Abstract
The taxonomic characterization of a bacterial community is difficult to combine with the monitoring of its temporal changes. None of the currently available identification techniques are able to visualize a “complete” community, whereas techniques designed for analyzing bacterial ecosystems generally display limited or labor-intensive identification potential. This paper describes the optimization and validation of a nested-PCR-denaturing gradient gel electrophoresis (DGGE) approach for the species-specific analysis of bifidobacterial communities from any ecosystem. The method comprises a Bifidobacterium-specific PCR step, followed by purification of the amplicons that serve as template DNA in a second PCR step that amplifies the V3 and V6-V8 regions of the 16S rRNA gene. A mix of both amplicons is analyzed on a DGGE gel, after which the band positions are compared with a previously constructed database of reference strains. The method was validated through the analysis of four artificial mixtures, mimicking the possible bifidobacterial microbiota of the human and chicken intestine, a rumen, and the environment, and of two fecal samples. Except for the species Bifidobacterium coryneforme and B. indicum, all currently known bifidobacteria originating from various ecosystems can be identified in a highly reproducible manner. Because no further cloning and sequencing of the DGGE bands is necessary, this nested-PCR-DGGE technique can be completed within a 24-h span, allowing the species-specific monitoring of temporal changes in the bifidobacterial community.
The genus Bifidobacterium consists of gram-positive bacteria with a G+C content above 50%, currently comprising over 30 species (9). The main habitat of bifidobacteria is the human and animal intestinal tract (3, 13, 24), although sewage (23), anaerobic digesters (4), and fermented milk (14) have also been reported as isolation sources of certain Bifidobacterium species. The past decade has witnessed a fast growing interest in bifidobacteria, mainly because of the health-promoting properties of certain species (2, 7). Because of their growing application in probiotic dairy products and dried food supplements (25), many recent studies emphasize only the intestinal bifidobacteria (13, 18, 21).
Until recently, routine identification of bifidobacteria was mainly based on phenotypic characterization, often leading to conflicting or doubtful results. Molecular techniques such as amplified ribosomal DNA restriction analysis (8, 27), 16S rRNA gene sequencing (9), fluorescent in situ hybridization (27), sodium dodecyl sulfate-polyacrylamide gel electrophoresis of cellular proteins, randomly amplified polymorphic DNA PCR, pulsed-field gel electrophoresis, dot blot hybridization (19), and rep-PCR (12) have been evaluated and optimized for identification of bifidobacterial pure cultures to the species or even the strain level. However, numerous situations call for the direct species-specific detection of bifidobacteria in microbial ecosystems in relation to temporal and environmental changes. Because of their culture-dependent nature, most of the techniques mentioned above are not suitable for this purpose (20). Therefore, culture-independent methods have been designed, of which denaturing gradient gel electrophoresis (DGGE) is the most commonly used technique. The DGGE principle relies on the electrophoretic separation of PCR amplicons of equal length in a sequence-specific manner (15, 16). When universal bacterial PCR primers are used, only the dominant microbiota of an ecosystem will be visualized on a DGGE gel (28), producing complex banding patterns. In case identification of these bands is desired, additional cloning and sequencing of the extracted bands are required (1, 5, 6, 21). However, these extra steps render the method laborious and time-consuming, impairing the potential of DGGE as a fast method for bacterial population fingerprinting. In this regard, the use of species- or genus-specific primers represents a major step forward, usually resulting in less complex DGGE banding patterns that only display the diversity of a specific bifidobacterial group within the targeted ecosystem (10, 11, 13, 18). Most of these types of studies reported the detection of a limited number of mainly intestinal bifidobacterial species, although cloning and sequencing steps were still needed in order to confirm the detection results.
In an attempt to decrease the operational time of culture-independent detection of bifidobacteria, this paper describes the optimization and validation of a nested-PCR-DGGE approach for the direct identification of all currently known bifidobacterial species present in ecosystems with variable degrees of complexity, including both artificial and natural samples.
MATERIALS AND METHODS
Strain collection.
All strains used for this study were obtained from the BCCM/LMG bacterial collection (Fig. 1) and were grown for 24 h at 37°C under anaerobic conditions (80% N2, 10% H2, and 10% CO2) on modified Columbia agar (MCA) comprising 23 g of special peptone (Oxoid, Basingstoke, United Kingdom), 1 g of soluble starch (Merck, Darmstadt, Germany), 5 g of NaCl, 0.3 g of cysteine-HCl-H2O (Sigma, Bornem, Belgium), 5 g of glucose (Vel, Leuven, Belgium), and 15 g of agar (Oxoid) dissolved in 1 liter of distilled water.
FIG. 1.
Dendrogram showing the normalized band positions of bifidobacterial reference strains. Numbers behind the species assignment indicate the BCCM/LMG strain numbers. T, type strain.
Total DNA preparation.
Cells (half of a loop) from a 24-h culture were washed in 500 μl of TE buffer (1 mM EDTA [pH 8.0], 10 mM Tris-HCl [pH 8.0]), followed by centrifugation for 2 min at 13,000 rpm (Eppendorf centrifuge, model 5804R). After removal of the supernatant, the resulting pellet was then frozen at −20°C for 1 h to facilitate the rupture of the gram-positive cell wall. The thawed pellet was suspended in 150 μl of lysozyme solution (5 mg of lysozyme [Serva, Heidelberg, Germany] in 150 μl of TE buffer) and subsequently incubated at 37°C for 40 min. The remaining steps of the procedure were performed according to the protocol of Pitcher and coworkers (17). The resulting DNA pellet was then dissolved in 200 μl of TE buffer overnight at 4°C, after which an RNA-digesting step was performed by adding 2 μl of RNase solution (10 mg of RNase [Sigma] dissolved in 1 ml of Milli-Q water) followed by a 90-min incubation step at 37°C. The presence of DNA was verified on a 1% agarose gel, followed by spectrophotometric measurements at 260, 280, and 234 nm to determine the quality of the DNA samples.
Upon collection of fecal samples from two healthy, adult volunteers (one male and one female) following a normal diet, 700 mg (wet weight) of the fecal samples was homogenized in 9.3 ml of physiological phosphate buffer. One milliliter of the fecal sample suspension was transferred to an Eppendorf tube and centrifuged for 5 min at 13,000 rpm (Eppendorf centrifuge, model 5804R). After removal of the supernatant, the pellet was resuspended in 1 ml of TE buffer and centrifuged for 5 min at 13,000 rpm. After removal of the supernatant, the pellet was resuspended in 150 μl of enzyme solution to degenerate the bacterial cell wall. Per sample, this enzyme mix consisted of 6 mg of lysozyme powder and 40 μl of mutanolysine dissolved in 110 μl of TE buffer. Further steps were performed according to the protocol of Pitcher and coworkers (17).
Nested PCR.
All PCRs were performed by use of a Taq polymerase kit (Applied BioSystems, Foster City, Calif.). The first PCR applied primers lm26-f (5′-GATTCTGGCTCAGGATGAACG-3′) and lm3-r (5′-CGGGTGCTICCCCACTTTCATG-3′) described by Kaufmann and coworkers (10), amplifying a 1,417-bp fragment of the bifidobacterial 16S rRNA gene. PCR volumes of 50 μl contained 8 μl of 10× PCR buffer, 3.5 μl of bovine serum albumin (0.1 mg/ml), 3.5 μl of deoxynucleoside triphosphates (2 mM each), 3 μl of each primer (5 μM), 0.35 μl of Taq polymerase (5 U/μl), 27.65 μl of sterile Milli-Q water, and 1 μl of 10-fold-diluted DNA solution. The following PCR program was used: initial denaturation at 94°C for 5 min; 3 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 2 min, and extension at 72°C for 1 min; 30 cycles of denaturation at 94°C for 20 s, annealing at 55°C for 1 min, and extension at 72°C for 1 min; and final extension at 72°C for 7 min followed by cooling to 4°C. The presence of PCR products was verified on a 1% agarose gel. In order to eliminate the remaining oligonucleotides and original template DNA, purification of the amplicons was performed by use of the QIAquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Subsequently, a second PCR was performed, using the amplicons of the first PCR as template DNA. Because of the length of the first amplicon (positions 15 to 1432), different primer pairs can be used for the second PCR, depending on the desired application. This study made use of two sets of primers. The first primer set (F357-GC and 518R) (5′-GC-clamp-GCCTACGGAGGCAGCAG-3′ and 5′-ATTACCGCGGCTGCTGG-3′, respectively) amplifies the V3 region of the bacterial 16S rRNA gene (15), whereas the second set of primers (U968F-GC and L1401R) (5′-GC-clamp-AACGCGAAGAACCTTAC-3′ and 5′-GCGTGTGTACAAGACCC-3′, respectively) targets the V6-to-V8 region of the bacterial 16S rRNA gene (28). In both cases, the forward primer contained a GC clamp (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGG-3′) to facilitate separation of the amplicons in a DGGE gel. For both primer sets, the PCR volumes of 50 μl contained 6 μl of 10× PCR buffer, 2.5 μl of bovine serum albumin (0.1 mg/ml), 2.5 μl of deoxynucleoside triphosphates (2 mM each), 2 μl of each primer (5 μM), 0.25 μl of Taq polymerase (5 U/μl), 33.75 μl of sterile Milli-Q water, and 1 μl of 10-fold-diluted DNA solution. The following PCR program was used: initial denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 20 s, annealing at 55°C for 45 s, and extension at 72°C for 1 min; and final extension at 72°C for 7 min, followed by cooling to 4°C.
DGGE.
PCR products were analyzed on DGGE gels based on the protocol of Muyzer and coworkers (15, 16), with modifications according to the work of Temmerman and coworkers (26). Because of the high G+C content of bifidobacteria, gels with a 50 to 70% denaturing gradient were used. Gels were stained with ethidium bromide for 15 min, allowing digital capturing of the DGGE band profiles under UV light.
Gel processing.
For direct identification of bands in a given DGGE profile, a database was created, by use of the BioNumerics (BN) software package, version 2.50 (Applied-Maths, St.-Martens-Latem, Belgium), that contains the V3 and V6-V8 band positions of bifidobacterial type and reference strains (Fig. 2). A reference pattern (consisting of four different type strain V3 amplicons) was included in every five lanes of each DGGE gel, facilitating digital normalization of banding patterns lying between reference lanes by alignment of each reference lane with the standard reference pattern defined in the BN database. This normalization enables comparison of banding patterns originating from different DGGE gels, provided that they comprise the same reference pattern run under identical electrophoretic conditions. After normalization, the identities of bifidobacteria were determined by comparing the band positions in the sample profiles with the BN database.
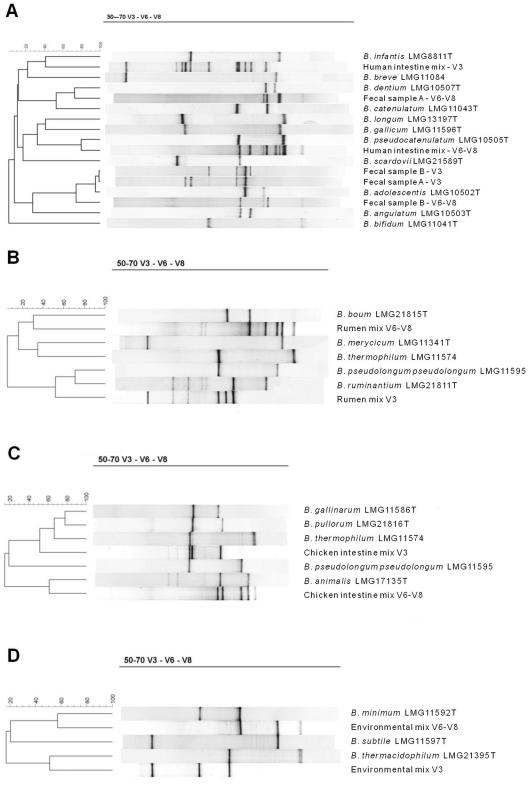
FIG. 2.
Use of the BN software to compare digitized V3 and V6-V8 profiles for the identification of bifidobacteria present in four artificial mixtures mimicking the human intestine (A), the rumen (B), the chicken intestine (C), and an environmental sample (D). Panel A also contains the V3 and V6-V8 profiles of two fecal samples. Fecal sample A contains B. adolescentis, B. bifidum, B. catenulatum, B. gallicum, and B. infantis, and fecal sample B contains B. adolescentis, B. angulatum, B. bifidum, and B. catenulatum.
RESULTS
Nested PCR.
The nested-PCR approach described in this study for the identification of bifidobacteria in various ecosystems applies a first PCR step using the genus-specific primers lm-3 and lm-26, resulting in an amplicon for all bifidobacterial reference strains. Also, a broad range of nontarget organisms was tested, and in contrast to the results of Kaufmann and coworkers (10), weak PCR signals for Propionibacterium freudenreichii and Gardnerella vaginalis were obtained. The bifidobacterial amplicons could not be analyzed directly on a DGGE gel, because their length of 1,417 bp by far exceeds the 500-bp limit for DGGE analysis. The advantage of generating amplicons of this length is that they can serve as template DNA for other 16S rRNA gene primers, such as the V3 primer combination F357-GC and 518-R, during the second PCR step. Because of the universal nature of these primers, a purification of the amplicons from the first PCR was performed in order to remove all remaining nonbifidobacterial template DNA. Analysis of the V3 amplicons on a DGGE gel showed that not all bifidobacterial species could be separated from each other, necessitating the additional use of a second universal primer set. For this purpose, we opted for the U968F-GC and L1401R primers targeting the V6-V8 region of the 16S rRNA gene. Because the same temperature program could be used, amplification of the V3 and V6-V8 regions during the second PCR step could be performed at the same time, with each reaction in a separate PCR tube (Fig. 1). For all bifidobacterial species, an amplicon was obtained for both the V3 and V6-V8 region (data not shown).
DGGE analysis of V3 and V6-V8 amplicons.
Because of the high G+C content of bifidobacteria, the conventional 35 to 70% denaturing gradient was replaced with a 50 to 70% denaturing gradient. For some species (e.g., Bifidobacterium lactis and B. pseudolongum subsp. globosum), the band position of the V3 amplicon was indistinguishable (Fig. 1, left-hand band in each lane), whereas for other bifidobacteria (e.g., B. longum and B. pseudocatenulatum), identical band positions were found for the V6-V8 amplicon (Fig. 1, right-hand band in each lane). However, clustering analysis of the combined DGGE profile of both amplicons (V3-V6-V8) allowed us to differentiate all bifidobacteria according to their (sub)species designation, except for the species B. indicum and B. coryneforme (Fig. 1). Although most V3-V6-V8 DGGE profiles consisted of two strong bands, one or more additional weak bands were noticed for B. adolescentis, B. pseudocatenulatum, B. animalis, and B. ruminantium. Furthermore, in the case of B. thermophilum and B. breve, two and three different combinations, respectively, of V3 and V6-V8 band positions exist, due to slightly different band positions among certain strains. Due to its low %G+C, the amplicon of G. vaginalis did not enter the 50 to 70% DGGE gel, whereas the band positions of P. freundenreichii could clearly be separated from those of all bifidobacteria (Fig. 1).
Artificial mixtures and fecal samples.
The discriminatory potential of the V3-V6-V8 DGGE technique was further validated by use of four artificial mixtures of bifidobacterial DNA (Table 1), mimicking a human intestine, chicken intestine, rumen, and environmental ecosystem (sewage), and by means of two human fecal samples. In order to avoid overlap of the V3 and V6-V8 band positions, both types of amplicons were loaded separately in two adjacent lanes on the DGGE gel. After normalization, band positions of both amplicon types were compared with the BN database of reference strains. Clustering analysis of mixed community profiles using the BN software enabled identification of all bifidobacterial species present in all mixtures. Species with highly similar or identical V3 band positions can be further differentiated by comparison with the band positions of the V6-V8 amplicons and vice versa. Figure 2 shows the resulting identification of bands for the four artificial mixtures mimicking the possible bifidobacterial microbiota of the human intestine (panel A), rumen (panel B), chicken intestine (panel C), and an environmental sample (panel D). These four mixtures clearly demonstrate the need to analyze both the V3 and V6-V8 amplicons. Overall, a detection limit of 104 CFU/ml was established, and no preferential amplification resulting from various bacterial concentrations in the mixtures was noticed. Finally, the method was also validated for characterization of bifidobacterial species present in two fecal samples originating from two volunteers (Fig. 2A). Fecal sample A contained five bifidobacterial species (B. adolescentis, B. bifidum, B. catenulatum, B. gallicum, and B. infantis), whereas fecal sample B contained four species (B. adolescentis, B. angulatum, B. bifidum, and B. catenulatum). Although some bands were less intense than those for the artificial mixtures or pure cultures, all bands could clearly be linked to Bifidobacterium species. Furthermore, there were no bands present in the fecal sample lanes which could not be assigned to any of the bifidobacterial species.
TABLE 1.
Microbial composition of four artificial mixtures mimicking bifidobacterial ecosystems
| Mix no. | Type of ecosystem | Species composition |
|---|---|---|
| 1 | Human intestine | B. angulatum, B. bifidum, B. adolescentis, B. infantis, B. longum, B. breve, B. catenulatum, B. dentium, B. gallicum, B. pseudocatenulatum, B. scardovii |
| 2 | Rumen | B. ruminantium, B. pseudolongum subsp. pseudolongum, B. merycicum, B. thermophilum, B. boum |
| 3 | Chicken intestine | B. pullorum, B. pseudolongum subsp. pseudolongum, B. animalis, B. thermophilum, B. gallinarium |
| 4 | Environment | B. thermacidophilum, B. minimum, B. subtile |
DISCUSSION
At present, DGGE is the most frequently applied technique to analyze bifidobacterial ecosystems, although cloning and sequencing of the DGGE bands are still necessary to obtain a reliable identification (5, 6, 21). Based on previous research on DGGE analysis of probiotic products (26), the present study describes the design and validation of a nested-PCR-DGGE method for the direct identification of currently known bifidobacteria present in natural or industrial ecosystems.
Until now, Bifidobacterium-specific primers suitable for DGGE which allow the direct identification of all bifidobacteria have not been described in the literature. Therefore, a nested-PCR approach was applied, combining a first genus-specific PCR step with a second universal PCR step. In between, a purification of the amplicons was necessary to remove small remaining fractions of nonbifidobacterial DNA. Amplification of the 16S rRNA gene V3 region alone did not allow the complete differentiation of all bifidobacteria, necessitating the combination of analysis of both the V3 and V6-V8 regions of the 16S rRNA gene. Because both primer sets can be used during the same PCR run, though in separate tubes, only a limited amount of extra work was required. For all bifidobacteria tested, both genus-specific and universal primers produced sufficient amounts of amplicon. Separation of the V3 and V6-V8 amplicons in a 50 to 70% gradient DGGE gel resulted in a clear identification of all bifidobacteria, except for B. coryneforme and B. indicum, which displayed identical band positions for both amplicons. The fact that these two species, originating from the intestinal tracts of two different species of bees (22), cannot be differentiated is in line with the rep-PCR data of Masco and coworkers (12). After analysis of six different B. breve reference strains, we found that three different combinations of V3 and V6-V8 band positions occurred. Likewise, two different combinations were observed among five B. thermophilum reference strains (Fig. 1). This is probably due to minor sequence variations within these species. As none of the combinations coincided with other species, this did not impair the identification potential of the method. This observation, together with the fact that both subspecies of B. pseudolongum could be readily distinguished, indicates that DGGE has an identification potential up to the subspecies level. Another observation was that B. adolescentis, B. animalis, B. pseudocatenulatum, and B. ruminantium displayed additional weak bands for both 16S rRNA gene regions, possibly as a result of operon heterogeneity, as previously observed for different genera (15, 28). This heterogeneity does not impair the identification potential of DGGE, because these additional bands are consistent among different strains of a specific taxon and are readily recognizable. As members of our laboratory already observed during a previous study (26), the V3 and V6-V8 amplicons of B. animalis and B. lactis have completely different band positions than B. animalis also showing operon heterogeneity, which indicates that both taxa probably do not belong to the same species, as confirmed by Masco and coworkers (12).
Because this technique has been designed and optimized for the analysis of (complex) mixtures of bifidobacteria, it was validated by means of four representative artificial mixtures of bifidobacteria and two fecal samples. For these mixtures, perfect identification of all bifidobacteria present was possible. Because some band positions of the V3 region coincide with the V6-V8 band positions of other bifidobacteria, it was necessary to load the two different amplicons in two adjacent lanes, which also prevented the banding patterns from becoming too complex. Because most of the bifidobacterial ecosystems also contain nonbifidobacteria, great care should be taken in the evaluation of the specificity of the approach. In addition to the selectivity of the genus-specific PCR and the purification of the amplicons, the use of a 50 to 70% gradient DGGE gel also prevents amplicons from nonbifidobacteria with denaturation points between 35 and 50% denaturant from entering the gel. This was demonstrated by the fact that G. vaginalis did not produce any bands on the 50 to 70% denaturing gel because of its G+C content of <50%. The only nonbifidobacterial species besides G. vaginalis that is known so far to produce an amplicon when the Bifidobacterium-specific primers are used, namely P. freundenreichii, displayed clearly separated band positions on the DGGE gels, thereby not impairing the identification potential of the technique. The bifidobacterial specificity of the technique was further demonstrated by the analysis of two fecal samples. Besides the four to five bifidobacterial species detected, no other bands were present on the gel that could not be linked to a certain bifidobacterial species. The fact that some bands were less intense than those for the artificial mixtures was due to the fact that in natural ecosystems different bifidobacterial species are present in various concentrations. However, it cannot be guaranteed that the optimized nested-PCR-DGGE technique was capable of detecting all bifidobacteria present in the fecal samples. In this regard, optimization of certain procedural steps, such as DNA extraction, might be necessary, depending on the ecosystem analyzed.
The nested-PCR-DGGE approach described in this paper has the potential to analyze bifidobacterial communities to the subspecies level. From the methodological point of view, the main advantage of this technique is that a complete analysis of a bifidobacterial community can be performed within a 24-h time span. Provided that an identification match is obtained with the database, the fact that no further cloning and sequencing of the DGGE bands are necessary makes this technique very suitable for temporal analysis of bifidobacterial ecosystems. Although not yet verified, this approach also holds great promise if applied to other genera, provided that suitable primer sets are designed and that the intrageneric taxonomic structure is not too complex.
Acknowledgments
This research was financially supported by a Ph.D. grant from the Flemish Institute for the Promotion of Innovation by Science and Technology (IWT-Vlaanderen, Brussels, Belgium). G.H. is a postdoctoral fellow of the Fund for Scientific Research—Flanders (Belgium) (F.W.O.-Vlaanderen).
REFERENCES
- 1.Ampe, F., N. Ben Omar, C. Moizan, C. Wacher, and J. P. Guyot. 1999. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65:5464-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballongue, J., J. P. Grill, and P. Baratteeuloge. 1995. Effects of Bifidobacterium fermented milks on human intestinal flora. Lait 73:249-256. [Google Scholar]
- 3.Crociani, F., B. Biavati, A. Alessandrini, C. Chiarini, and V. Scardovi. 1996. Bifidobacterium inopinatum sp. nov. and Bifidobacterium denticolens sp. nov., two new species isolated from human dental caries. Int. J. Syst. Microbiol. 46:564-571. [DOI] [PubMed] [Google Scholar]
- 4.Dong, X. Z., Y. H. Xin, W. Y. Jian, X. L. Liu, and D. W. Ling. 2000. Bifidobacterium thermacidophilum sp. nov., isolated from an anaerobic digester. Int. J. Syst. Evol. Microbiol. 50:119-125. [DOI] [PubMed] [Google Scholar]
- 5.Ercolini, D., G. Moschetti, G. Blaiotta, and S. Coppola. 2001. The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water buffalo mozzarella cheese production: bias of culture-dependent and culture-independent analyses. Syst. Appl. Microbiol. 24:610-617. [DOI] [PubMed] [Google Scholar]
- 6.Favier, C. F., E. E. Vaughan, W. M. De Vos, and A. D. L. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldin, B. R., S. L. Gorbach, S. Saxelin, L. Barakat, L. Gualtieri, and S. Salminen. 1992. Survival of Lactobacillus species (strain GG) in the human gastrointestinal tract. Dig. Dis. Sci. 37:121-128. [DOI] [PubMed] [Google Scholar]
- 8.Hall, V., T. Lewis-Evans, and B. I. Duerden. 2001. Identification of actinomyces, propionibacteria, lactobacilli and bifidobacteria by amplified 16S rDNA restriction analysis. Anaerobe 7(2):55-57. [Google Scholar]
- 9.Hoyles, L., E. Inganas, E. Falsen, M. Drancourt, N. Weiss, A. L. McCartney, and M. D. Collins. 2002. Bifidobacterium scardovii sp. nov., from human sources. Int. J. Syst. Evol. Microbiol. 52:995-999. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann, P., A. Pfefferkorn, M. Teuber, and L. Meile. 1997. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl. Environ. Microbiol. 63:1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kok, R. G., A. de Waal, F. Schut, G. W. Welling, G. Weenk, and K. J. Hellingwerf. 1996. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl. Environ. Microbiol. 62:3668-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masco L., G. Huys, D. Gevers, L. Verbrugghen, and J. Swings. Identification of Bifidobacterium species using rep-PCR fingerprinting. Syst. Appl. Microbiol., in press. [DOI] [PubMed]
- 13.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meile, L., W. Ludwig, U. Rueger, C. Gut, P. Kaufmann, G. Dasen, S. Wenger, and M. Teuber. 1997. Bifidobacterium lactis sp. nov, a moderately oxygen tolerant species isolated from fermented milk. Syst. Appl. Microbiol. 20:57-64. [Google Scholar]
- 15.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 17.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 18.Requena, T., J. Burton, T. Matsuki, K. Munro, M. A. Simon, R. Tanaka, K. Watanabe, and G. W. Tannock. 2002. Identification, detection, and enumeration of human Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 68:2420-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuter, G., G. Klein, and M. Goldberg. 2002. Identification of probiotic cultures in food samples. Food Res. Int. 35:117-124. [Google Scholar]
- 20.Roy, D. 2001. Media for the isolation and enumeration of bifidobacteria in dairy products. Int. J. Food Microbiol. 69:167-182. [DOI] [PubMed] [Google Scholar]
- 21.Satokari, R. M., E. E. Vaughan, A. D. L. Akkermans, M. Saarela, and W. M. De Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scardovi, V., and L. D. Trovatelli. 1969. New species of bifid bacteria from Apis mellifica L. and Apis indica F. A contribution to the taxonomy and biochemistry of the genus Bifidobacterium. Zentbl. Bakteriol. Parasitenkd. Abt. 2 123:64-88. [PubMed] [Google Scholar]
- 23.Scardovi, V., L. D. Trovatelli, B. Biavati, and G. Zani. 1979. Bifidobacterium cuniculi, Bifidobacterium choerinum, Bifidobacterium boum, and Bifidobacterium pseudocatenulatum—four new species and their deoxyribonucleic acid homology relationships. Int. J. Syst. Bacteriol. 29:291-311. [Google Scholar]
- 24.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanton C., G. Gardiner, H. Meehan, K. Collins, G. Fitzgerald, P. B. Lynch, and R. P. Ross. 2001. Market potential for probiotics. Am. J. Clin. Nutr. 73:476S-483S. [DOI] [PubMed] [Google Scholar]
- 26.Temmerman, R., I. Scheirlinck, G. Huys, and J. Swings. 2003. Culture-independent analysis of probiotic products by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:220-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ventura, M., M. Elli, R. Reniero, and R. Zink. 2001. Molecular microbial analysis of Bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ARDRA). FEMS Microbiol. Ecol. 36:113-121. [DOI] [PubMed] [Google Scholar]
- 28.Zoetendal, E. G., A. D. L. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]