X-Gluc. In total, 23 pools were tested this way.
Histochemical analysis of rice root colonization.
For histochemical analysis, rice roots were washed in PBS and transferred to 0.1 M phosphate buffer (pH 7), containing 0.5 mg of X-Gluc/ml, 0.5 mM K3Fe(CN)6 and 0.5 mM K4Fe(CN)6 as oxidation catalysts, and 10 mM EDTA to mitigate the partial inhibition of the enzyme by the oxidation catalysts. Roots were stained overnight at 37°C in this buffer, rinsed with PBS, and analyzed microscopically with Nikon Optiphot-2.
Nucleotide sequence accession numbers.
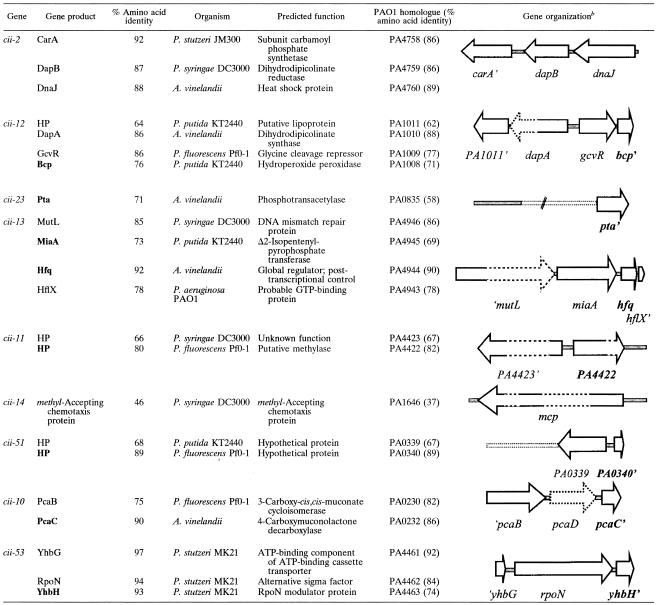
The nucleotide sequence determined here for the pFAJ2936 insert, containing dnaK (partial), dnaJ, dapB, and carA (partial), has been deposited in the GenBank database under accession number AY344804.
RESULTS
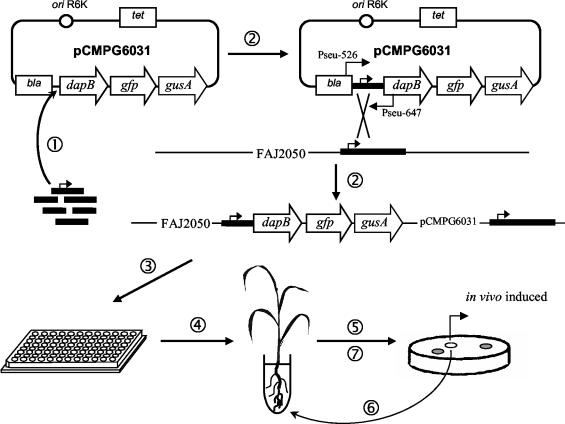
In this study, we adapted IVET to study the interaction between P. stutzeri A15 and rice. As outlined in Fig. 1, this adaptation of IVET to our system requires a mutant strain with a knocked out dapB gene and an IVET vector (pIVET) carrying a promoterless copy of the wild-type dapB gene transcriptionally fused to a promoterless reporter gene.
FIG. 1.
Selection for genes showing rhizosphere-specific induction. The strategy is based upon random insertion into the IVET vector (pCMPG6031) of genomic DNA fragments in front of a promoterless dapB gene (step 1), which is integrated into the chromosome of the P. stutzeri A15 dapB mutant (FAJ2050) (step 2). Pools of recombinant strains (step 3) are used to inoculate rice seedlings (step 4). Only in strains that carry a promoter active under these conditions can the dapB mutation be complemented. After 2 to 3 weeks, bacteria are reisolated and spread on X-Gluc-containing minimal medium to distinguish constitutive promoters from specifically in vivo-induced promoters (step 5). White colonies bearing a putative cii fusion are subjected to a second IVET screening to eliminate false-positive results (steps 6 and 7).
Isolation of the P. stutzeri A15 dapB gene.
The dapB gene, encoding l-2,3-dihydrodipicolinate reductase, is required for the biosynthesis of meso-DAP (3). DAP is an essential component of peptidoglycan and also serves as a direct precursor of lysine. A dapB-containing cosmid clone (pFAJ2935) was isolated from a P. stutzeri A15 genomic library. The presence of a functional dapB gene in pFAJ2935 was demonstrated by complementation of a P. fluorescens SBW25 dapB mutant (17).
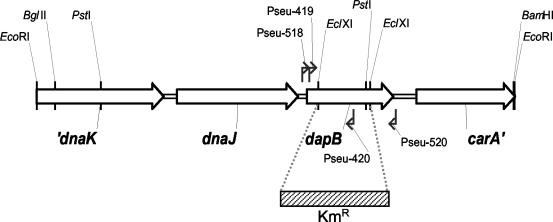
A 4.4-kb EcoRI restriction fragment containing dapB was subcloned, yielding pFAJ2936, and sequence analysis revealed four open reading frames (ORFs), including dapB (Fig. 2). The region upstream of dapB contains two ORFs (′dnaK and dnaJ) encoding heat shock proteins. Downstream of dapB, a partial ORF (carA′) was found. The carA gene encodes the small subunit of carbamoylphosphate synthetase. This enzyme provides carbamoylphosphate, which is required for the de novo synthesis of arginine and pyrimidine nucleotides. Inspection of finished (Microbial Genome DataBase) (65) and unfinished (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/bact.html) microbial genome sequences revealed that the gene organization dnaK-dnaJ-dapB-carA is conserved among Pseudomonas species and some other γ-proteobacteria. The carAB operon (encoding subunits of carbamoylphosphate synthetase) is located downstream of dapB in nearly all sequenced γ-proteobacterial genomes.
FIG. 2.
Physical and genetic maps of the dapB region in P. stutzeri A15. Primer-binding sites and relevant restriction sites are indicated. FAJ2050 was constructed by replacement of the EclXI restriction fragment with the Kmr cassette.
In silico analysis revealed stem-loop structures that may function as transcriptional terminators 18 bp downstream of the dnaJ stop codon (ΔG, −112.1 kJ/mol) and 36 bp downstream of the dapB stop codon (ΔG, −98.3 kJ/mol). Northern analysis with an internal dapB DNA fragment as a probe resulted in a signal of approximately 940 nucleotides (data not shown), which led to the conclusion that dapB in P. stutzeri A15 does not form a transcriptional unit with its flanking genes. These results are in line with those of the transcriptional analysis of the carAB operon in P. aeruginosa, which revealed no transcriptional linkage with the upstream dapB gene (30).
Construction and characterization of a P. stutzeri A15 dapB mutant.
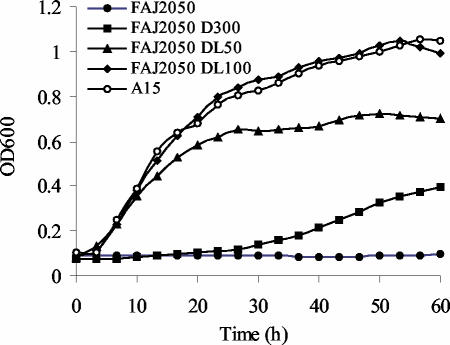
A P. stutzeri A15 dapB mutant (FAJ2050) was constructed by replacement of an internal fragment with a Kmr cassette (Fig. 2). Because the growth of FAJ2050 must be abolished in the absence of DAP to make it useful in an IVET screening, the growth characteristics of the mutant were analyzed under different conditions. In rich media (LB and D), the mutant showed a prolonged lag phase of about 15 h (data not shown). In minimal MMAB medium, the growth of FAJ2050 was completely abolished. The addition of DAP only partially restored the growth of the mutant. Even at relatively high DAP concentrations (300 μg/ml), the mutant showed a prolonged lag phase and a reduced growth rate (Fig. 3). When both DAP and lysine were added to the medium at 50 μg/ml, growth was restored, although a slightly lower growth rate and a lower final cell density compared to those of the wild type were still observed. Only when DAP and lysine were added at a sufficiently high concentration (100 μg/ml) was the growth of the mutant comparable to wild-type growth (Fig. 3). We speculate that the transport of DAP across the cytoplasmic membrane is inefficient, while lysine, for which DAP is the precursor, is readily taken up by the cells. These data are consistent with observations made with P. fluorescens SBW25, where relatively large amounts of DAP and lysine were necessary to complement a dapB mutation (17). When the medium was supplemented with higher concentrations of DAP and lysine (300 or 500 μg/ml), a lower growth rate, a longer lag phase, and a lower final cell density were observed for FAJ2050 (data not shown). The insertion of the Kmr cassette did not result in a polar effect on carAB, as no arginine or pyrimidine auxotrophy due to impaired synthesis of carbamoylphosphate was observed. This result is consistent with the transcription of dapB as a single gene, as observed by Northern analysis.
FIG. 3.
Growth characteristics of the dapB mutant (FAJ2050) in minimal MMAB medium compared to those of the wild type. DAP and lysine were each added at 50 μg/ml (DL50) and 100 μg/ml (DL100). Growth was also investigated when only DAP was added at 300 μg/ml (D300). Data points represent the means of five experiments.
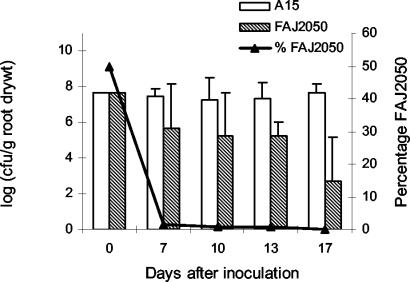
To examine the stringency of in situ selection, we verified that the dapB mutation could not be extracellularly complemented in the rhizosphere by plant exudates or the colonizing wild-type strain. This notion was assessed in a competition experiment by inoculating rice plants with a 1:1 mixture of a spontaneous rifampin-resistant mutant of P. stutzeri A15 (FAJ2062) and the P. stutzeri A15 dapB mutant (FAJ2050). Figure 4 shows clearly that FAJ2050 is severely impaired in root colonization. After 10 and 17 days, the proportions of the mutants relative to the total bacterial population dropped below 1 and 0.001%, respectively. Similar results were obtained with a P. fluorescens SBW25 dapB mutant (17). The large error bars for the mutant were due to the fact that in some replicates, the mutant became extinct.
FIG. 4.
Competitive colonization of the rice rhizosphere by P. stutzeri A15 (white bars) and the dapB mutant (FAJ2050) (hatched bars). The proportion of FAJ2050 relative to the total bacterial population is indicated. The experiment was carried out seven times. The error bars represent standard deviations.
Construction of a P. stutzeri A15 fusion library in pCMPG6031.
The IVET vector used in our study (pCMPG6031) was constructed by starting from pGFPGUS as described in Materials and Methods. This vector contains a promoterless P. stutzeri A15 dapB gene which is transcriptionally fused to promoterless gfp and gusA reporter genes (Fig. 1). Upstream of dapB, a multiple cloning site is present. Furthermore, the IVET vector carries two antibiotic resistance genes (bla and tet, for ampicllin and tetracycline resistance, respectively) and an oriR6K origin of replication for stable maintenance in λpir strains (23). A fusion library was constructed by inserting genomic P. stutzeri A15 DNA fragments (∼1 to 5 kb long) into the unique BglII restriction site of pCMPG6031. Subsequently, this fusion library was transferred to FAJ2050. Since pCMPG6031 is unable to replicate in FAJ2050, the pIVET fusion constructs integrate, upon selection for tetracycline, into the genome at sites homologous to the cloned genomic fragment.
Isolation of P. stutzeri A15 genes induced in the rice rhizosphere.
The fusion library pools were used to inoculate rice seedlings (100 recombinants/plant). After 2 to 3 weeks, bacteria were reisolated from rice plants and spread on MMAB medium (in vitro conditions) containing (i) DAP and lysine to complement the metabolic deficiencies of the mutant, (ii) kanamycin and tetracycline for the selection of dapB mutant strains carrying a pCMPG6031 fusion, and (iii) X-Gluc to assess β-glucuronidase activity in vitro. The majority of inoculated strains are expected not to multiply in the rice rhizosphere (in vivo conditions), since they lack a functional promoter in front of the dapB gene. In principle, only clones carrying a promoter that can drive dapB expression during the interaction with the rice plant can survive the in vivo screening. To eliminate fusions with a constitutive promoter, recovered bacteria were plated on X-Gluc-containing minimal medium to screen for β-glucuronidase activity. White colonies lacking in vitro gusA expression were subjected to a second IVET selection round to eliminate possible false-positive results. To this end, rice seedlings were inoculated with individual selected clones. The corresponding pCMPG6031 fusions were considered probable cii fusions when (i) the number of subsequently recovered cells for the possible positive IVET clone was comparable (>106 CFU/mg of root dry weight) to the number of wild-type cells reisolated from the inoculated plants; (ii) the in vivo activity of the cii promoters was confirmed by histochemical analysis of the inoculated rice roots; and (iii) the switching off of the promoter under in vitro conditions was verified by the inability to grow on MMAB medium. Subsequently, the cii fusions were recovered by conjugative cloning (50). Following transfer of the fusions into E. coli, plasmid DNA was isolated, and both ends of the insert DNA were sequenced partially with forward (Pseu-526) and reverse (Pseu-647) primers (Fig. 1). With these primers, the distal (5′) and proximal (3′) ends of the insert, relative to the promoterless dapB gene, were sequenced unidirectionally. For some clones, additional sequence information was obtained from subclones of the insert. Homology searches were conducted to identify ORFs and possible regulatory regions.
Identification of P. stutzeri A15 genes induced in the rice rhizosphere.
The present IVET system should enable the isolation of cii fusions activated during the P. stutzeri A15-rice interaction. In this study, we focused on bacterial genes that are specifically induced during the colonization of plant roots, exemplified here with rice. In the initial screening of approximately 2,300 clones, 9 cii fusions were isolated and further characterized (Table 2).
TABLE 2.
P. stutzeri A15 genes expressed specifically in the rice rhizospherea
Predicted cii genes and gene products are indicated in bold type. HP, hypothetical protein.
Broken lines indicate fragments that were not sequenced.
(i) cii-2.
Sequence analysis of transcriptional fusion cii-2 revealed the presence of a genomic dapB region (see construction of the dapB mutant) containing the carA′, dapB, and dnaJ genes orientated opposite the promoterless dapB gene of pCMPG6031. The fact that this fragment, containing the dapB region, was isolated validates the IVET approach used here, that is, relying on the expression of the insert-borne functional dapB gene.
(ii) cii-12.
The cii-12 fusion was sequenced only partially (Table 2). The 3′ end of the insert contained a bcp homologue, encoding a putative bacterioferritin comigratory protein (Bcp), with the same upstream gene organization as in P. aeruginosa PAO1 (PA1011-dapA-gcvR-bcp). gcvR and dapA, respectively, encode the glycine cleavage repressor and dihydrodipicolinate synthase. The latter enzyme catalyzes the first step in DAP biosynthesis. It remains to be analyzed which promoter(s) in cii-12 drives dapB expression in vivo. Studies with E. coli have demonstrated that bcp and gcvR do not form an operon (19), so that the bcp promoter likely is responsible for dapB transcription. The E. coli Bcp enzyme reduces hydroperoxides by using thioredoxin as a reducing agent (25). This peroxiredoxin is classified as a member of the thiol-specific antioxidant protein/alkyl hydroperoxide peroxidase family (TSA/AhpC) (73).
(iii) cii-23.
The 3′ end of the cii-23 fusion encodes a possible phosphotransacetylase (Pta), while no significant homology could be found for the 5′ region. Pta catalyzes the reversible conversion of acetyl phosphate to acetyl coenzyme A. Another key enzyme of acetate metabolism is acetate kinase, which produces acetyl phosphate from acetate. The coding region of this enzyme (ack) often is found immediately upstream of pta, e.g., in several γ-proteobacteria (including P. aeruginosa PAO1), or immediately downstream of pta. In most bacteria, the expression of ack and pta is directed from the same promoter (reference 52 and references therein). In contrast to the presence of ack in P. aeruginosa PAO1, no ack homologue is found in P. putida KT2440, P. fluorescens Pf0-1, P. syringae pv. tomato DC3000, or the closely related Azotobacter vinelandii. Apparently, pta is not clustered with ack (if at all present) in P. stutzeri A15.
(iv) cii-13.
The 5′ end of fusion cii-13 contains a partial ORF (′mutL) encoding a DNA mismatch repair protein. The 3′ DNA sequence contained the N-terminal coding region (80 bp) of HflX, a putative GTPase of unknown function. In addition, miaA and hfq homologues were identified. MiaA is a Δ2-isopentenylpyrophosphate transferase that catalyzes isopentenyl modification of some tRNAs. Such base modifications contribute to the efficiency and accuracy of translation (66). The hfq gene encodes so-called host factor I (HF-I, Hfq). In E. coli, Hfq is necessary for the efficient translation of rpoS (stationary-phase sigma factor) mRNA, affects the in vivo stability of several other mRNAs (including those of miaA and hfq), and also is necessary for the regulation by untranslated RNA (such as DsrA) of two global transcription regulators (RpoS and H-NS) (59, 63). Therefore, disruption of the hfq gene results in pleiotropic phenotypes, including decreased growth rate, increased cell size, osmosensitivity, increased oxidation of carbon sources, and increased sensitivity to UV (64).
The gene order mutL-miaA-hfq-hflX of P. stutzeri is found in various other γ-proteobacteria. Actually, in E. coli this region is part of a superoperon (amiB-mutL-miaA-hfq-hflX-hflK-hflC) controlling diverse cellular functions. Regulation of this superoperon involves multiple Eσ70- and Eσ32-dependent promoters, located upstream of mutL, miaA, and hfq (63). By analogy, it is likely that this P. stutzeri fusion contains multiple promoters and that in vivo expression of the dapB gene was driven by an internal miaA promoter, hfq promoter, or both.
(v) cii-11.
Fusion cii-11 contains coding regions for homologues of the hypothetical P. aeruginosa PAO1 proteins PA4422 and PA4423 (partial). PA4422 belongs to a family of tetrapyrrole methylases but has no functionally characterized homologue. The PA4422 coding sequence is followed by a noncoding DNA sequence of 300 bp. This region is strongly conserved (96% nucleotide identity) among several Pseudomonas species and precedes a large conserved operon involved in cell division and cell envelope biosynthesis (39). Such an operon is expected to be expressed in vitro; because the 300-bp region does not contain an active promoter, the PA4422 promoter would be active in vivo.
(vi) cii-14.
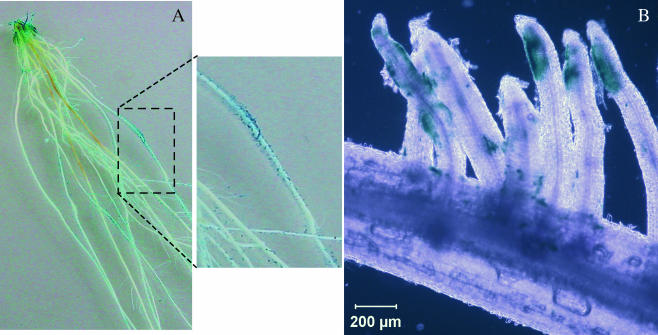
Fusion cii-14 contains a single ORF encoding a putative chemotaxis transducer protein and orientated in the direction opposite that of dapB. The remaining 200-bp DNA region should contain the active promoter. Glucuronidase-based histochemical staining of rice roots colonized by FAJ2050 carrying gusA fusion cii-14 revealed an interesting phenotype (Fig. 5). Mainly the colonized root hairs showed blue spots, while noninoculated roots remained white. Roots inoculated with other IVET-positive clones were stained along their entirety (data not shown). These results indicate the presence of a promoter driving root hair-specific gene expression.
FIG. 5.
Histochemical staining of rice roots colonized by FAJ2050 carrying transcriptional fusion cii-4. The cii promoter drives β-glucuronidase expression, which results in blue staining. (A) An image of a whole root system suggests a specific colonization pattern. (B) A microscopic image of a root with root hairs shows that mostly the tips of root hairs are colonized.
(vii) cii-51.
The 3′-end DNA sequence of transcriptional fusion cii-51 revealed the presence of two divergent ORFs encoding homologues of the P. aeruginosa conserved hypothetical proteins PA0339 and PA0340. The product of the properly oriented PA0340 sequence is a putative membrane protein. The intergenic region of only 10 bp between the PA0340 gene and the downstream lgt gene (PA0341) in P. aeruginosa PAO1 suggests that these two genes may be cotranscribed from the PA0340 promoter. PA0341 is a putative prolipoprotein diacylglyceryl transferase involved in the posttranslational lipid modification of proteins (48).
(viii) cii-10.
ORFs identified in fusion cii-10 showed the highest similarity to pcaB and pcaC, which are involved in the degradation of aromatic compounds through the protocatechuate pathway. PcaC is a γ-carboxymuconolactone decarboxylase, while pcaB encodes a 3-carboxy-cis, cis-muconate cycloisomerase. In several other pseudomonads, pcaBDC genes are clustered (26). Therefore, it is likely that a third ORF, pcaD, which encodes a β-ketoadipate enol-lactone hydrolase, is present in the cii-10 fusion. Several bacteria are able to degrade aromatic compounds through the β-ketoadipate pathway (ortho-cleavage pathway), which consists of two branches, i.e., the protocatechuate branch (encoded by pca genes) and the catechol branch (encoded by cat genes) (26). The sequential enzymatic activity of pca gene products results in the cleavage of protocatechuate to intermediates of the tricarboxylic acid cycle.
(ix) cii-53.
The cii-53 fusion carries three ORFs, designated yhbG, rpoN, and yhbH based on the nomenclature of the E. coli homologues. The yhbG gene encodes a putative ATP-binding subunit of an ATP-binding cassette transporter, while the rpoN gene encodes the σ54 sigma factor (40, 53). YhbH was originally designated the RpoN modulator protein (41). In several α-, β-, and γ-proteobacteria, an rpoN-yhbH-ptsN gene order has been observed, with ptsN encoding a component of a phosphotransferase system, indicating a high degree of synteny. However, yhbH homologues are also found in microorganisms without rpoN, suggesting that YhbH fulfills a more general regulatory role, beyond the regulation of nitrogen metabolism. In Bacillus subtilis, yhbH is induced during phosphate deprivation (47), and Bacillus cereus YhbH is expressed during the transition to the biofilm mode of growth (45). Maki et al. (37) demonstrated that YhbH accumulates in the stationary phase in E. coli and binds to ribosomes, enabling them to be stored in an inactive form. Upon transfer to fresh medium, most of the YhbH molecules are released.
There are indications that the expression of yhbH is directed from a promoter different from the rpoN promoter. Based on complementation analysis with P. aeruginosa, it was postulated that yhbH is cotranscribed with downstream ptsN (27). For E. coli, it was shown that yhbH was induced after the addition of autoinducer AI-2, in contrast to rpoN, which was downregulated under the same conditions (11).
DISCUSSION
In this article, we describe the use of IVET to study the interaction between plant growth-promoting nitrogen-fixing P. stutzeri A15 and rice. IVET has many attractive features, as outlined in the introduction, but some possible drawbacks have to be considered in the interpretation of the resulting data. First, with IVET it is not possible to isolate host-repressed promoters. Second, the subset of genes that are identified depends on the strength of in situ selection. If the selection is too strong, weakly or transiently expressed promoters will not be identified. This can be circumvented by using the recombinase-based IVET (RIVET) technique (58). On the other hand, in situ selection that is too weak will lead to false-positive results. In our experimental system, the strength of the in situ selection can easily be adjusted by changing the time of reisolation. Rainey and Preston (51) argued that the selection of promoters using dapB auxotrophy is not too stringent because of the fact that such mutation is lethal for actively growing cells, whereas nongrowing cells remain viable for long periods. However, if the cells are harvested for periods beyond that viability period, highly stringent conditions are generated and the number of identified nonconstitutive genes is reduced. Recently, the successful application of an analogous dapB-based system was demonstrated in the study of sugar beet colonization by P. fluorescens SBW25 (17). We devised an analogous dapB-based IVET strategy to isolate P. stutzeri A15 genes that are induced during colonization and infection (cii genes) of rice roots. The prerequisite to apply such IVET system is that the mutation cannot be complemented by metabolites from the plant or associated microorganisms. The probability that a dapB mutation would be complemented by DAP of plant origin is very low, due to the strict regulation mechanisms in the biosynthetic pathway of lysine (6, 18). Therefore, the dapB-based IVET system should also be suitable for isolating genes important for infection of rice roots and survival within the plant.
A first screening of the fusion library enabled us to identify several transcriptional cii fusions that are specifically induced in the rice rhizosphere and are therefore of possible importance for root colonization by P. stutzeri. The proportion of IVET-positive clones (0.4%) is comparable to those found in other IVET screenings, but obviously more clones need to be screened to obtain a more comprehensive picture of specifically in vivo induced P. stutzeri A15 genes. The majority of the cii fusions revealed extensive homology and synteny with P. aeruginosa genes (61).
When bacteria colonize plant roots, they need to optimize their gene expression to suit this particular environment. In this study, transcriptional fusions that carried genes encoding proteins with possible roles in metabolism, chemotaxis, stress response, and adaptation to the rhizosphere environment were isolated. Although for the sugar beet colonizer P. fluorescens SBW25, genes of the same classes were isolated (49), different genes were identified here. These data illustrate the complexity of the complement of rhizosphere-inducible genes.
It is known that plant cells induce a series of defense responses against pathogens, including the generation of reactive oxygen species such as O2− and H2O2 (32). The induction of bcp expression during colonization of rice roots by P. stutzeri A15 may represent a response to this oxidative stress in the rhizosphere. Among other stress-related proteins, a Bcp protein was induced in Frankia by root exudates of its symbiotic host, Alnus glutinosa (21).
The YhbH, Hfq, and MiaA proteins may be involved in adaptation to rhizosphere conditions similar to those faced during stationary growth in culture media. Under starvation for a limiting nutrient or exposure to extreme environmental conditions, bacteria shift from balanced growth (exponential growth phase) into another physiological state (stationary growth phase), which often coincides with an increased resistance to environmental stress, such as nutritional deprivation, oxidative stress, and low pH.
Hfq is necessary for the efficient translation of the alternative sigma factor RpoS, which controls the expression of several genes in the stationary phase. However, Hfq probably has a more general regulatory role, because it can affect the stability of mRNAs which are involved in DNA damage repair (mutS) or modification of the outer membrane (ompA). It has also been demonstrated that Hfq plays an important role in the interaction of certain pathogenic bacteria with their hosts. The Hfq homologue of Yersinia enterocolitica positively regulates the expression of an enterotoxin gene (yst), being involved in the regulation of expression of virulence factors (43). It has also been demonstrated that an hfq homologue (brg) in phytopathogen Erwinia carotovora is necessary for the synthesis of low-molecular-weight bacteriocins (9), again suggesting an important role of hfq for bacterial survival and enhanced competitiveness in the rhizosphere. Hfq is also of importance in the persistence of spleen infection by the animal pathogen Brucella. Brucella mutants that lack the hfq gene are more sensitive to acidic conditions and oxidative stress (55).
The E. coli miaA and hfq genes are part of a superoperon with multiple heat shock-dependent promoters (63). By modifying tRNAs, the isopentenyl transferase MiaA plays a role in improving reading frame maintenance by the protein translation machinery (66). Inefficient translation of the virulence-related regulator VirF was observed in the absence of MiaA in S. flexneri (12) showing that MiaA can play a role during the interaction of a bacterial pathogen with its host. In addition to its involvement in maintaining efficient protein translation under stress conditions, MiaA is involved in the low-level production of the cytokinin phytohormone trans-zeatin, which is frequently found in nonpathogenic plant-associated bacteria. In plant leaf surface-colonizing Methylobacterium spp., zeatin was found to originate from the turnover of isopentenylated tRNA rather than from de novo synthesis (29).
The ability to degrade and assimilate aromatic compounds present in rice exudates confers a selective advantage in a rhizosphere environment. In addition, these aromatic compounds are often toxic and can induce a stress response in certain bacteria, e.g., E. coli (42). The specific expression of a P. stutzeri A15 pca gene cluster in rice rhizosphere may add to the rhizosphere fitness of the strain by assimilation and detoxification of aromatic compounds (26).
Our study also suggests a possible role for pta-encoded phosphotransacetylase activity during rice root colonization by P. stutzeri. Pta is a key enzyme in the metabolism of acetyl phosphate. Different functions can be assigned to acetyl phosphate. First, acetyl phosphate is a major secondary source of phosphoryl groups. It has been shown that several response regulator proteins can be phosphorylated by acetyl phosphate (28, 38). Since pta, together with ack (encoding acetate kinase), can influence the size of the acetyl phosphate pool, pta and ack can indirectly modulate some regulatory pathways. Second, acetyl phosphate can serve as an energy source. In Bradyrhizobium japonicum bacteroids and in A. vinelandii, acetyl phosphate is used as an energy source to support nitrogen fixation (4, 46). Concerning the regulation of pta, not much is known to date. It has been shown that phosphate stress induces ack and pta expression in S. meliloti (52, 62). In addition, both ack and pta seem to be induced in several bacterial species when grown on acetate as a carbon source. Interestingly, it has been shown that acetate is an important product of anaerobic cellulose degradation in flooded rice fields (10, 20). The Vibrio cholerae pta gene was previously isolated by using the signature-tagged transposon mutagenesis technique to search for genes required for colonization of the host intestine (7). In E. coli, biofilm formation is affected by altered acetyl phosphate metabolism (72).
For some of the transcriptional fusions it was not possible to determine unequivocally the in vivo-induced promoter and/or the corresponding gene, as was the case with cii-14. Two other transcriptional fusions, cii-51 and cii-11, contained genes that encode hypothetical proteins of unknown functions but with an apparent role in root colonization. It is worth noting that some of these transcriptional fusions were isolated in the IVET screening of different pools, a result which can be considered an independent confirmation of the in vivo induction of the corresponding genes, as was the case with clones cii-7 and cii-51, clones cii-11 and cii-101, and clones cii-14 and cii-15. The repeated isolation of these genes suggests a significant role in seedling colonization. As validation of the strategy used, we isolated a clone that contained the functional dapB gene with its promoter in the orientation opposite that of the pIVET-borne promoterless dapB gene.
In conclusion, we have demonstrated here the successful application of the dapB-based IVET selection strategy for the identification of candidate P. stutzeri genes that are involved in the colonization of rice roots. In future work, this approach can be used for the identification of P. stutzeri genes that are expressed during the endophytic stage of the interaction with the rice host. Moreover, the possible applications of this IVET system are not limited to the study of host-microbe interactions. IVET can be used to study bacterial behavior in various specific ecological niches, as demonstrated recently for the study of P. aeruginosa biofilm formation (14).
Acknowledgments
H. Rediers is indebted to the Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen for a predoctoral fellowship.
We thank Geert Schoofs for sequencing.
REFERENCES
- 1.Bartoleschi, C., M. C. Pardini, C. Scaringi, M. C. Martino, C. Pazzani, and M. L. Bernardini. 2002. Selection of Shigella flexneri candidate virulence genes specifically induced in bacteria resident in host cell cytoplasm. Cell. Microbiol. 4:613-626. [DOI] [PubMed] [Google Scholar]
- 2.Boch, J., V. Joardar, L. Gao, T. L. Robertson, M. Lim, and B. N. Kunkel. 2002. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 44:73-88. [DOI] [PubMed] [Google Scholar]
- 3.Born, T. L., and J. S. Blanchard. 1999. Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall biosynthesis. Curr. Opin. Chem. Biol. 3:607-613. [DOI] [PubMed] [Google Scholar]
- 4.Bresters, T. W., J. Krul, P. C. Scheepens, and C. Veeger. 1972. Phosphotransacetylase associated with the pyruvate dehydrogenase complex from the nitrogen fixing Azotobacter vinelandii. FEBS Lett. 22:305-309. [DOI] [PubMed] [Google Scholar]
- 5.Camilli, A., and J. J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee, S. P., B. K. Singh, and C. Gilvarg. 1994. Biosynthesis of lysine in plants: the putative role of meso-diaminopimelate dehydrogenase. Plant Mol. Biol. 26:285-290. [DOI] [PubMed] [Google Scholar]
- 7.Chiang, S. L., and J. J. Mekalanos. 1998. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol. Microbiol. 27:797-805. [DOI] [PubMed] [Google Scholar]
- 8.Chiang, S. L., J. J. Mekalanos, and D. W. Holden. 1999. In vivo genetic analysis of bacterial virulence. Annu. Rev. Microbiol. 53:129-154. [DOI] [PubMed] [Google Scholar]
- 9.Chuang, D. Y., A. G. Kyeremeh, Y. Gunji, Y. Takahara, Y. Ehara, and T. Kikumoto. 1999. Identification and cloning of an Erwinia carotovora subsp. carotovora bacteriocin regulator gene by insertional mutagenesis. J. Bacteriol. 181:1953-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad, R., M. Klose, and P. Claus. 2002. Pathway of CH4 formation in anoxic rice field soil and rice roots determined by 13C-stable isotope fractionation. Chemosphere 47:797-806. [DOI] [PubMed] [Google Scholar]
- 11.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durand, J. M., B. Dagberg, B. E. Uhlin, and G. R. Bjork. 2000. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol. Microbiol. 35:924-935. [DOI] [PubMed] [Google Scholar]
- 13.Eggermont, K., I. J. Goderis, and W. F. Broekaert. 1996. High throughput RNA extraction from plant samples based on homogenization by reciprocal shaking in the presence of a mixture of sand and glass beads. Plant Mol. Biol. Rep. 14:273-279. [Google Scholar]
- 14.Finelli, A., C. V. Gallant, K. Jarvi, and L. L. Burrows. 2003. Use of in-biofilm expression technology to identify genes involved in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:2700-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuller, T. E., R. J. Shea, B. J. Thacker, and M. H. Mulks. 1999. Identification of in vivo induced genes in Actinobacillus pleuropneumoniae. Microb. Pathog. 27:311-327. [DOI] [PubMed] [Google Scholar]
- 16.Gahan, C. G., and C. Hill. 2000. The use of listeriolysin to identify in vivo induced genes in the Gram-positive intracellular pathogen Listeria monocytogenes. Mol. Microbiol. 36:498-507. [DOI] [PubMed] [Google Scholar]
- 17.Gal, M., G. M. Preston, R. C. Massey, A. J. Spiers, and P. B. Rainey. Genes encoding a cellulosic polymer contribute toward ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol. Ecol., in press. [DOI] [PubMed]
- 18.Galili, G. 2002. New insights into the regulation and functional significance of lysine metabolism in plants. Annu. Rev. Plant Biol. 53:27-43. [DOI] [PubMed] [Google Scholar]
- 19.Ghrist, A. C., and G. V. Stauffer. 1998. Promoter characterization and constitutive expression of the Escherichia coli gcvR gene. J. Bacteriol. 180:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glissmann, K., S. Weber, and R. Conrad. 2001. Localization of processes involved in methanogenic degradation of rice straw in anoxic paddy soil. Environ. Microbiol. 3:502-511. [DOI] [PubMed] [Google Scholar]
- 21.Hammad, Y., J. Marechal, B. Cournoyer, P. Normand, and A. M. Domenach. 2001. Modification of the protein expression pattern induced in the nitrogen-fixing actinomycete Frankia sp. strain ACN14a-tsr by root exudates of its symbiotic host Alnus glutinosa and cloning of the sodF gene. Can. J. Microbiol. 47:541-547. [DOI] [PubMed] [Google Scholar]
- 22.Handfield, M., and R. C. Levesque. 1999. Strategies for isolation of in vivo expressed genes from bacteria. FEMS Microbiol. Rev. 23:69-91. [DOI] [PubMed] [Google Scholar]
- 23.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James, E. K., P. Gyaneshwar, N. Mathan, W. L. Barraquio, and J. K. Ladha. 2000. Endophytic diazotrophs associated with rice, p. 119-140. In J. K. Ladha and P. M. Reddy (ed.), The quest for nitrogen fixation in rice. International Rice Research Institute, Makati City, Philippines.
- 25.Jeong, W., M. K. Cha, and I. H. Kim. 2000. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 275:2924-2930. [DOI] [PubMed] [Google Scholar]
- 26.Jimenez, J. I., B. Minambres, J. L. Garcia, and E. Diaz. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4:824-841. [DOI] [PubMed] [Google Scholar]
- 27.Jin, S., K. Ishimoto, and S. Lory. 1994. Nucleotide sequence of the rpoN gene and characterization of two downstream open reading frames in Pseudomonas aeruginosa. J. Bacteriol. 176:1316-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S. B., B. S. Shin, S. K. Choi, C. K. Kim, and S. H. Park. 2001. Involvement of acetyl phosphate in the in vivo activation of the response regulator ComA in Bacillus subtilis. FEMS Microbiol. Lett. 195:179-183. [DOI] [PubMed] [Google Scholar]
- 29.Koenig, R. L., R. O. Morris, and J. C. Polacco. 2002. tRNA is the source of low-level trans-zeatin production in Methylobacterium spp. J. Bacteriol. 184:1832-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon, D. H., C. D. Lu, D. A. Walthall, T. M. Brown, J. E. Houghton, and A. T. Abdelal. 1994. Structure and regulation of the carAB operon in Pseudomonas aeruginosa and Pseudomonas stutzeri: no untranslated region exists. J. Bacteriol. 176:2532-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai, Y. C., H. L. Peng, and H. Y. Chang. 2001. Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect. Immun. 69:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamb, C., and Dixon, R. A. 1997. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:251-275. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S. W., and D. A. Cooksey. 2000. Genes expressed in Pseudomonas putida during colonization of a plant-pathogenic fungus. Appl. Environ. Microbiol. 66:2764-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loper, J. E., T. V. Suslow, and M. N. Schroth. 1984. Lognormal distribution of bacterial populations in rhizosphere. Phytopathology 74:1454-1460. [Google Scholar]
- 35.Lowe, A. M., D. T. Beattie, and R. L. Deresiewicz. 1998. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol. Microbiol. 27:967-976. [DOI] [PubMed] [Google Scholar]
- 36.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 37.Maki, Y., H. Yoshida, and A. Wada. 2000. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 5:965-974. [DOI] [PubMed] [Google Scholar]
- 38.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 269:31567-31572. [PubMed] [Google Scholar]
- 39.Mengin-Lecreulx, D., J. Ayala, A. Bouhss, J. van Heijenoort, C. Parquet, and H. Hara. 1998. Contribution of the Pmra promoter to expression of genes in the Escherichia coli mra cluster of cell envelope biosynthesis and cell division genes. J. Bacteriol. 180:4406-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merrick, M. J. 1993. In a class of its own—the RNA polymerase sigma factor σ54 (σN). Mol. Microbiol. 10:903-909. [DOI] [PubMed] [Google Scholar]
- 41.Merrick, M. J., and J. R. Coppard. 1989. Mutations in genes downstream of the rpoN gene (encoding σ54) of Klebsiella pneumoniae affect expression from σ54-dependent promoters. Mol. Microbiol. 3:1765-1775. [DOI] [PubMed] [Google Scholar]
- 42.Miché, L., S. Belkin, R. Rozen, and J. Balandreau. 2003. Rice seedling whole exudates and extracted alkylresorcinols induce stress-response in Escherichia coli biosensors. Environ. Microbiol. 5:403-411. [DOI] [PubMed] [Google Scholar]
- 43.Nakao, H., H. Watanabe, S. Nakayama, and T. Takeda. 1995. yst gene expression in Yersinia enterocolitica is positively regulated by a chromosomal region that is highly homologous to Escherichia coli host factor 1 gene (hfq). Mol. Microbiol. 18:859-865. [DOI] [PubMed] [Google Scholar]
- 44.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 45.Oosthuizen, M. C., B. Steyn, J. Theron, P. Cosette, D. Lindsay, A. Von Holy, and V. S. Brozel. 2002. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl. Environ. Microbiol. 68:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peterson, J. B., and T. A. LaRue. 1982. Soluble aldehyde dehydrogenase and metabolism of aldehydes by soybean bacteroids. J. Bacteriol. 151:1473-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pragai, Z., and C. R. Harwood. 2002. Regulatory interactions between the Pho and σB-dependent general stress regulons of Bacillus subtilis. Microbiology 148:1593-1602. [DOI] [PubMed] [Google Scholar]
- 48.Qi, H. Y., K. Sankaran, K. Gan, and H. C. Wu. 1995. Structure-function relationship of bacterial prolipoprotein diacylglyceryl transferase: functionally significant conserved regions. J. Bacteriol. 177:6820-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 50.Rainey, P. B., D. M. Heithoff, and M. J. Mahan. 1997. Single-step conjugative cloning of bacterial gene fusions involved in microbe-host interactions. Mol. Gen. Genet. 256:84-87. [DOI] [PubMed] [Google Scholar]
- 51.Rainey, P. B., and G. M. Preston. 2000. In vivo expression technology strategies: valuable tools for biotechnology. Curr. Opin. Biotechnol. 11:440-444. [DOI] [PubMed] [Google Scholar]
- 52.Reinscheid, D. J., S. Schnicke, D. Rittmann, U. Zahnow, H. Sahm, and B. J. Eikmanns. 1999. Cloning, sequence analysis, expression and inactivation of the Corynebacterium glutamicum pta-ack operon encoding phosphotransacetylase and acetate kinase. Microbiology 145:503-513. [DOI] [PubMed] [Google Scholar]
- 53.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Retallack, D. M., G. S. Deepe, Jr., and J. P. Woods. 2000. Applying in vivo expression technology (IVET) to the fungal pathogen Histoplasma capsulatum. Microb. Pathog. 28:169-182. [DOI] [PubMed] [Google Scholar]
- 55.Roop, R. M., G. T. Robertson, G. P. Ferguson, L. E. Milford, M. E. Winkler, and G. C. Walker. 2002. Seeking a niche: putative contributions of the hfq and bacA gene products to the successful adaptation of the brucellae to their intracellular home. Vet. Microbiol. 90:349-363. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 57.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 58.Slauch, J. M., and A. Camilli. 2000. IVET and RIVET: use of gene fusions to identify bacterial virulence factors specifically induced in host tissues. Methods Enzymol. 326:73-96. [DOI] [PubMed] [Google Scholar]
- 59.Sledjeski, D. D., C. Whitman, and A. Zhang. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staib, P., M. Kretschmar, T. Nichterlein, G. Kohler, S. Michel, H. Hof, J. Hacker, and J. Morschhauser. 1999. Host-induced, stage-specific virulence gene activation in Candida albicans during infection. Mol. Microbiol. 32:533-546. [DOI] [PubMed] [Google Scholar]
- 61.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 62.Summers, M. L., M. C. Denton, and T. R. McDermott. 1999. Genes coding for phosphotransacetylase and acetate kinase in Sinorhizobium meliloti are in an operon that is inducible by phosphate stress and controlled by phoB. J. Bacteriol. 181:2217-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsui, H. C., G. Feng, and M. E. Winkler. 1996. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K-12 from clustered Eσ32-specific promoters during heat shock. J. Bacteriol. 178:5719-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsui, H. C., H. C. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13:35-49. [DOI] [PubMed] [Google Scholar]
- 65.Uchiyama, I. 2003. MBGD: microbial genome database for comparative analysis. Nucleic Acids Res. 31:58-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Urbonavicius, J., Q. Qian, J. M. Durand, T. G. Hagervall, and G. R. Bjork. 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vermeiren, H. 2002. Pseudomonas stutzeri A15-Oryza sativa root interactions: a genetic and histochemical study. Ph.D. thesis. Catholic University of Leuven, Leuven, Belgium.
- 68.Vermeiren, H., J. Vanderleyden, and W. L. Hai. 1998. Colonization and nifH expression on rice roots by Alcaligenes faecalis A15, p. 167-177. In K. A. Malik, M. S. Mirza, and J. K. Ladha (ed.), Nitrogen fixation with non-legumes. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 69.Vermeiren, H., A. Willems, G. Schoofs, R. de Mot, V. Keijers, W. Hai, and J. Vanderleyden. 1999. The rice inoculant strain Alcaligenes faecalis A15 is a nitrogen-fixing Pseudomonas stutzeri. Syst. Appl. Microbiol. 22:215-224. [DOI] [PubMed] [Google Scholar]
- 70.von Mering, C., M. Huynen, D. Jaeggi, S. Schmidt, P. Bork, and B. Snel. 2003. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31:258-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, J., A. Mushegian, S. Lory, and S. Jin. 1996. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. USA 93:10434-10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolfe, A. J., D. E. Chang, J. D. Walker, J. E. Seitz-Partridge, M. D. Vidaurri, C. F. Lange, B. M. Pruss, M. C. Henk, J. C. Larkin, and T. Conway. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 48:977-988. [DOI] [PubMed] [Google Scholar]
- 73.Wood, Z. A., E. Schroder, H. J. Robin, and L. B. Poole. 2003. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28:32-40. [DOI] [PubMed] [Google Scholar]
- 74.Wu, Y., S. W. Lee, J. D. Hillman, and A. Progulske-Fox. 2002. Identification and testing of Porphyromonas gingivalis virulence genes with a pPGIVET system. Infect. Immun. 70:928-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 76.Yoshida, S., D. A. Formo, and J. H. Cock. 1976. Laboratory manual for physiological studies of rice. International Rice Research Institute, Los Banos, Philippines.
- 77.You, C. B., M. Lin, and X. J. Fang. 1995. Field release of genetically engineered associative diazotrophs and its risk assessment. J. Agric. Biotechnol. 1:34-41. [Google Scholar]
- 78.You, C. B., W. Song, H. X. Wang, J. P. Li, M. Lin, and W. L. Hai. 1983. Associative nitrogen fixation of Alcaligenes faecalis with rice plant. Biol. Nitrogen Fix. Newsl. Sydney. Univ. 11:92-103. [Google Scholar]
- 79.You, C. B., and F. Zhou. 1989. Non-nodular endorhizospheric nitrogen fixation in wetland rice. Can. J. Microbiol. 35:403-408. [Google Scholar]
- 80.Young, G. M., and V. L. Miller. 1997. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol. Microbiol. 25:319-328. [DOI] [PubMed] [Google Scholar]