Abstract
The vaoA gene from Penicillium simplicissimum CBS 170.90, encoding vanillyl alcohol oxidase, which also catalyzes the conversion of eugenol to coniferyl alcohol, was expressed in Escherichia coli XL1-Blue under the control of the lac promoter, together with the genes calA and calB, encoding coniferyl alcohol dehydrogenase and coniferyl aldehyde dehydrogenase of Pseudomonas sp. strain HR199, respectively. Resting cells of the corresponding recombinant strain E. coli XL1-Blue(pSKvaomPcalAmcalB) converted eugenol to ferulic acid with a molar yield of 91% within 15 h on a 50-ml scale, reaching a ferulic acid concentration of 8.6 g liter−1. This biotransformation was scaled up to a 30-liter fermentation volume. The maximum production rate for ferulic acid at that scale was 14.4 mmol per h per liter of culture. The maximum concentration of ferulic acid obtained was 14.7 g liter−1 after a total fermentation time of 30 h, which corresponded to a molar yield of 93.3% with respect to the added amount of eugenol. In a two-step biotransformation, E. coli XL1-Blue(pSKvaomPcalAmcalB) was used to produce ferulic acid from eugenol and, subsequently, E. coli(pSKechE/Hfcs) was used to convert ferulic acid to vanillin (J. Overhage, H. Priefert, and A. Steinbüchel, Appl. Environ. Microbiol. 65:4837-4847, 1999). This process led to 0.3 g of vanillin liter−1, besides 0.1 g of vanillyl alcohol and 4.6 g of ferulic acid liter−1. The genes ehyAB, encoding eugenol hydroxylase of Pseudomonas sp. strain HR199, and azu, encoding the potential physiological electron acceptor of this enzyme, were shown to be unsuitable for establishing eugenol bioconversion in E. coli XL1-Blue.
Vanillin is a frequently used aromatic flavor compound in the food and cosmetics industries. The most intensively studied process for producing vanillin by biotransformation, which then can be designated “natural,” is based on the substrate ferulic acid (22, 24). Previously, the gram-positive microorganisms Amycolatopsis sp. strain HR167 and Streptomyces setonii were identified as being able to convert ferulic acid to vanillin and which exhibited a very high tolerance towards this highly reactive aromatic aldehyde (2, 14; B. Müller, T. Münch, A. Muheim, and M. Wetli, European Patent Office patent application EP0885968, 1998; J. Rabenhorst and R. Hopp, European Patent Office patent application EP0761817, 1997). When these bacteria were used in ferulic acid bioconversions, vanillin concentrations of more than 10 g liter−1 were obtained (Müller et al., patent application; Rabenhorst and Hopp, patent application). The major drawback of the Amycolatopsis- and S. setonii-based microbial production of vanillin is the relatively high price of the substrate ferulic acid.
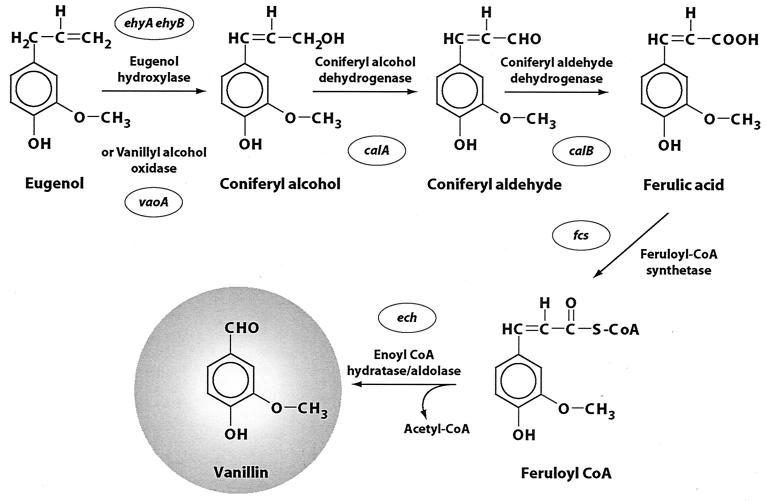
A new strategy for producing ferulic acid and vanillin is the enzymatic oxidative conversion of eugenol (Fig. 1). Eugenol is a cheap “natural” substrate that can be isolated from the essential oil of the clove tree Syzygium aromaticum on an industrial scale. The genes encoding proteins of the oxidative eugenol catabolism pathway have been investigated in detail in different Pseudomonas strains (1, 5, 13, 15, 16, 17, 18, 20, 21). The genes ech and fcs of Pseudomonas sp. strain HR199, encoding feruloyl-coenzyme A (CoA) synthetase and enoyl-CoA hydratase/aldolase, respectively, were used to engineer Escherichia coli strains able to catalyze the conversion of ferulic acid to vanillin (18). The genes ehyAB, encoding the eugenol hydroxylase of Pseudomonas sp. strain HR199 (12, 20), were expressed in Ralstonia eutropha H16 to establish the key steps of eugenol biotransformation, including the initial conversion of eugenol to coniferyl alcohol, in this recombinant strain (19).
FIG. 1.
Biotransformation of eugenol to vanillin.
In this study we expressed the genes ehyAB, calA, and calB of Pseudomonas sp. strain HR199 in E. coli to establish eugenol biotransformation to ferulic acid in this strain. Since no bioconversion was achieved by this strategy, the genes ehyAB were replaced by vaoA from Penicillium simplicissimum CBS 170.90, encoding vanillyl alcohol oxidase (8). This enzyme also catalyzes the conversion of eugenol to coniferyl alcohol (3, 9, 10), and its primary structure shares extensive regions of homology with the amino acid sequence of the flavoprotein subunit of eugenol hydroxylases (5, 20). However, it lacks a cytochrome c subunit. By using this gene, a highly efficient biotransformation of eugenol to ferulic acid was established with recombinant strains of E. coli on a technical scale.
MATERIALS AND METHODS
Strains, plasmids, and cultivation conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli was cultivated in complex media (Luria-Bertani [LB] or Terrific broth [TB]) (25) at 30 or 37°C in Erlenmeyer flasks with baffles on a rotary water bath shaker (G76; New Brunswick Scientific Co, Edison, N.J.) at 150 rpm. P. simplicissimum CBS170.90 was grown in a minimal medium as described previously (8). Ampicillin was used at a final concentration of 100 μg/ml for recombinant strains of E. coli. Eugenol was directly added to the medium at final concentrations of 0.01 to 0.1% (vol/vol). Growth was monitored by measuring the turbidity of the cultures at 600 nm.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant chracteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 λ−lac [F′ proAB lacIqZΔM15 Tn10(Tcr)] | 6 |
| P. simplicissimum CBS 170.90 | ATCC 90172 | |
| Plasmids | ||
| pBluescript SK− | AprlacPOZ, T7 and T3 promoter | Stratagene, San Diego, Calif. |
| pSKcalA | pBluescript SK(−) harboring the PCR product comprising calA as a NotI-SpeI fragment | 19 |
| pSKcalAm | pBluescript SK(−) harboring the PCR product comprising calA as a NotI-SpeI fragment with the EcoRI restriction site eliminated | This study |
| pSKcalB | pBluescript SK(−) harboring the PCR product comprising calB as a SpeI-BamHI fragment | 19 |
| pSKcalAmcalB | pBluescript SK(−) harboring the PCR products comprising calAm and calB as a NotI-BamHI fragment | This study |
| pSKvao | pBluescript SK(−) harboring the PCR product comprising vao as a SacI-NotI fragment | This study |
| pSKvaom | pBluescript SK(−) harboring the PCR product comprising vao as SacI-NotI fragment with the EcoRI and BamHI restriction sites eliminated | This study |
| pSKvaomPcalAmcalB | pBluescript SK(−) harboring the PCR products comprising vaom, PlacZ, calA, and calB | This study |
| pSKechE/Hfcs | pBluescript SK(−) harboring the PCR product comprising ech as an EcoRI-HindIII fragment and fcs as a PstI fragment | 18 |
Analytical methods.
Culture supernatants were analyzed for excreted intermediates of eugenol catabolism by liquid chromatography without prior extraction, using a Knauer (Berlin, Germany) high-pressure liquid chromatography (HPLC) apparatus. Intermediates were separated by reverse-phase chromatography on a Nucleosil-100 C18 column (particle size, 5 μm; column, 250 by 4.0 mm) with a gradient of 0.1% (vol/vol) formic acid (eluant A) and acetonitrile (eluant B) in a range of 27 to 100% (vol/vol) eluant B and at a flow rate of 1 ml/min. For quantification, all intermediates were calibrated with external standards. The compounds were identified by their retention times, as well as the corresponding spectra, which were recorded with a diode array detector (WellChrom Diodenarray-Detektor K-2150; Knauer).
Isolation, manipulation, analysis, and transfer of DNA.
All genetic techniques were performed as described by Sambrook et al. (25). DNA sequences of constructed hybrid plasmids were confirmed by DNA sequencing according to the chain termination method, using an automatic sequencer (Li-Cor model 4000L; MWG-Biotech, Ebersberg, Germany).
Isolation of total RNA and cloning of vanillyl alcohol oxidase gene vaoA.
Total RNA was isolated from P. simplicissimum CBS170.90 cells after 48 h of growth on minimal medium containing veratryl alcohol as inducer of vanillyl alcohol oxidase expression using a Qiagen RNeasy minikit. The vaoA comprising cDNA that encoded vanillyl alcohol oxidase was amplified in a reverse transcription-PCR using the primers vaoU (5′-AAAAGAGCTCTAAGGAGGTGACAACATGTCCAAGACACAGGAATTTAGGCC-3′) and vaoD (5′-AAAAGCGGCCGCTTACAGTTTCCAAGTAACATGACTG-3′), which were designed from the published vaoA sequence (3). These primers were used together with the isolated total RNA applying a Qiagen OneStep reverse transcription-PCR kit. For an enhanced expression of the vaoA gene in E. coli, a consensus Shine-Dalgarno sequence (TAAGGAGGTGA) was introduced four nucleotides from the vaoA start codon in the upstream primer vaoU. The PCR product was digested with SacI (site underlined in the upstream primer) and NotI (site underlined in the downstream primer) and cloned in pBluescript SK(−). In the resulting hybrid plasmid pSKvao, the vaoA gene was colinear to and downstream of the lacZ promoter of the vector.
PCR-directed mutagenesis for elimination of interfering restriction sites.
The vaoA gene contained an EcoRI restriction site at position 70 (referring to the published coding region) (3) and a BamHI site at position 1608, while calA contained an EcoRI site at position 462. For further subcloning of vaoA and calA, it was necessary to eliminate these interfering restriction sites. Because of the terminal positions of the EcoRI and BamHI restriction sites in vaoA, these two sites were eliminated using the primers vaoU-m (5′-AAAAGAGCTCTAAGGAGGTGAAACATGTCCAAGACACAGGAATTTAGGCCTTTGACACTGCCACCCAAGCTGTCGTTAAGTGACTTCAATGAGTTCATC-3′) and vaoD-m (5′-AAAAGCGGCCGCTTACAGTTTCCAAGTAACATGACTGTATTGACTCGGCCAAACACCAGACTTTCCCGGGGCAATGATGCCATTAGGGTCCACCG-3′) in a PCR with pSKvao as the template DNA. The recognition sequences of the restriction enzymes were mutated by silent nucleotide exchanges in the primers used (underlined). The PCR product was digested with SacI and NotI and cloned in pBluescript SK(−), resulting in pSKvaom.
The EcoRI restriction site in calA was eliminated by using a unique site elimination mutagenesis kit from Amersham Pharmacia (Freiburg, Germany) with the primers calAm (5′-CCCCGAGTTCTGCTACCAGTATTT-3′) and Pmut (5′-GAACTCGATATCAAGCTTATCGGTAC-3′), resulting in hybrid plasmid pSKcalAm.
Construction of hybrid plasmid pSKvaomPcalAmcalB.
The coniferyl aldehyde dehydrogenase gene calB was isolated from SpeI-BamHI-digested pSKcalB, which has been described recently (19), and was subsequently cloned in SpeI-BamHI-digested pSKcalAm, resulting in hybrid plasmid pSKcalAmcalB. In order to increase the activities of the coniferyl alcohol and coniferyl aldehyde dehydrogenases in corresponding recombinant E. coli strains, an additional promoter was integrated upstream of calAm. This was achieved by amplification of calAmcalB together with the lacZ promoter in a PCR using the primers lacZU (5′-AAAAGGATCCACCGAGCGCAGCGAGTCAGTG-3′) and calBD2 (5′-AAAAAGAATTCCCACTACCAACGGTTCTAACACTCCG-3′). The PCR product was digested with BamHI (site underlined in the upstream primer) and EcoRI (site underlined in the downstream primer) and cloned in BamHI-EcoRI-digested pSKvaom, resulting in hybrid plasmid pSKvaomPcalAmcalB.
Enzyme assays.
Cells of recombinant strains of E. coli were grown for 12 h at 37°C in LB or TB in the presence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested by centrifugation and resuspended in a suitable buffer with respect to the subsequent enzyme assay. Disruption of the cells was performed either by a twofold French press passage at 96 MPa or by sonication (1 min/ml of cell suspension with an amplitude of 40 μm) with a Bandelin Sonopuls GM200 ultrasonic disintegrator. The soluble fractions of crude extracts were obtained by centrifugation at 100,000 × g at 4°C for 1 h. Vanillyl alcohol oxidase, coniferyl alcohol dehydrogenase, and coniferyl aldehyde dehydrogenase were measured in optical enzymatic tests as described previously (1, 3, 19). The amount of soluble protein was determined as described by Bradford (4).
Cultivation on a 30-liter scale.
A Biostat DL30 stainless steel reactor (B. Braun Biotech International, Melsungen, Germany) with a total volume of 42 liters (28-cm inner diameter and 71-cm height) and a relation of stirrer diameter to vessel diameter of 0.375 was used for cultivations on the 30-liter scale. This bioreactor was equipped with three stirrers, each containing six paddles and a Funda-Foam mechanical foam destroyer (B. Braun Biotech International). In addition, sterilizable probes were used to measure dissolved oxygen (pO2) (model 25; Mettler-Toledo GmbH, Steinbach, Switzerland), pH (model Pa/25; Mettler-Toledo GmbH), foam (model L300/Rd. 28; B. Braun Biotech International), temperature (pt 100 electrode; M. K. Juchheim GmbH, Fulda, Germany), and optical density at 850 nm (model CT6; Sentex/Monitek Technology Inc.). The fermentation process was controlled and recorded by a digital control unit in combination with the MFCS/win software package (B. Braun Biotech International).
Carbon dioxide and oxygen concentrations in the exhaust gas were measured with a URAS 10 P NDIR spectrophotometer (Mannesmann, Hartmann and Braun, Frankfurt, Germany) or a Magnos 6 G oxygen analyzer (Mannesmann, Hartmann and Braun), respectively. Cultivations were performed at 30°C and at a pO2 range of 0 to 100% air saturation of the medium, which was controlled by agitation rates between 200 and 500 rpm and aeration rates of 0.4 and 1.0 volume per volume per minute. The pH in the medium was controlled between 7.1 and 7.3 by addition of 4 N HCl or NaOH. If the mechanical foam destroyer insufficiently removed foam, the antifoam agent Silikon Antischaum Emulsion SLE (Wacker, Darwin Vertriebs GmbH, Ottobrunn, Germany) was added. Small cell-free samples of the culture fluid for analytical purposes were prepared by 10 min of centrifugation at 3,500 × g.
Materials.
Restriction endonucleases, T4 DNA ligase, lambda DNA, and substrates used in the enzyme assays were obtained either from Roche Molecular Biochemicals (Mannheim, Germany) or from Invitrogen (Karlsruhe, Germany). Agarose type NA was purchased from Amersham Pharmacia Biotech (Freiburg, Germany). Synthetic oligonucleotides were purchased from MWG-Biotech. All other chemicals were from Merck Eurolab (Darmstadt, Germany), Serva Feinbiochemica (Heidelberg, Germany), or Sigma-Aldrich Fine Chemicals (Deisenhofen, Germany).
Nucleotide sequence accession numbers.
The nucleotide sequence accession numbers of the calA (named CADH in the patent), calB, and vao genes are A92130, AJ006231, and Y15627, respectively.
RESULTS
Tolerance of E. coli XL1-Blue towards different eugenol concentrations.
E. coli is able neither to use the aromatic substrates eugenol or ferulic acid as the carbon source for growth nor to convert or degrade ferulic acid. Thus, it is an eligible host for establishing a biochemical pathway enabling biotransformation of eugenol to ferulic acid.
Eugenol is a highly toxic substance for most bacteria (11). To investigate the eugenol tolerance of the selected host E. coli XL1-Blue, several 50-ml cultures in 250-ml Erlenmeyer flasks without baffles containing LB medium and different concentrations of eugenol ranging from 0.01% (vol/vol) up to 0.1% (vol/vol) were inoculated with an overnight preculture. The cultures were incubated for 48 h at 37°C, and the optical densities of the cultures at 600 nm were determined daily. As shown in Table 2, growth of E. coli XL1-Blue was only slightly affected by eugenol concentrations of up to 0.025% (vol/vol). There was a notable inhibition of E. coli XL1-Blue at a eugenol concentration of 0.05% (vol/vol), and the strain was no longer able to grow at concentrations higher than 0.075% (vol/vol).
TABLE 2.
Tolerance by E. coli XL1-Blue of different eugenol concentrationsa
| Eugenol concn (% [vol/vol]) | OD600 after an incubation period ofb:
|
||
|---|---|---|---|
| 0 h | 24 h | 48 h | |
| 0 | 0.07 | 3.5 | 3.4 |
| 0.010 | 0.08 | 3.1 | 2.9 |
| 0.025 | 0.08 | 2.6 | 2.4 |
| 0.050 | 0.08 | 0.07 | 0.7 |
| 0.075 | 0.07 | 0.06 | 0.05 |
| 0.100 | 0.07 | 0.05 | 0.05 |
Cells of E. coli XL1-Blue were precultured overnight in LB medium. The cells were used for inoculation of a series of 50-ml cultures in 250-ml unbaffled Erlenmeyer flasks containing LB medium and different concentrations of eugenol.
Data are means for three independent determinations. OD600, optical density at 600 nm.
Cloning and functional expression of the vanillyl alcohol oxidase gene (vaoA) from P. simplicissimum CBS 170.90 in E. coli XL1-Blue.
In initial experiments, ehyAB, calA, and calB, encoding eugenol hydroxylase, coniferyl alcohol dehydrogenase, and coniferyl aldehyde dehydrogenase in Pseudomonas sp. strain HR199, respectively, were expressed in E. coli to establish conversion of eugenol to ferulic acid. However, no biotransformation was achieved with this strain, although the corresponding enzyme activities were detectable in protein extracts of the recombinant cells in vitro (data not shown). Even the coexpression of azu from Pseudomonas sp. strain OPS1, which encodes the potential physiological electron acceptor of the eugenol hydroxylase (5), was in vain (data not shown). Thus, the initial reaction leading from eugenol to coniferyl alcohol could not be established by expression of the eugenol hydroxylase genes in E. coli.
Since the hydroxylation of eugenol to coniferyl alcohol is also catalyzed by vanillyl alcohol oxidase (VaoA) from P. simplicissimum CBS 170.90 (8, 10), the corresponding gene vaoA was chosen to establish the initial eugenol biotransformation step in E. coli. vaoA was amplified as cDNA and cloned in pBluescript SK(−), and site-specific silent mutations for elimination of an EcoRI and a BamHI restriction site in the gene were introduced as described in Materials and Methods. The resulting hybrid plasmid pSKvaom, harboring vaoA colinear to and downstream of the lacZ promoter, conferred vanillyl alcohol oxidase activity to recombinant strains of E. coli XL1-Blue (Table 3). Furthermore, with IPTG-induced resting cells of E. coli XL1-Blue(pSKvaom), a conversion of 3.25 mM eugenol (0.05% [vol/vol]) to coniferyl alcohol was obtained within 1 h. In addition, coniferyl aldehyde was detected in the medium, which was most probably due to an unspecific alcohol dehydrogenase activity of the host cells.
TABLE 3.
Organization of genes in hybrid plasmids and in vitro enzyme activitiesa
| Hybrid plasmid | Organization of genesb | In vitro activity (U/mg of protein) of:
|
Reference | ||
|---|---|---|---|---|---|
| Vanillyl alcohol oxidase | Coniferyl alcohol dehydrogenase | Coniferyl aldehyde dehydrogenase | |||
| pSKvao | 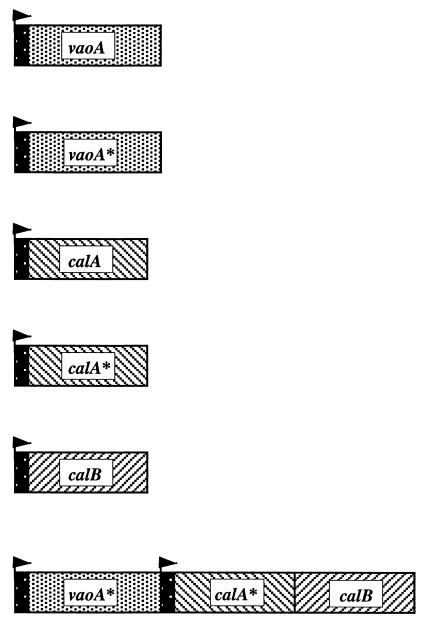 |
0.02 ± 0.01 | NDc | ND | This study |
| pSKvaom | 0.02 ± 0.01 | ND | ND | This study | |
| pSKcalA | ND | 1.16 ± 0.05 | ND | 19 | |
| pSKcalAm | ND | 1.16 ± 0.05 | ND | This study | |
| pSKcalB | ND | ND | 0.06 ± 0.01 | 19 | |
| pSKvaomPcalAmcalB | 0.02 ± 0.01 | 3.7 ± 0.15 | 0.28 ± 0.02 | This study | |
Cells were grown and enzyme activities were determined as described in Materials and Methods. Experiments and enzyme assays were done in triplicate, and the values are means ± deviations calculated by using standard statistics (P < 0.05).
Dark areas with arrows represent lac promoters.
ND, not determined.
Since the expression of vaoA enabled E. coli XL1-Blue(pSKvaom) to convert eugenol to coniferyl alcohol, coexpression of vaoA, calA, and calB was expected to enable E. coli to convert eugenol to ferulic acid efficiently. For this reason, the hybrid plasmid pSKvaomPcalAmcalB was constructed. Recombinant cells of E. coli XL1-Blue harboring pSKvaomPcalAmcalB were grown at 30°C, and expression of the introduced genes was induced by IPTG addition as described above. Protein extracts derived from these cells exhibited vanillyl alcohol oxidase, coniferyl alcohol dehydrogenase, and coniferyl aldehyde dehydrogenase activities, as determined by applying the corresponding enzyme assays (Table 3).
Biotransformation of eugenol to ferulic acid in 50-ml cultures.
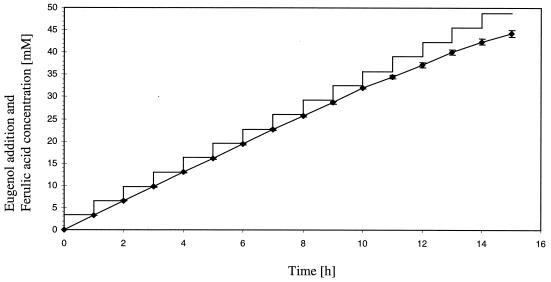
To investigate the biotransformation of eugenol to ferulic acid, cells of E. coli XL1-Blue(pSKvaomPcalAmcalB) were grown overnight at 30°C in 50 ml of TB medium containing tetracycline, ampicillin, and IPTG. When the culture entered the stationary growth phase, 25-μl portions of eugenol (final concentration, 0.05% [vol/vol]) were added hourly during a 15-h biotransformation phase, and the appearance of coniferyl alcohol, coniferyl aldehyde, and ferulic acid in the culture supernatant was analyzed by HPLC. The time course of the production of ferulic acid from eugenol is summarized in Fig. 2. Within 15 h a ferulic acid concentration of 8.6 g liter−1 was obtained, corresponding to a molar yield of 90.8% with respect to the added amount of eugenol. Cultures of E. coli XL1-Blue harboring only the vector pBluescript SK(−), which were treated in the same way as a negative control, showed no conversion of eugenol.
FIG. 2.
Production of ferulic acid from eugenol by E. coli XL1-Blue(pSKvaomPcalAmcalB). Cells were grown in 50 ml of liquid TB medium in the presence of tetracycline, ampicillin, and IPTG overnight at 30°C. Portions (25 μl) of eugenol (final concentration, 0.05% [vol/vol]) were added to the culture hourly. Samples were taken, and ferulic acid concentrations were determined by HPLC. Data are means for three independent experiments. The variance is indicated by error bars. —, total amount of added eugenol; ♦, ferulic acid concentration.
Technical-scale production of ferulic acid from eugenol.
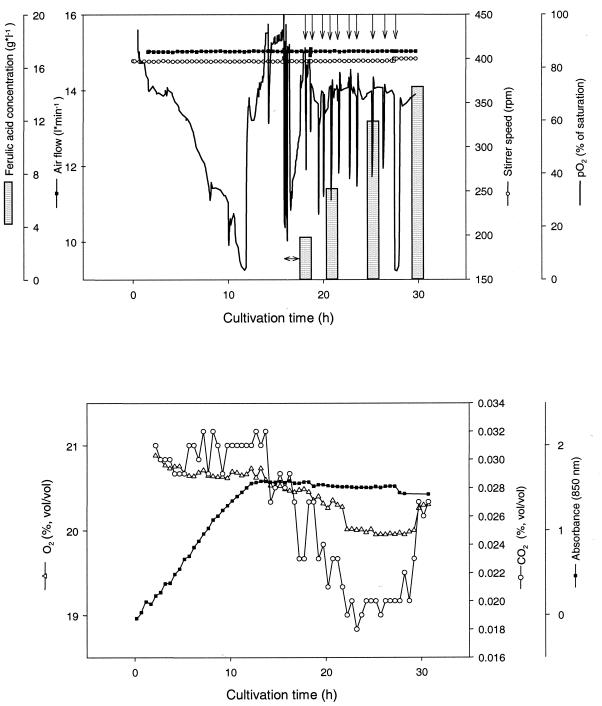
To test the capability of E. coli XL1-Blue(pSKvaomPcalAmcalB) to transform eugenol to ferulic acid in a technical-scale production process, 28 liters of TB medium containing tetracycline, ampicillin, and IPTG was inoculated with a 2-liter preculture of this strain grown overnight in the same medium. The time course of this fermentation is shown in Fig. 3. After 16 h, when the cells had been in the stationary growth phase for approximately 4 h, 100 ml of eugenol was fed to the culture continuously over 2 h, maintaining a final eugenol concentration of approximately 0.1% (vol/vol) in the medium. Subsequently, eugenol was fed in 10 pulses of 30 ml to the culture as indicated in Fig. 3. To ensure optimal conditions for the coniferyl aldehyde dehydrogenase, the temperature was decreased from 30 to 28°C at the beginning of the biotransformation. In this experiment a maximum production rate of 14.4 mmol of ferulic acid per h per liter of culture was observed in the biotransformation phase in the time frame from 0 to 90 min, when eugenol was added continuously to the medium. The maximum concentration of ferulic acid obtained was 14.7 g liter−1 after a total fermentation time of 30 h, which corresponded to a total amount of 441 g and a molar yield of 93.3% with respect to the amount of eugenol added.
FIG. 3.
Fed-batch fermentation of E. coli XL1-Blue(pSKvaomPcalAmcalB) on the 30-liter scale in TB medium. The cultivation was done in a 42-liter stirred-tank reactor, and the cultivation parameters were pH 7.2, aeration at 0.8 to 1.0 volume per volume per minute, and agitation at 200 to 400 rpm. Aeration and agitation were adjusted according to the oxygen demand of the culture. The temperature was decreased from 30 to 28°C at the beginning of the biotransformation process. Eugenol (100 ml) was added to the culture continuously over 2 h (↔), maintaining a final concentration of approximately 0.1% (vol/vol) in the medium. Subsequently, eugenol was added in pulses of 30 ml to the medium (↓).
Two-step biotransformation of eugenol to vanillin with recombinant strains of E. coli.
As described above, the quantitative conversion of eugenol to ferulic acid was achieved with E. coli XL1-Blue(pSKvaomPcalAmcalB). In a recent study, it was shown that the coexpression of fcs and ech, encoding feruloyl-CoA synthetase and enoyl-CoA hydratase/aldolase, respectively, enabled recombinant E. coli XL1-Blue strains to convert ferulic acid to vanillin (18). Thus, we established a two-step biotransformation of eugenol to vanillin with recombinant E. coli XL1-Blue strains harboring the hybrid plasmids pSKvaomPcalAmcalB and pSKechE/Hfcs (18), respectively.
Cells of E. coli XL1-Blue(pSKvaomPcalAmcalB) were incubated on the 50-ml scale as described above until a ferulic acid concentration of 5 g liter−1 was reached. Subsequently, washed cells of a 50-ml culture of E. coli XL1-Blue(pSKechE/Hfcs), which had been grown to the late exponential growth phase and induced by the addition of IPTG, were added to the culture. In order to inactivate the calB gene product, which besides coniferyl aldehyde dehydrogenase also possesses vanillin dehydrogenase activity (17), the temperature was raised to 37°C. Further incubation of this mixed culture and analysis of culture supernatants via HPLC revealed a production of 0.3 g of vanillin liter−1 after 2 h. At this time point, 4.6 g of ferulic acid liter−1 and 0.1 g of vanillyl alcohol liter−1 were detected in the culture supernatant. During further incubation, however, the vanillin produced was completely reduced to vanillyl alcohol and no more ferulic acid was converted to vanillin, indicating the loss of biotransformation capability of this mixed culture.
DISCUSSION
With an increasing interest in natural products, biotechnological production of vanillin has gained more interest in recent years, since it offers an alternative way to produce this aromatic substance as a “natural” flavor compound. Until today, only the biotransformation of ferulic acid to vanillin had been developed to an economically feasible process. Using Amycolatopsis sp. strain HR167, 11.5 g of vanillin liter−1 was produced from 19.9 g of ferulic acid liter−1 within 32 h on a 10-liter scale, corresponding to a molar yield of 77.8% (Rabenhorst and Hopp, patent application). A biotransformation with similar efficiency based on the same substrate using S. setonii as a biocatalyst was developed (14; Müller et al., patent application). However, these biotransformations depend on the use of the expensive substrate ferulic acid.
A different source for a natural ferulic acid feedstock is eugenol, which is a commercially available natural raw material extracted from clove tree leaves. The current market price is about $5 kg−1 (22). Eugenol can be enzymatically converted to ferulic acid via coniferyl alcohol and coniferyl aldehyde. The genes ehyAB, calA, and calB, which are involved in this bioconversion, were investigated in detail in Pseudomonas sp. strain HR199 and Pseudomonas sp. strain OPS1 (1, 5, 20). Pseudomonas sp. strain HR199 has been used to produce natural ferulic acid (23); however, the yield of 6.7 g of ferulic acid liter−1 from 13.9 g of eugenol liter−1 within 78 h (R. Hopp and J. Rabenhorst, European Patent Office patent application EP0583687, 1994) was too low for a commercial process. To achieve higher yields, the said genes of Pseudomonas sp. strain HR199 were expressed in R. eutropha H16, which is not able to degrade ferulic acid (19). With this recombinant strain, 3.5 g of ferulic acid liter−1 from 3.2 g liter−1 eugenol was obtained within only 20 h, which corresponded to a molar yield of 93.8% (19). The major difficulty of this biotransformation process was the sensitivity of R. eutropha H16 to eugenol, demanding a stringent eugenol addition mode to keep the final concentration of eugenol in the medium below 3.25 mM (corresponding to 0.05% [vol/vol]).
In the present study, we used E. coli XL1-Blue as an alternative host strain for gene expression, since it exhibited a significantly higher eugenol tolerance than R. eutropha H16 (Table 2) (19). However, the expression of the Pseudomonas sp. strain HR199 genes ehyAB, calA, and calB in this strain did not lead to a suitable biocatalyst, even after coexpression of azu, encoding the putative physiological electron acceptor of eugenol hydroxylase (7). A suitable alternative for ehyAB was found in the gene vaoA from P. simplicissimum CBS 170.90, encoding vanillyl alcohol oxidase (8). This well-characterized flavoprotein has a wide substrate specificity and is able to catalyze the hydroxylation of eugenol to coniferyl alcohol in addition to the oxidation of vanillyl alcohol to vanillin and other reactions (8, 10). This enzyme has even been used for the in vitro enzymatic synthesis of vanillin (26). By expressing the vaoA gene together with calA and calB in E. coli XL1-Blue, we were able to obtain 8.6 g of ferulic acid liter−1 within 15 h at a molar yield of 90.8% with respect to the substrate eugenol. This biotransformation process was successfully transferred to the 30-liter scale, where we obtained a ferulic acid concentration of 14.7 g liter−1 after a total fermentation time of 30 h. The molar yield of this biotransformation was 93.3% and even higher than at the 50-ml scale. Thus, E. coli XL1-Blue(pSKvaomPcalAmcalB) is a highly efficient biocatalyst for producing natural ferulic acid from eugenol.
Based on this successful conversion, we established a two-step biotransformation process leading from eugenol to vanillin. In a recent study it was shown that the coexpression of the fcs and ech genes of Pseudomonas sp. strain HR199, encoding feruloyl-CoA synthetase and enoyl-CoA hydratase/aldolase, respectively, enabled recombinant E. coli strains to convert ferulic acid to vanillin (18). The sequential application of the two recombinant E. coli strains harboring the hybrid plasmids pSKvaomPcalAmcalB and pSKechE/Hfcs initially led to the production of 0.3 g of vanillin liter−1. However, during further incubation vanillin was reduced to vanillyl alcohol and no more ferulic acid was converted to vanillin. The reduction of vanillin represents a detoxification mechanism, which was also observed with E. coli XL1-Blue cells during incubation with vanillin (17).
Acknowledgments
Skillful assistance by Simone Diniz during fermentation and Christian Ewering for computer problems is gratefully acknowledged.
REFERENCES
- 1.Achterholt, S., H. Priefert, and A. Steinbüchel. 1998. Purification and characterization of the coniferyl aldehyde dehydrogenase from Pseudomonas sp. strain HR199 and molecular characterization of the gene. J. Bacteriol. 180:4387-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achterholt, S., H. Priefert, and A. Steinbüchel. 2000. Identification of Amycolatopsis sp. strain HR167 genes, involved in the bioconversion of ferulic acid to vanillin. Appl. Microbiol. Biotechnol. 54:799-807. [DOI] [PubMed] [Google Scholar]
- 3.Benen, J. A. E., P. Sanchez-Torres, M. J. M. Wagemaker, M. W. Fraaije, W. J. van Berkel, and J. Visser. 1998. Molecular cloning, sequencing, and heterologous expression of the vaoA gene from Penicillium simplicissimum CBS 170.90 encoding vanillyl-alcohol oxidase. J. Biol. Chem. 273:7865-7872. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Brandt, K., S. Thewes, J. Overhage, H. Priefert, and A. Steinbüchel. 2001. Characterization of the eugenol hydroxylase genes (ehyA/ehyB) from the new eugenol degrading Pseudomonas sp. strain OPS1. Appl. Microbiol. Biotechnol. 56:724-730. [DOI] [PubMed] [Google Scholar]
- 6.Bullock, W. O., J. M. Fernandez, and J. M. Stuart. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 7.Causer, M. J., D. J. Hopper, W. S. McIntire, and T. P. Singer. 1984. Azurin from Pseudomonas putida: an electron acceptor for p-cresol methylhydroxylase. Biochem. Soc. Trans. 12:1131-1132. [Google Scholar]
- 8.De Jong, E., W. J. van Berkel, R. P. van der Zwan, and J. A. de Bont. 1992. Purification and characterization of vanillyl-alcohol oxidase from Penicillium simplicissimum. A novel aromatic alcohol oxidase containing covalently bound FAD. Eur. J. Biochem. 208:651-657. [DOI] [PubMed] [Google Scholar]
- 9.Fraaije, M. W., and W. J. van Berkel. 1997. Catalytic mechanism of oxidative demethylation of 4-(methoxymethyl)phenol by vanillyl-alcohol oxidase. Evidence for formation of a p-quinone methide intermediate. J. Biol. Chem. 272:18111-18116. [DOI] [PubMed] [Google Scholar]
- 10.Fraaije, M. W., C. Veeger, and W. J. van Berkel. 1995. Substrate specificity of flavin-dependent vanillyl alcohol oxidase from Penicillium simplicissimum. Evidence for the production of 4-hydroxycinnamyl alcohols from 4-allylphenols. Eur. J. Biochem. 234:271-277. [DOI] [PubMed] [Google Scholar]
- 11.Friedman, M., P. R. Henika, and R. E. Mandrell. 2002. Bactericidal activities of plant essential oils and some of their isolated constituents against Campylobacter jejuni, Escherichia coli, Listeria monocytogenes, and Salmonella enterica. J. Food Prot. 65:1545-1560. [DOI] [PubMed] [Google Scholar]
- 12.Furukawa, H., M. Wieser, H. Morita, T. Sugo, and T. Nagasawa. 1998. Purification and characterization of eugenol dehydrogenase from Pseudomonas fluorescens E118. Arch. Microbiol. 171:37-43. [DOI] [PubMed] [Google Scholar]
- 13.Gasson, M. J., Y. Kitamura, W. R. McLauchlan, A. Narbad, A. J. Parr, E. L. Parsons, J. Payne, M. J. Rhodes, and N. J. Walton. 1998. Metabolism of ferulic acid to vanillin. A bacterial gene of the enoyl-SCoA hydratase/isomerase superfamily encodes an enzyme for the hydration and cleavage of a hydroxycinnamic acid SCoA thioester. J. Biol. Chem. 237:4163-4170. [DOI] [PubMed] [Google Scholar]
- 14.Muheim, A., and K. Lerch. 1999. Towards a high-yield bioconversion of ferulic acid to vanillin. Appl. Microbiol. Biotechnol. 51:456-461. [Google Scholar]
- 15.Narbad, A., and M. J. Gasson. 1998. Metabolism of ferulic acid via vanillin using a novel CoA dependent pathway in a newly-isolated strain of Pseudomonas fluorescens. Microbiology 144:1397-1405. [DOI] [PubMed] [Google Scholar]
- 16.Overhage, J., A. U. Kresse, H. Priefert, H. Sommer, G. Krammer, J. Rabenhorst, and A. Steinbüchel. 1999. Molecular characterization of the genes pcaG and pcaH, encoding protocatechuate 3,4-dioxygenase, which are essential for vanillin catabolism in Pseudomonas sp. strain HR199. Appl. Environ. Microbiol. 65:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overhage, J., H. Priefert, J. Rabenhorst, and A. Steinbüchel. 1999. Biotransformation of eugenol to vanillin by a mutant of Pseudomonas sp. strain HR199 constructed by disruption of the vanillin dehydrogenase (vdh) gene. Appl. Microbiol. Biotechnol. 52:820-828. [DOI] [PubMed] [Google Scholar]
- 18.Overhage, J., H. Priefert, and A. Steinbüchel. 1999. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl. Environ. Microbiol. 65:4837-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Overhage J., A. Steinbüchel, and H Priefert. 2002. Biotransformation of eugenol to ferulic acid by a recombinant strain of Ralstonia eutropha H16. Appl. Environ. Microbiol. 68:4315-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priefert, H., J. Overhage, and A. Steinbüchel. 1999. Identification and molecular characterization of the eugenol hydroxylase genes (ehyA/ehyB) of Pseudomonas sp. strain HR199. Arch. Microbiol. 172:354-363. [DOI] [PubMed] [Google Scholar]
- 21.Priefert, H., J. Rabenhorst, and A. Steinbüchel. 1997. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 179:2595-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Priefert, H., J. Rabenhorst, and A. Steinbüchel. 2001. Biotechnological production of vanillin. Appl. Microbiol. Biotechnol. 56:296-314. [DOI] [PubMed] [Google Scholar]
- 23.Rabenhorst, J. 1996. Production of methoxyphenol type natural aroma chemicals by biotransformation of eugenol with a new Pseudomonas sp. Appl. Microbiol. Biotechnol. 46:470-474. [Google Scholar]
- 24.Rosazza, J. P. N., Z. Huang, L. Dostal, T. Volm, and B. Rousseau. 1995. Review. Biocatalytic transformations of ferulic acid: an abundant aromatic natural product. J. Ind. Microbiol. 15:457-471. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.van de Heuvel, R. H., M. W. Fraaije, C. Laane, and W. J. van Berkel. 2001. Enzymatic synthesis of vanillin. J. Agric. Food Chem. 49:2954-2958. [DOI] [PubMed] [Google Scholar]