Abstract
The guts of soil-feeding macroinvertebrates contain a complex microbial community that is involved in the transformation of ingested soil organic matter. In a companion paper (T. Lemke, U. Stingl, M. Egert, M. W. Friedrich, and A. Brune, Appl. Environ. Microbiol. 69:6650-6658, 2003), we show that the gut of our model organism, the humivorous larva of the cetoniid beetle Pachnoda ephippiata, is characterized by strong midgut alkalinity, high concentrations of microbial fermentation products, and the presence of a diverse, yet unstudied microbial community. Here, we report on the community structure of bacteria and archaea in the midgut, hindgut, and food soil of P. ephippiata larvae, determined with cultivation-independent techniques. Clone libraries and terminal restriction fragment length polymorphism analysis of 16S rRNA genes revealed that the intestines of P. ephippiata larvae contain a complex gut microbiota that differs markedly between midgut and hindgut and that is clearly distinct from the microbiota in the food soil. The bacterial community is dominated by phylogenetic groups with a fermentative metabolism (Lactobacillales, Clostridiales, Bacillales, and Cytophaga-Flavobacterium-Bacteroides [CFB] phylum), which is corroborated by high lactate and acetate concentrations in the midgut and hindgut and by the large numbers of lactogenic and acetogenic bacteria in both gut compartments reported in the companion paper. Based on 16S rRNA gene frequencies, Actinobacteria dominate the alkaline midgut, while the hindgut is dominated by members of the CFB phylum. The archaeal community, however, is less diverse. 16S rRNA genes affiliated with mesophilic Crenarchaeota, probably stemming from the ingested soil, were most frequent in the midgut, whereas Methanobacteriaceae-related 16S rRNA genes were most frequent in the hindgut. These findings agree with the reported restriction of methanogenesis to the hindgut of Pachnoda larvae.
Saprophagous macroinvertebrates, such as earthworms, termites, and many coleopteran and dipteran larvae, play a major role in the degradation and stabilization of soil organic matter (SOM) and strongly influence important physical and chemical soil parameters (34, 61). The transformation of SOM during passage through the alkaline guts of humivorous insects has so far been investigated only with soil-feeding termites (for reviews see references 5, 6, and 8). It has been shown that the conditions in the anterior hindgut of Cubitermes spp. (high alkalinity and oxygen influx) enhance the extraction of organic matter from the inorganic matrix, cause chemical oxidation of humic substances, and lead to a decrease of the molecular weight of the organic matter (30). The resulting increase in solubility renders the organic matter accessible for digestion in subsequent, less-alkaline compartments (27, 28, 31). Although the complex microbial community in the guts of humivorous macroinvertebrates is believed to participate in the transformation of ingested SOM (10, 29), detailed information on the composition and activities of the gut microbiota is lacking.
To elucidate the contribution of gut microorganisms to digestion of SOM and host nutrition, information about structure and function of the microbial gut community has to be linked to gut morphology, the physicochemical conditions in different gut compartments, and the spatial distribution of the microorganisms (8, 9). Due to difficulties in simulating the microenvironment of an insect gut system, which is characterized by steep and overlapping O2, H2, pH, and redox gradients (8), culture-based approaches do not adequately reflect the microbial diversity in the guts of soil feeders, and it is necessary to study the structure of microbial gut communities with culture-independent techniques (2). To date, such studies have been restricted to only a few arthropods, mostly termites (7, 19, 25, 32, 37, 44, 45, 53), where especially the combination of clonal and terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes has provided a deeper insight into the gut microbial community of soil-feeding termites and its dependence on axial gut differentiation (19, 51, 52).
The gut of our model organism, the humivorous larva of the cetoniid beetle Pachnoda ephippiata (Coleoptera: Scarabaeidae), resembles the termite gut with respect to strong midgut alkalinity, high concentrations of microbial fermentation products, and methane emission from the hindgut (see the companion paper [35]). Like the termite gut, the Pachnoda gut is also characterized by a dense and diverse microbial community (11, 35), which is obviously involved in digestion, but whose exact composition and role remain to be studied.
In order to gain more insight into the role of the gut microbiota of scarab beetles in the transformation of SOM, we used a culture-independent approach, combining a clonal analysis and T-RFLP fingerprinting analysis of 16S rRNA genes, to investigate the bacterial and archaeal community structure in midgut and hindgut of P. ephippiata larvae and to compare it to that in the soil consumed by the larvae.
MATERIALS AND METHODS
DNA extraction and purification.
Larvae of P. ephippiata (second instars) were taken from a controlled breeding facility at the University of Konstanz and dissected as described in the companion paper (35). Guts were separated into midgut and hindgut and stored frozen at −20°C until further analysis. To minimize the influence of differences between individual larvae, DNA was extracted from four pooled gut sections each and food soil (0.5 g), according to a direct lysis protocol that has been previously described in detail (23). DNA was purified from the supernatant by consecutive ammonium acetate, isopropanol, and ethanol precipitation steps. To remove humic substances, extracts were passed through spin columns filled with polyvinylpolypyrrolidone as described previously (48). The protocol was further improved by using a swinging bucket rotor (A-8-11 for Eppendorf centrifuge 5417 R; Eppendorf, Hamburg, Germany) so that the amount of polyvinylpolypyrrolidone suspension necessary for sufficient purification could be reduced to ≤500 μl. Extraction efficiency and quality of extracted DNA were verified by standard gel electrophoresis.
PCR amplification of bacterial and archaeal 16S rRNA genes.
16S rRNA genes were specifically amplified from gut DNA extracts with the primer combination 27f (5′-AGA-GTT-TGA-TCC-TGG-CTC-AG-3′) (17) and 1492r (5′-TAC-GGY-TAC-CTT-GTT-ACG-ACT-T-3′) (33) targeting Bacteria and Ar109f (5′-ACK-GCT-CAG-TAA-CAC-GT-3′) (20) and Ar912r (5′-CTC-CCC-CGC-CAA-TTC-CTT-TA-3′) (40) targeting Archaea. The reaction mixture contained, in a total volume of 50 μl, 1× PCR buffer II (Applied Biosystems, Weiterstadt, Germany), 1.5 mM MgCl2, 50 μM concentrations of each of the four deoxynucleoside triphosphates (Amersham Pharmacia Biotech, Freiburg, Germany), 0.5 μM concentrations of each primer (MWG Biotech, Ebersberg, Germany), 1.25 U of AmpliTaq DNA polymerase (Applied Biosystems), and 1 μl of a 1:30 dilution of the gut DNA extract. All reaction mixtures were prepared at 4°C in 0.2-ml reaction tubes to avoid nonspecific priming. Amplification was started by placing the reaction tubes immediately into the preheated (94°C) block of a GeneAmp 9700 Thermocycler (Applied Biosystems). The standard thermal profiles for the amplification of 16S rRNA genes were as follows: initial denaturation (94°C, 3 min); 32 cycles of denaturation (94°C, 30 s), annealing (55°C, 30 s), and extension (72°C, 60 s) for Bacteria; and 35 cycles of denaturation (94°C, 45 s), annealing (52°C, 45 s), and extension (72°C, 90 s) for Archaea. After terminal extension (72°C, 5 to 7 min), samples were stored at 4°C until further analysis. Aliquots (5 μl) of 16S rRNA gene amplicons were analyzed by gel electrophoresis on 1% agarose gels and visualized after staining with ethidium bromide. PCR products were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany).
16S rRNA gene libraries.
Bacterial and archaeal 16S rRNA gene amplicons were cloned in Escherichia coli JM109 with the pGEM-T Easy Vector system (Promega, Mannheim, Germany) according to the manufacturer's instructions. Randomly selected clones were checked for correct insert size via standard vector-targeted PCR and gel electrophoresis. DNA was sequenced on an ABI Prism 377 DNA sequencer with Big Dye terminator chemistry as specified by the manufacturer (Applied Biosystems). Bacterial clones were designated PeM (midgut section; 60 clones) and PeH (hindgut section; 53 clones), and archaeal clones were designated PeMAr (midgut section; 24 clones) and PeHAr (hindgut section; 44 clones). Clones from food soil (only Archaea) were named FSAr (24 clones).
T-RFLP analysis.
For T-RFLP analysis, 16S rRNA genes were PCR amplified as described above, except that for bacterial clones, 6-carboxyfluorescein-labeled 27f forward primer, nonlabeled 907r (5′-CCG-TCA-ATT-CCT-TTR-AGT-TT-3′) (42) backward primer, and an annealing temperature of 52°C were used. For archaeal clones, primer Ar912r was 6-carboxyfluorescein labeled. DNA concentrations in the PCR products were determined photometrically. For digestion, ∼75 ng of DNA, 2.5 U of restriction enzyme (MspI for Bacteria and TaqI or AluI for Archaea; Promega), 1 μl of the appropriate 10× incubation buffer (Promega), and 1 μg of bovine serum albumin were combined in a total volume of 10 μl and digested for 3 h at 65°C (TaqI) or 37°C (MspI and AluI). Fluorescently labeled terminal restriction fragments (T-RFs) were size separated on an ABI 373A automated sequencer (Applied Biosystems) with an internal size standard (GeneScan-1000 ROX; Applied Biosystems). T-RFLP electropherograms were analyzed with GeneScan 2.1 software (Applied Biosystems). To determine the relative 16S rRNA gene frequency, the fluorescence intensity of each individual peak, i.e., a single T-RF, expressed as single peak height, was compared to the total fluorescence intensity of all T-RF peaks, expressed as total peak height.
Phylogenetic analysis.
Sequence data were analyzed and trees were constructed by using the ARB software package with its database (version 2.5b; O. Strunk and W. Ludwig, Technische Universität München, Munich, Germany; http://www.arb-home.de). 16S rRNA gene sequences were added to the database and aligned with the Fast Aligner tool (version 1.03). Alignments were corrected manually if necessary. All clonal 16S rRNA gene sequences were compared to sequences in public databases by using BLAST (1), and closely related sequences from databases were retrieved and added to the alignment. Trees were constructed using the neighbor-joining algorithm (50) and base frequency filters (50 to 100% similarity) for Bacteria or Archaea provided with the ARB package. For bacterial and archaeal tree construction, 1,198 sequence positions (E. coli numbering 59 to 1454) and 659 sequence positions (E. coli numbering 109 to 894), respectively, were used. Sequences of species closely related to the gut clones but significantly shorter than 1,500 bp (Bacteria) or 900 bp (Archaea) were added to the tree by using the ARB parsimony tool, which allows the addition of short sequences to phylogenetic trees without changing global tree topologies (39). The terminal sequence positions at the 5′ and 3′ ends of the 16S rRNA gene sequences (450 bp for archaeal clones and 750 bp for bacterial clones) were also subjected to a separate treeing analysis (“fractional treeing” [38]); significant differences in the phylogenetic placement of a fragment pair were considered indicative of chimera formation, and chimeric rRNA gene clones were excluded from further phylogenetic analysis.
Ecological indices.
Bacterial and archaeal species richness in midgut and hindgut clone libraries was estimated using Chao1 (13) as a nonparametric indicator, calculated with EstimateS (version 5.0.1; R. Colwell, University of Connecticut [http://viceroy.eeb.uconn.edu/estimates]) as described in reference 26. For this purpose, a “species” was defined as a group of 16S rRNA gene clones with ≥97% sequence similarity (55). To determine the similarity of the compositions of different microbial communities based on T-RFLP analysis, Morisita community similarity indices (IM) were calculated as described by Dollhopf et al. (16). For this purpose, only T-RFs with a relative peak height of ≥1% of the total electropherogram peak height were used. IM values range from 0 to 1, with 1 indicating complete identity of the two compared communities.
Nucleotide sequence accession number.
16S rRNA gene sequences of clones from the gut of P. ephippiata larvae and food soil are accessible under numbers AJ576118 to AJ576204 (archaeal clones) and AJ576323 to AJ576428 (bacterial clones). Clones PeM75, PeH59, and PeMAr04 are accessible under numbers AJ538350 to AJ538352, respectively.
RESULTS
16S rRNA gene clone libraries.
Four separate clone libraries were generated from the 16S rRNA gene fragments amplified from midgut and hindgut DNA with Bacteria-specific and Archaea-specific primer pairs, respectively. An additional archaeal clone library was generated using the DNA extracted from the food soil.
From the bacterial clone libraries, randomly selected clones from the midgut (60 clones) and hindgut (53 clones) were sequenced. Phylogenetic analysis revealed that most clones were affiliated with known taxa of the Bacteria. Four midgut clones and one hindgut clone were identified as chimeras and were excluded from further analysis.
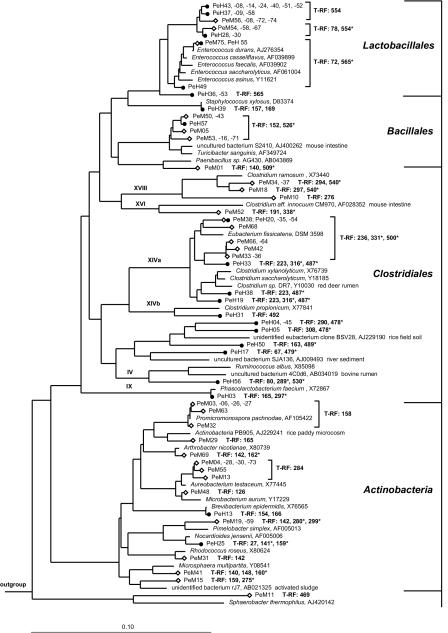
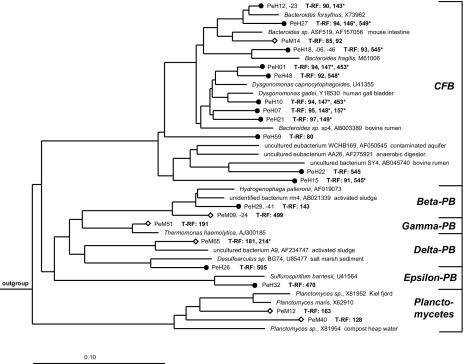
The remaining 56 clones from the midgut clone library could be assigned to 10 distinct phylogenetic groups (Fig. 1 and 2). Most clones (36%) were affiliated with Actinobacteria, followed by clones related to Clostridiales, Lactobacillales, and Bacillales (Table 1). Six midgut clones were closely related (96% sequence similarity) to Turicibacter sanguinis, a recently described strictly anaerobic gram-positive bacterium isolated from a blood sample of a human with acute appendicitis (4). Some of the midgut clones clustered with Proteobacteria from the β, γ, and δ subdivisions; clones related to members of the Cytophaga-Flavobacterium-Bacteroides (CFB) phylum, Planctomycetales, and Sphaerobacter thermophilus were rare (Table 1). Clone PeM47, not displayed in the figures, grouped with a human mouth clone (BS003) from the TM7 phylum (46). The 52 hindgut clones were mostly affiliated with Lactobacillales, Clostridiales, and members of the CFB phylum (Fig. 1 and 2). Clones related to Proteobacteria (β, δ, and ɛ subdivisions), Actinobacteria, and Bacillales were recovered less frequently (Table 1).
FIG. 1.
Phylogenetic tree showing the positions of bacterial 16S rRNA gene sequences affiliated with gram-positive bacteria, recovered from the midgut (⋄) and hindgut (•) of P. ephippiata larvae. Scale bar represents 10% sequence difference. Accession numbers of reference sequences are indicated. Species used as the outgroup were Thermus thermophilus (M26923), Fervidobacterium gondwanense (Z49117), and Thermotoga maritima (M21774). Roman numerals indicate clostridial subgroups sensu (14). Lengths of T-RFs result from in vitro digestion of clonal 16S rRNA gene amplicons with MspI; pseudo-T-RFs are marked with asterisks.
FIG. 2.
Phylogenetic tree showing the positions of bacterial 16S rRNA gene sequences affiliated with the CFB phylum, the Proteobacteria (PB), and the Planctomycetales, recovered from the midgut (⋄) and hindgut (•) of P. ephippiata larvae. Scale bar represents 10% sequence difference. Accession numbers of reference sequences are indicated. Species used as the outgroup were Thermus thermophilus (M26923), Fervidobacterium gondwanense (Z49117), and Thermotoga maritima (M21774). Lengths of T-RFs result from in vitro digestion of clonal 16S rRNA gene amplicons with MspI; pseudo-T-RFs are marked with asterisks.
TABLE 1.
Relative abundance (percent) of major phylogenetic groups in the midgut and hindgut of P. ephippiata larvae, based on the frequencies of 16S rRNA genes in 16S rRNA gene clone libraries and on T-RFLP analysisa
| Phylogenetic group | Midgut
|
Hindgut
|
||
|---|---|---|---|---|
| Clone library | T-RFLP | Clone library | T-RFLP | |
| Actinobacteria | 35.7 | 36.9-64.0 | 3.8 | 2.0-10.4 |
| Bacillales | 12.5 | 9.4-28.1 | 3.8 | 1.3-5.9 |
| Lactobacillales | 14.3 | 7.0 | 30.8 | 16.4-20.9 |
| Clostridiales | 21.4 | 5.4-9.1 | 26.9 | 21.8-28.9 |
| CFB phylum | 1.8 | 1.7 | 26.9 | 33.7-44.0 |
| Planctomycetales | 3.6 | 0-7.3 | ND | ND |
| β-Proteobacteria | 3.6 | 0-2.1 | 3.8 | 0-4.5 |
| γ-Proteobacteria | 1.8 | 0-0.5 | ND | ND |
| δ-Proteobacteria | 1.8 | NA | 1.9 | NA |
| ɛ-Proteobacteria | ND | ND | 1.9 | NA |
| Sphaerobacter related | 1.8 | NA | ND | ND |
| TM7 phylum | 1.8 | NA | ND | ND |
For an explanation of the T-RFLP-based frequency ranges, see the text. ND, not detected; NA, not assignable.
By using an arbitrarily defined limit of 97% sequence similarity, the 16S rRNA gene clones in the bacterial midgut and hindgut clone libraries can be grouped into 29 or 33 different species, respectively. The total species richness of the midgut and hindgut communities was calculated using the data from the clone libraries and Chao1 as a nonparametric richness estimator (26). The estimated numbers of bacterial species were 96 for the midgut and 118 for the hindgut community, which means that the clonal approach detected about one-third of bacterial species actually present in the midgut and hindgut communities. Since the corresponding 95% confidence intervals were 57 and 186 for the midgut community and 72 and 214 for the hindgut community, species richness in the hindgut is not significantly higher than that in the midgut (26).
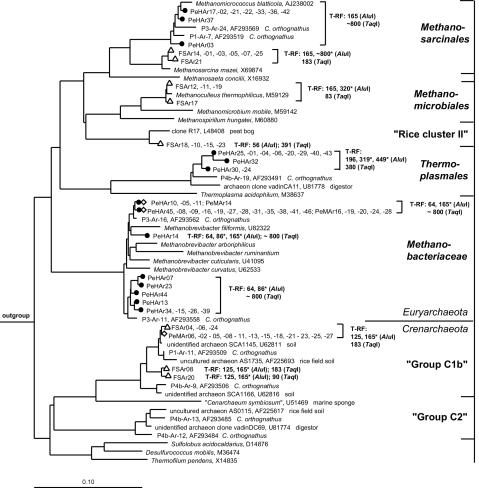
For the archaeal clone libraries, 30 randomly selected clones from the midgut, 45 clones from the hindgut, and 24 clones from the food soil were sequenced. Phylogenetic analysis revealed no chimera, but six midgut clones, one hindgut clone, and four soil clones were not affiliated with the Archaea and were excluded from further analyses. The remaining clones were all affiliated with known taxa of Euryarchaeota and Crenarchaeota (Fig. 3). Seventy-five percent of the midgut clones were affiliated with mesophilic Crenarchaeota (group C1b sensu DeLong and Pace [15]), and the remaining clones clustered with the Methanobacteriaceae (Fig. 3). In contrast, the hindgut library was dominated by Methanobacteriaceae-related clones (55%), and the remaining clones were affiliated almost in equal shares with Thermoplasmales and Methanosarcinales. The clones related to the Euryarchaeota clustered with methanogenic species isolated from other insect guts, such as Methanomicrococcus blatticola (96 to 97% sequence similarity), Methanobrevibacter filiformis (96% similarity), and Methanobrevibacter curvatus (95 to 96% similarity). Moreover, all archaeal clones obtained from the P. ephippiata gut clustered with clones obtained from the hindgut of Cubitermes orthognathus, a soil-feeding termite (19).
FIG. 3.
Phylogenetic tree showing the positions of archaeal 16S rRNA gene sequences affiliated with Euryarchaeota and Crenarchaeota, recovered from the midgut (⋄) and hindgut (•) of P. ephippiata larvae and from the soil fed to the larvae (▵). Scale bar represents 10% sequence difference. Accession numbers of reference sequences are indicated. Species used as outgroup were Nitrospira marina (X82559), Chloroflexus aurantiacus (M34116), Holophaga foetida (X77215), Rhodothermus marinus (X77140), Streptomyces coelicolor (X60514), and Acidobacterium capsulatum (D26171). Lengths of T-RFs result from in vitro digestion of clonal 16S rRNA gene amplicons with AluI or TaqI; pseudo-T-RFs are marked with asterisks.
Using a 97% sequence similarity threshold, the archaeal midgut clone library and the hindgut clone library contained two and five different species, respectively. Species richness estimation with Chao1 yielded the same result, and 95% confidence intervals of zero provided strong evidence that all archaeal species from the P. ephippiata intestine detectable with the applied primer system were represented in the clone library and that archaeal species richness in the hindgut can be regarded as being significantly higher than that in the midgut.
Thirty-five percent of the soil clones were affiliated with members of the Methanosarcinales, but in contrast to the gut clones, they were closely related to Methanosarcina thermophila. Four soil clones each (20%) were affiliated with the Methanomicrobiales and uncultured archaea from rice field soil (rice cluster II [21]). Five soil clones (25%) were affiliated with mesophilic Crenarchaeota and clustered with the clones obtained from the P. ephippiata and C. orthognathus guts.
T-RFLP analyses.
Structure and diversity of the microbial communities in the two P. ephippiata gut sections and in the food soil were also assessed directly by T-RFLP analysis of bacterial and archaeal 16S rRNA gene fragments. In silico determination of the expected size of T-RFs allowed us to assign T-RFs in the fingerprints of gut and soil communities to distinct phylogenetic groups (Fig. 4 and 5). In the context of this study, we observed that several of the clones exhibited additional peaks in the T-RFLP analysis, which we called “pseudo-T-RFs” (18). Pseudo-T-RFs are restriction fragments longer than the expected T-RF of a clone and form an additional, albeit smaller, peak in the T-RFLP profile. They are caused by partially single-stranded 16S rRNA gene amplicons, formed during the PCR, where the terminal restriction site is not accessible for the restriction endonucleases (18). Therefore, all clones were tested for their in vitro T-RF formation pattern to detect those showing pseudo-T-RF formation and to interpret the corresponding peaks in the community T-RFLP profiles (Fig. 1 to 3).
FIG. 4.
T-RFLP profiles of bacterial 16S rRNA genes amplified from DNA extracts of the midgut and hindgut of P. ephippiata larvae and food soil. MspI was used for restriction digestion. All major T-RFs (≥2% of total peak height; triangles), assignable minor T-RFs (dots), and peaks probably influenced by pseudo-T-RF formation (asterisks) are marked. Assignable phylogenetic groups are given in parentheses with the following abbreviations: AB, Actinobacteria; B, Bacillales; CFB, CFB phylum; C, Clostridiales, LB, Lactobacillales; P, Planctomycetales; βPB, β-Proteobacteria; n.a., not assignable. Boldface T-RFs in the soil profiles indicate major T-RFs without a corresponding major T-RF in the gut profiles.
FIG. 5.
Relative frequencies of distinct archaeal groups in the midgut, hindgut, and food soil of P. ephippiata larvae, based on composition of 16S rRNA gene clone libraries and T-RFLP analysis, with TaqI and AluI. T-RF frequencies were calculated by comparing the individual heights of assignable T-RFs to the sum of all peak heights in the electropherograms. T-RF lengths in base pairs are given in parentheses; T-RFs affected by formation of pseudo-T-RFs are marked with an asterisk (see text). C, Crenarchaeota; MB, Methanobacteriaceae; MM, Methanomicrobiales; MS, Methanosarcinales; RCII, rice cluster II; TP, Thermoplasmales, n.a., not assignable.
Since many T-RFs were shared by more than one phylogenetic group (Fig. 4 and 5), the frequency of certain phylogenetic groups in the P. ephippiata gut, when based on T-RFLP data, can be expressed only as frequency ranges (Table 1). Minimum frequencies were calculated by considering only those T-RFs that were unique for a certain group, whereas for maximum frequencies those T-RFs shared with others were also taken into account.
In the T-RFLP profile of bacterial 16S rRNA genes from the midgut, all major T-RFs (≥2% of total peak height) and 12 out of 23 minor T-RFs (<2% of total peak height) could be assigned to phylogenetic groups from the midgut clone library (Fig. 4). In total, the peaks assigned to clones represented 90% of the total peak height. All phylogenetic groups in the midgut clone library, except those affiliated with the δ-Proteobacteria, S. thermophilus, and the TM7 phylum, were represented by T-RFs in the electropherogram. In the T-RFLP profile of bacterial 16S rRNA genes from the hindgut sample, 15 out of 17 major and 12 out of 18 minor T-RFs were assignable to phylogenetic groups from the hindgut clone library (Fig. 4). Eighty-eight percent of the total peak height could be assigned, and all phylogenetic groups from the clone library except δ-Proteobacteria and ɛ-Proteobacteria were represented by T-RFs.
16S rRNA gene frequencies estimated from the T-RFLP profiles (Table 1) generally corroborated the results obtained with the clone libraries. Differences were found with respect to the frequencies of 16S rRNA genes related to Clostridiales and Lactobacillales, which seem to be overrepresented in the midgut clone library, whereas those of Actinobacteria were apparently underrepresented. Also in the hindgut clone library, 16S rRNA genes related to Lactobacillales appeared to be overrepresented compared to the T-RFLP data, whereas genes related to the CFB phylum were apparently underrepresented. The Morisita index of community similarity (IM) was 0.40 for the comparison of midgut with hindgut, underlining the clear differences in the bacterial community composition between the major gut sections; this difference was also displayed by the clone libraries.
As some archaeal clones were lacking TaqI restriction sites, T-RFLP analysis of archaeal community structure was performed also with AluI to resolve these peaks further. In the T-RFLP profiles of the midgut, all major T-RFs (≥5% of total peak height) could be assigned to clones in the archaeal clone library of the midgut, and for all phylogenetic groups from the clone library, the corresponding T-RFs were detected (Fig. 5). In total, 94 and 91% of the total electropherogram peak height obtained using TaqI or AluI, respectively, could be assigned to clones. Also in the archaeal T-RFLP profiles of the hindgut, all major T-RFs, representing 99% (TaqI) or 95% (AluI) of total peak height, could be assigned to clones (Fig. 5).
The results of the T-RFLP analysis also affirmed the clone frequencies observed in the clone libraries (Fig. 5). 16S rRNA genes of Crenarchaeota, which have the highest relative abundance in the midgut, were also restricted to this section, whereas those affiliated with Thermoplasmales were located exclusively in the hindgut. Clones closely related to Methanomicrococcus blatticola (no TaqI restriction site, 165-bp T-RF with AluI) were restricted to the hindgut, as shown by their lack of appearance in the midgut clone library and by the 165-bp T-RF in the midgut profile proving to be a pseudo-T-RF caused by clones related to Crenarchaeota and Methanobacteriaceae (18). An IM value of 0.48 (calculated for AluI-derived T-RF patterns) underlines the idea that the Pachnoda midgut and hindgut differ considerably in their archaeal community composition.
The relative gene frequencies deduced from the results of the T-RFLP analysis of the hindgut DNA were in good accordance with the 16S rRNA gene frequencies in the hindgut clone library, whereas the clones related to Crenarchaeota appeared to be slightly overrepresented in the midgut library.
T-RFLP profiles of bacterial 16S rRNA genes of the food soil extract (Fig. 4) differed clearly from the gut profiles. Morisita indices of community similarity (IM) were 0.28 for the comparison of soil and midgut and 0.33 for the comparison of soil and hindgut. Therefore, no bacterial 16S rRNA gene clone library was created from the soil DNA extract. However, T-RFLP profiles of archaeal 16S rRNA genes showed relatively high similarities between food soil and midgut (IM = 0.69) and food soil and hindgut (IM = 0.61), particularly with respect to the occurrence of T-RFs of 183 bp (TaqI) and 125 and 165 bp (AluI). Since these T-RFs all represent Crenarchaeota, the latter appear to be the only archaeal group occurring both in the soil and in the (mid)gut. The clone library data indicate an increase in the proportion of this group among the archaea from the soil (25%) to the midgut (75%); unfortunately, T-RFLP data cannot be used to corroborate this observation since all Crenarchaeota clones shared T-RFs with other archaeal clones. However, the T-RFLP profiles clearly document that 16S rRNA genes related to Methanomicrobiaceae and rice cluster II are both restricted to the soil. T-RFLP profiles of the gut samples lacked the corresponding TaqI T-RFs (82 and 392 bp [Fig. 5]). Clones related to Methanobacteriaceae and Thermoplasmales were found only in the gut samples. Neither their corresponding TaqI (∼800 and 380 bp, respectively) nor AluI (64, 196, 321, and 449 bp, respectively) T-RFs occurred in the soil electropherograms. As the 165-bp AluI T-RF in the midgut electropherogram was a pseudo-T-RF (18), it can be excluded that Methanosarcinales species from the soil occur in the midgut. It can also be excluded that the Methanosarcinales present in the hindgut are ingested with the food soil since there was no ∼800-bp T-RF in the soil electropherogram.
DISCUSSION
This is the first report on the intestinal microbiota of a beetle larva analyzed with cultivation-independent techniques. Our results showed that the morphological and physicochemical heterogeneity of the gut, described in the companion paper (35), is reflected in a high microbial diversity and pronounced axial differences in the composition of the gut microbiota, which is also clearly different from that in the soil fed to the larvae. Because in highly complex microbial communities a cloning approach is prone to the risk of undersampling (51), we employed T-RFLP analysis to corroborate the results of the 16S rRNA gene clone libraries and to document differences in community structures among the investigated samples with the necessary resolution.
Bacterial community.
The bacterial community in the intestine of P. ephippiata larvae is highly diverse, which is typical for any gut system studied so far with culture-independent techniques (e.g., references 25, 36, 45, 56, and 60). Most clones obtained from the midgut and hindgut were affiliated with well-described phylogenetic groups commonly found in other intestinal tracts (e.g., Lactobacillales, Bacillales, Clostridiales, CFB phylum, and Proteobacteria; see literature cited above and references 8 and 9 for reviews about termites).
However, except for the clones related to Promicromonospora pachnodae (see below), only one clone obtained in this study clustered with clones or isolates from other arthropod guts, e.g., from termites (see studies cited in references 9 and 25). Clone PeH05, affiliated with the Clostridiales, was closely related (94% sequence similarity) to clone Rs-B65, obtained from the gut of the wood-feeding termite Reticulitermes speratus (25). Clones related to spirochetes, possibly accounting for up to 50% of all prokaryotes in some termites (e.g., references 25 and 47), were not detected at all, not even with a specific primer system (37) (data not shown). By contrast, the closest relatives of several clones stemmed from the intestines of higher animals (ruminants and rodents) or from human specimens, i.e., Dysgonomonas species (24) (90 to 95% sequence similarity) and T. sanguinis (96% similarity) (4). Obviously the gut microbiota of P. ephippiata larvae differs clearly from that of other soil arthropods, such as termites.
The apparent dominance of fermenting bacteria (Lactobacillales, Clostridiales, members of the CFB phylum, and clones related to T. sanguinis) in the gut of Pachnoda larvae corroborates the high lactate and acetate concentrations in midgut and hindgut and the large numbers of lactogenic and acetogenic bacteria in both compartments (35). The dramatic changes in the physicochemical conditions between the midgut and hindgut of scarabaeid beetle larvae (35) are in good agreement with the major differences in the bacterial community structure: the relative dominance of 16S rRNA genes of Actinobacteria in the midgut and of members of the CFB phylum in the hindgut. The combined results of clonal and T-RFLP analysis also document that Bacillales, Planctomycetales, and γ-Proteobacteria occur mainly in the midgut, whereas β-Proteobacteria are present in the midgut and hindgut. As T-RFLP analysis revealed no T-RFs corresponding to clones affiliated with the δ and ɛ subgroups of Proteobacteria, the TM7 phylum, and S. thermophilus, these groups are obviously overrepresented in the clone libraries, which contain only a limited number of clones. Also the high frequency of Clostridiales-related 16S rRNA genes in the midgut library was not supported by the T-RFLP profiles, whereas the high proportion of Clostridiales in the hindgut and the twofold increase in frequency of Lactobacillales between midgut and hindgut were corroborated by both methods.
The most prominent among the environmental factors possibly determining major differences in bacterial community structure between the midgut and hindgut of P. ephippiata larvae are the availability of oxygen and the intestinal pH. Although both compartments are largely anoxic, the influx of oxygen via the gut epithelium should be considerably larger in the case of the midgut because of its tubular shape (35). This might explain why many of the midgut clones grouped among Actinobacteria, which are capable of aerobic metabolism, and why many of the hindgut clones were affiliated with Bacteroides species or the Clostridiales, whose members are known as obligate anaerobes.
In view of the high midgut alkalinity, it is reasonable to assume that at least some of the bacteria in the midgut are alkaliphilic. One clone (PeM01) was distantly related (92%) to an alkaliphilic Bacillus species that was isolated from soil (43). Six midgut clones were closely related (>98%) to P. pachnodae, a facultatively aerobic, hemicellulolytic bacterium isolated from the hindgut of Pachnoda marginata larvae (12). Interestingly, all Promicromonospora-related clones obtained in the present study were recovered exclusively from the P. ephippiata midgut, as were the vast majority of the clones representing Actinobacteria. However, it is not possible to infer the pH tolerance of the bacteria represented by the clones from the phylogenetic analysis, but it is tempting to speculate that bacilli and Actinobacteria might participate in the hydrolysis of polysaccharides in this gut compartment.
Archaeal community.
Our data document clear differences between the archaeal communities in the midgut and hindgut of P. ephippiata larvae, which were also less diverse than the bacterial communities in the respective gut sections. In contrast to the bacterial clones, all archaeal clones clustered with clones or isolates previously retrieved from the guts of other insects, in particular from the soil-feeding termite C. orthognathus (19). As in the case of this termite, also the different archaeal populations in the gut of P. ephippiata larvae are highly specific for the respective gut compartment: while Crenarchaeota-like clones were absent from the hindgut, no clones affiliated with Methanosarcinales and Thermoplasmales were recovered from the midgut. Also the clones affiliated with Methanobacteriaceae and Methanosarcinaceae were more frequent in the hindgut. The apparent dominance of methanogenic archaea in the hindgut agrees with the restriction of methanogenesis to this compartment of Pachnoda larvae (22, 35). The absence of methane emission from the midgut indicates either that the absolute number of methanogens in this section is very small and/or that they are inactive. Cultivation-based enumeration showed that the numbers of methanogens in the midgut are about 2 orders of magnitude lower than those in the hindgut (35). It is likely that the presence of methanogens (and possibly also of other hindgut bacteria) in the midgut is merely the result of a reflux of hindgut content into the midgut, which has been described as a part of the digestion process in scarabaeid beetle larvae (49).
The close relationship of our clones to Methanobrevibacter and Methanomicrococcus species (95 to 97% sequence similarity) allows us to speculate cautiously that, besides H2 and formate, methanol could also be a substrate for methanogenesis in the Pachnoda hindgut (54), as has already been assumed for the hindgut of C. orthognathus (19). Indeed, Lemke et al. (35) have shown an approximately fourfold stimulation of methanogenesis in the Pachnoda hindgut upon addition of methanol (5 mM) to isolated hindgut sections.
Besides methanogens, nonmethanogenic archaea affiliated with Thermoplasmales inhabit insect (hind)guts, as has been shown for termites (19, 53) and here for Pachnoda larvae. It was not possible to determine the phylogenetic relationship to clones from the wood-feeding termite R. speratus (53), since the overlapping region of sequences (∼20 bp) was too small. As there are no cultured representatives of these mesophilic Thermoplasmales, the function of this group, comprising approximately 25% of the archaeal 16S rRNA genes in the P. ephippiata hindgut, remains to be elucidated. Clones affiliated with Crenarchaeota have been obtained from gut systems of fish (58), Holothuria (41), and soil-feeding termites (19). The clones obtained in this study were closely related to clones from the “terrestrial cluster” of Crenarchaeota and clustered with clones from C. orthognathus (97 to 98% sequence similarity to clone P1-Ar-11). Since they were the only archaeal group also occurring in the food soil, they cannot be considered as a specific gut flora, at least in P. ephippiata larvae.
Conclusions.
The results of this study represent first and important insights into microbial community structure in the intestinal tract of humivorous beetle larvae. As with all PCR-based methods, it should be kept in mind that the relative abundance of clones for the respective community is prone to considerable bias, since several factors can affect relative rRNA gene amplicon frequencies in PCR products from mixed-template reactions (3, 57; for a review see reference 59). In a recent study with soil-feeding termites (51, 52), the results obtained with the same PCR-based methods as those used in the present study were generally in good agreement with those obtained by fluorescence in situ hybridization with group-specific probes, but there were also remarkable discrepancies, e.g., with respect to the relative abundance of Planctomycetales. We are presently employing fluorescence in situ hybridization to determine the absolute abundance of the members of the gut microbiota and the spatial organization of the microbial community within the P. ephippiata gut sections, i.e., their exact radial and axial distribution. Such information will not only serve to corroborate the results of the present study but will also help us to infer possible functions of the different phylogenetic groups detected in this study.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (DFG) and by the Max Planck Society.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet, R., A. Suau, J. Dore, G. R. Gibson, and M. D. Collins. 2002. Differences in rDNA libraries of faecal bacteria derived from 10-and 25-cycle PCRs. Int. J. Syst. E vol. Microbiol. 52:757-763. [DOI] [PubMed] [Google Scholar]
- 4.Bosshard, P. P., R. Zbinden, and M. Altwegg. 2002. Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, Gram-positive bacterium. Int. J. Syst. E vol. Microbiol. 52:1263-1266. [DOI] [PubMed] [Google Scholar]
- 5.Brauman, A. 2000. Effect of gut transit and mound deposit on soil organic matter transformations in the soil feeding termite: a review. Eur. J. Soil Biol. 36:117-125. [Google Scholar]
- 6.Brauman, A., D. E. Bignell, and I. Tayasu. 2000. Soil-feeding termites: biology, microbial associations and digestive mechanisms, p. 233-259. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbiosis, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 7.Brauman, A., J. Dore, P. Eggleton, D. Bignell, J. A. Breznak, and M. D. Kane. 2001. Molecular phylogenetic profiling of prokaryotic communities in guts of termites with different feeding habits. FEMS Microbiol. Ecol. 35:27-36. [DOI] [PubMed] [Google Scholar]
- 8.Brune, A. 1998. Termite guts: the world's smallest bioreactors. Trends Biotechnol. 16:16-21. [Google Scholar]
- 9.Brune, A., and M. Friedrich. 2000. Microecology of the termite gut: structure and function on a microscale. Curr. Opin. Microbiol. 3:263-269. [DOI] [PubMed] [Google Scholar]
- 10.Cazemier, A. E., J. H. P. Hackstein, H. J. M. Op den Camp, J. Rosenberg, and C. van der Drift. 1997. Bacteria in the intestinal tract of different species of arthropods. Microb. Ecol. 33:189-197. [DOI] [PubMed] [Google Scholar]
- 11.Cazemier, A. E., H. J. M. Op den Camp, J. H. P. Hackstein, and G. D. Vogels. 1997. Fibre digestion in arthropods. Comp. Biochem. Physiol. A 118:101-109. [Google Scholar]
- 12.Cazemier, A. E., J. C. Verdoes, F. A. G. Reubsaet, J. H. P. Hackstein, C. van der Drift, and H. J. M. Op den Camp. 2003. Promicromonospora pachnodae sp. nov., a member of the (hemi)cellulolytic hindgut flora of larvae of the scarab beetle Pachnoda marginata. Antonie Leeuwenhoek 83:135-148. [DOI] [PubMed] [Google Scholar]
- 13.Chao, A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 14.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. E. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 15.DeLong, E. E., and N. R. Pace. 2001. Environmental diversity of Bacteria and Archaea. Syst. Biol. 50:470-478. [PubMed] [Google Scholar]
- 16.Dollhopf, S. L., S. A. Hashsham, and J. M. Tiedje. 2001. Interpreting 16S rDNA T-RFLP data: application of self-organizing maps and principal component analysis to describe community dynamics and convergence. Microb. Ecol. 42:495-505. [DOI] [PubMed] [Google Scholar]
- 17.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes—characterization of a gene coding for 16S-ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedrich, M. W., D. Schmitt-Wagner, T. Lueders, and A. Brune. 2001. Axial differences in community structure of Crenarchaeota and Euryarchaeota in the highly compartmentalized gut of the soil-feeding termite Cubitermes orthognathus. Appl. Environ. Microbiol. 67:4880-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosskopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grosskopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackstein, J. H. P., and C. K. Stumm. 1994. Methane production in terrestrial arthropods. Proc. Natl. Acad. Sci. USA 91:5441-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofstad, T., I. Olsen, E. R. Eribe, E. Falsen, M. D. Collins, and P. A. Lawson. 2000. Dysgonomonas gen. nov. to accommodate Dysgonomonas gadei sp. nov., an organism isolated from a human gall bladder, and Dysgonomonas capnocytophagoides (formerly CDC group DF-3). Int. J. Syst. E vol. Microbiol. 50:2189-2195. [DOI] [PubMed] [Google Scholar]
- 25.Hongoh, Y., M. Ohkuma, and T. Kudo. 2003. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiol. Ecol. 44:231-242. [DOI] [PubMed] [Google Scholar]
- 26.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji, R., and A. Brune. 2001. Transformation and mineralization of C-14-labeled cellulose, peptidoglycan, and protein by the soil-feeding termite Cubitermes orthognathus. Biol. Fertil. Soils 33:166-174. [Google Scholar]
- 28.Ji, R., A. Kappler, and A. Brune. 2000. Transformation and mineralization of synthetic 14C-labeled humic model compounds by soil-feeding termites. Soil Biol. Biochem. 32:1281-1291. [Google Scholar]
- 29.Kane, M. D. 1997. Microbial fermentation in insect guts, p. 231-265. In R. I. Mackie and B. A. White (ed.), Gastrointestinal ecosystems and fermentations. Chapman and Hall, New York, N.Y.
- 30.Kappler, A., and A. Brune. 1999. Influence of gut alkalinity and oxygen status on mobilization and size-class distribution of humic acids in the hindgut of soil-feeding termites. Appl. Soil Ecol. 13:219-229. [Google Scholar]
- 31.Kappler, A., R. Ji, and A. Brune. 2000. Synthesis and characterization of specifically 14C-labeled humic model compounds for feeding trials with soil-feeding termites. Soil Biol. Biochem. 32:1271-1280. [Google Scholar]
- 32.Kudo, T., M. Ohkuma, S. Moriya, S. Noda, and K. Ohtoko. 1998. Molecular phylogenetic identification of the intestinal anaerobic microbial community in the hindgut of the termite, Reticulitermes speratus, without cultivation. Extremophiles 2:155-161. [DOI] [PubMed] [Google Scholar]
- 33.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acids techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 34.Lavelle, P., D. Bignell, M. Lepage, V. Wolters, P. Roger, P. Ineson, O. W. Heal, et al. 1997. Soil function in a changing world: the role of invertebrate ecosystem engineers. Eur. J. Soil Biol. 33:159-193. [Google Scholar]
- 35.Lemke, T., U. Stingl, M. Egert, M. W. Friedrich, and A. Brune. 2003. Physicochemical conditions and microbial activities in the highly alkaline gut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69:6650-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lilburn, T. G., T. M. Schmidt, and J. A. Breznak. 1999. Phylogenetic diversity of termite gut spirochaetes. Environ. Microbiol. 1:331-345. [DOI] [PubMed] [Google Scholar]
- 38.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, et al. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 39.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, et al. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 40.Lueders, T., and M. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McInerney, J. O., M. Wilkinson, J. W. Patching, T. M. Embley, and R. Powell. 1995. Recovery and phylogenetic analysis of novel archaeal rRNA sequences from a deep-sea deposit feeder. Appl. Environ. Microbiol. 61:1646-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel-electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 43.Nogi, Y., and K. Horikoshi. 1990. A thermostable alkaline beta-1,3-glucanase produced by alkalophilic Bacillus sp. AG-430. Appl. Microbiol. Biotechnol. 32:704-707. [Google Scholar]
- 44.Ohkuma, M., T. Iida, and T. Kudo. 1999. Phylogenetic relationships of symbiotic spirochetes in the gut of diverse termites. FEMS Microbiol. Lett. 181:123-129. [DOI] [PubMed] [Google Scholar]
- 45.Ohkuma, M., and T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paster, B. J., F. E. Dewhirst, S. M. Cooke, V. Fussing, L. K. Poulsen, and J. A. Breznak. 1996. Phylogeny of not-yet-cultured spirochetes from termite guts. Appl. Environ. Microbiol. 62:347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramakrishnan, B., T. Lueders, R. Conrad, and M. Friedrich. 2000. Effect of soil aggregate size on methanogenesis and archaeal community structure in anoxic rice field soil. FEMS Microbiol. Ecol. 32:261-270. [DOI] [PubMed] [Google Scholar]
- 49.Rössler, M. E. 1961. Ernährungsphysiologische Untersuchungen an Scarabaeidenlarven (Oryctes nasicornes L., Melolontha melolontha L.). J. Insect Physiol. 6:62-80. [Google Scholar]
- 50.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Axial dynamics, stability, and interspecies similarity of bacterial community structure in the highly compartmentalized gut of soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69:6018-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tracts of two soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69:6007-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shinzato, N., T. Matsumoto, I. Yamaoka, T. Oshima, and A. Yamagishi. 1999. Phylogenetic diversity of symbiotic methanogens living in the hindgut of the lower termite Reticulitermes speratus analyzed by PCR and in situ hybridization. Appl. Environ. Microbiol. 65:837-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sprenger, W. W., M. C. van Belzen, J. Rosenberg, J. H. P. Hackstein, and J. T. Keltjens. 2000. Methanomicrococcus blatticola gen. nov., sp nov., a methanol- and methylamine-reducing methanogen from the hindgut of the cockroach Periplaneta americana. Int. J. Syst. E vol. Microbiol. 50:1989-1999. [DOI] [PubMed] [Google Scholar]
- 55.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 56.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Maarel, M. J. E. C., R. R. E. Artz, R. Haanstra, and L. J. Forney. 1998. Association of marine archaea with the digestive tracts of two marine fish species. Appl. Environ. Microbiol. 64:2894-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 60.Whitford, M. F., R. J. Forster, C. E. Beard, J. H. Gong, and R. M. Teather. 1998. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe 4:153-163. [DOI] [PubMed] [Google Scholar]
- 61.Wolters, V. 2000. Invertebrate control of soil organic matter stability. Biol. Fertil. Soils 31:1-19. [Google Scholar]