Abstract
An ultrasensitive bioassay system for the detection of N-acylhomoserine lactones (AHLs) was constructed in Agrobacterium tumefaciens by using the T7 expression system to overproduce the AHL receptor TraR. This strain detected many diverse AHLs, some at extremely low concentrations. We used this strain to detect for the first time AHLs made by Mesorhizobium huakuii, which symbiotically fixes nitrogen in association with the legume Astragalus sinicus, a source of green manure throughout eastern Asia.
A number of bacterial processes are regulated by the exchange of chemical signals that permit a bacterial community to coordinate its responses to novel environmental challenges or opportunities. These responses include pathogenesis, symbiosis, antibiotic production, motility, genetic competence, and biofilm formation (25). Diffusible signals are generally thought to help a bacterial community to take a census of its population size, a phenomenon denoted quorum sensing (15). It is also possible that signaling between bacterial cells may be done for purposes other than census taking. The exchange of these signals has the effect of decentralizing a decision-making process, enabling all of the individuals in a community to participate in reaching a consensus (10, 11, 13, 15, 42).
In many proteobacteria, these chemical signals consist of a set of diffusible N-acylhomoserine lactones (AHLs; also called autoinducers). AHL-mediated signaling was once thought to be restricted to a few species of bioluminescent marine bacteria but has now been described in diverse groups of proteobacteria (25). The key regulatory components of these signaling systems are LuxI-type proteins, which act as AHL synthases, and LuxR-type proteins, which serve as AHL receptors and AHL-dependent transcription factors (11). The N-terminal domains of LuxR-type proteins are involved in AHL binding, while the C-terminal domains are required for DNA binding and transcriptional activation (7, 8, 38). All of the AHLs described to date contain invariant homoserine lactone moieties and highly variable fatty acyl groups. These acyl groups range in length from 4 to 18 carbon atoms. The C-3 carbon can be fully reduced or can bear hydroxyl or ketone substituents. The homoserine lactone moiety is derived from S-adenosylmethionine, while the acyl chains are derived from acyl-acyl carrier protein (26, 28, 34).
The TraI and TraR proteins of Agrobacterium tumefaciens are members of this family. TraI synthesizes the autoinducer N-3-oxooctanoyl-HSL (OOHL), while TraR is an OOHL-dependent activator of Ti plasmid tra, trb, and rep genes (14, 27, 29). TraR has also been reported to detect several diketopiperazines (18). Interaction of TraR with OOHL promotes TraR protein stability, TraR dimerization, and DNA binding (30, 46, 47). TraR protein synthesized in the absence of OOHL is rapidly targeted for proteolysis, while OOHL increases the stability of the protein at least 20-fold. Two recent reports of the X-ray crystal structure of TraR-OOHL-DNA ternary complexes showed that OOHL is deeply buried within the N-terminal domain of the protein, suggesting that folding of this domain might be impaired in the absence of ligand (40, 44).
Novel cell-cell signaling systems are continually being described, and these studies often include the detection and identification of the cognate signal molecule. The detection of AHLs has been facilitated by the development of a variety of bioassay strains. Such strains contain an easily assayable reporter gene and lack all AHL synthases, such that reporter activity requires exogenous AHLs. Various reporter genes have been described, including lacZ, gfp, lux, and the production of an endogenous pigment (1). One limitation is that most bioassay strains detect only a narrow range of AHLs. In such cases, one can use a variety of different bioassay strains having different AHL specificities, although this can be rather labor-intensive. For example, Chromobacterium violaceum reporter strain CV026 cannot detect any of the 3-hydroxy derivatives and lacks sensitivity to most 3-oxo derivatives (3, 24). LuxR-based reporters detect most of the 3-oxo and alkanoyl standards but not 3-hydroxy forms (3, 43). A gfp-based reporter strain was recently described, although its reported sensitivity is rather low (36). The use of radiotracers (incorporation of 14C label from [14C]methionine into AHLs) (32) is very sensitive and has broad specificity, especially for long-acyl-chain AHLs (23, 33), but requires the use of radiolabeled substrates. Furthermore, this technique might also radiolabel other compounds that have no bioactivity as AHLs.
Two A. tumefaciens bioassay strains were reported to allow detection of a broad range of AHL derivatives and to show great sensitivity (3, 45). In both strains, TraR was overexpressed. In one study, overexpression of TraR was shown to greatly increase the sensitivity of the assay and to broaden the variety of autoinducers detected. Unfortunately, these strains did not efficiently detect short-chain AHLs. In this study, we attempted to create a bioassay strain with even greater AHL sensitivity and even broader substrate specificity. This was done by adapting the bacteriophage T7 expression system to express TraR in A. tumefaciens. This protein expression system is widely used in Escherichia coli to overexpress proteins (39), but there are very few reports of its use in nonenteric bacteria (2, 37).
To construct a T7 expression system in A. tumefaciens, we subcloned a DNA fragment containing a fusion between the T7 promoter and traR, from pJZ358 (46), into spectinomycin-resistant broad-host-range vector pPZP201 (16), creating plasmid pJZ384. The T7 RNA polymerase gene of bacteriophage T7 and the cI857 gene of bacteriophage lambda were subcloned from pGP1-2 (39) into gentamicin resistance-encoding plasmid pBBR1MCS5 (21), creating pJZ410. A traI-lacZ reporter fusion was subcloned from pCF372 (12) into tetracycline resistance-encoding plasmid pSW213 (4), resulting in pJZ372. These plasmids were introduced into A. tumefaciens strain KYC55, a derivative of strain R10 that lacks the Ti plasmid and therefore cannot produce detectable autoinducers (6).
To determine whether the T7 expression system functions in A. tumefaciens, we used immunodetection (Western blot) assays of serially diluted cell extracts expressing the PT7-traR fusion. Cells were harvested after being cultured to late log phase in AT minimal medium (14). The TraR content of this strain was compared to that of one previously described (45), after overnight growth in the presence of 100 nM OOHL. The strain with the PT7-traR fusion accumulated at least threefold more TraR than the strain with the PtetR-traR fusion (Fig. 1A).
FIG. 1.
TraR expression in a T7 system in A. tumefaciens. (A) TraR protein abundance in PtetR-traR and PT7-traR strains. Strains WCF47(pCF218)(pCF372) and KYC55(pJZ372)(pJZ384)(pJZ410) were cultured overnight in the presence of 100 nM OOHL, diluted serially in fourfold increments, size fractionated by SDS-PAGE, and immunodetected with rabbit polyclonal antiserum. (B) Pulse-labeling of TraR. Strain KYC55(pJZ372)(pJZ384)(pJZ410) was cultured to mid-logarithmic phase in AT medium in the absence or presence of 100 nM OOHL at 28°C, incubated with rifampin for 30 min, and then pulse-labeled with [35S]methionine for 5 min. The cultures were then chased with nonradiolabeled methionine and terminated by freezing at −80°C at various intervals as indicated. Cleared lysates were size fractionated by SDS-PAGE, and gels were analyzed with a Storm B840 PhosphorImager (Molecular Dynamics).
We have previously shown that TraR is stabilized against proteolysis by OOHL (46, 47). When TraR is strongly overexpressed in E. coli in the presence of OOHL, some of it is soluble, while the remainder forms insoluble inclusion bodies. When strongly overexpressed in the absence of OOHL, all detectable TraR is insoluble. We interpreted this to mean that TraR is made in two forms, a soluble form that is stabilized by OOHL and an unfolded, insoluble form whose stability does not require OOHL. To determine whether two similar pools of TraR are made in A. tumefaciens, we pulse-labeled TraR with radioactive methionine (47). Strain KYC55(pJZ410)(pJZ384)(pJZ372) was cultured in AT minimal medium (14) to mid-logarithmic phase in the presence or absence of 1 μM OOHL. Rifampin was added (200-μg/ml final concentration) to inactivate bacterial RNA polymerase. After 30 min, the culture was pulse-labeled with 5 μCi of [35S]methionine per ml. After 5 min, excess nonlabeled methionine was added and samples were taken at various time intervals. Cell lysates were prepared with a French pressure cell and cleared by ultracentrifugation. Both soluble and insoluble fractions were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). A major 24-kDa band (the size of a TraR monomer) was easily visualized in the soluble fractions both in the absence and in the presence of OOHL (Fig. 1B). In the absence of OOHL, soluble TraR was rapidly degraded (Fig. 1B, left lanes), while in the presence of OOHL, TraR was far more stable (Fig. 1B, right lanes). In contrast, equally large amounts of TraR accumulated in the insoluble fractions in the presence or absence of OOHL (data not shown).
The T7 RNA polymerase gene cloned in pJZ410 is under the control of the PL of bacteriophage lambda, which is repressed by the CI repressor. The thermosensitive cI857 allele of the lambda repressor was also included on pJZ410. Our intention had been to render this system heat inducible (39). However, thermoinduction at 42°C for 30 min did not cause elevated TraR accumulation (data not shown). Nevertheless, these data suggest that expression of TraR with the T7 promoter could enable us to strongly overexpress TraR in its native host.
As described above, overproduction of TraR causes increased ability to detect autoinducers and broadens the array of detectable autoinducers (45), as well as a small number of other compounds (18). To determine whether expression of TraR at extremely high levels might allow detection of even lower autoinducer concentrations, we compared this strain with a bioassay strain described previously, WCF47(pCF218)(pCF372) (45). The major difference between these strains is that the traR gene is under the control of the tetR promoter in the latter strain. Both strains have essentially identical plasmid-borne PtraI-lacZ fusions, and both strains lack their own traI genes. Strong overproduction of TraR caused only a slight increase in the basal level of β-galactosidase activity (Fig. 2). The newly described strain expressed elevated levels of β-galactosidase with as little as 3 pM OOHL (equivalent to 3 fmol per ml of broth culture), while the previously described strain required 100-fold more OOHL for a detectable response. Both reporter strains expressed approximately the same amount of β-galactosidase when fully induced.
FIG. 2.
3-Oxooctanoyl-HSL dose-response curves of A. tumefaciens strains KYC55 (pJZ372)(pJZ384)(pJZ410) (squares) and WCF47(pCF218)(pCF372) (diamonds). Synthetic 3-oxooctanoyl-HSL was diluted as indicated and added to 2 ml of AT medium supplemented with approximately 107 bacterial cells, incubated with aeration for 12 h, and assayed for β-galactosidase specific activity.
We compared the abilities of these two bioassay strains to detect a set of 32 synthetic AHLs with various chain lengths, saturation levels, and oxidation states (see reference 45 for structures). The strain containing a PtetR-traR fusion detected low concentrations of many AHLs but did not sensitively detect all AHLs, especially those having four-carbon acyl moieties, such as butanoyl-HSL, 2-butenoyl-HSL, and 2-butynoyl-HSL (Table 1). In contrast, the strain containing a PT7-traR fusion was much more sensitive to all of the compounds. It was able to detect low concentrations of many AHLs that were not detected by the former strain, such as 3-oxopentanoyl-HSL, pentanoyl-HSL, 3-hydroxydodecanoyl-HSL, 2-hexenoyl-HSL, 2-decenoyl-HSL, di-HSL-3,12-dioxotetradecandioate, and O-hexyl-N-HSL. Compounds having four-carbon acyl moieties were also readily detectable when provided at low micromolar concentrations. These results indicate that increased expression of TraR greatly enhances its ability to detect diverse AHL signals.
TABLE 1.
Abilities of PT7-traR and PtetR-traR strains to respond to different AHLsa
| Compound | β-Galactosidase activity
|
|||
|---|---|---|---|---|
| PT7-traR
|
Ptet-traR
|
|||
| 5 μM | 5 nM | 5 μM | 5 nM | |
| 3-Oxobutanoyl-HSL | 712.5 | 9.9 | 30.9 | 2.3 |
| 3-Oxopentanoyl-HSL | 1,418.2 | 402.9 | 932.6 | 2.5 |
| 3-Oxohexanoyl-HSL | 1,687.9 | 1,326.9 | 1,607.8 | 407.8 |
| 3-Oxoheptanoyl-HSL | 1,185.2 | 15.5 | 248.9 | 1.6 |
| 3-Oxooctanoyl-HSL | 1,907.9 | 1,767.2 | 1,702.5 | 920 |
| 3-Oxoundecanoyl-HSL | 1,509.2 | 1,143.7 | 1,466.1 | 423.8 |
| 3-Oxodocecanoyl-HSL | 1,476.3 | 1,294.5 | 1,255.7 | 91.5 |
| 4-Oxa-3-oxohexanoyl-HSL | 1,571.1 | 195.8 | 626.3 | 3.8 |
| Butanoyl-HSL | 279 | 23.1 | 19.8 | 2.2 |
| Pentanoyl-HSL | 1,170.8 | 169.8 | 362.7 | 2.9 |
| Hexanoyl-HSL | 1,316.9 | 37 | 672.8 | 2.4 |
| Heptanoyl-HSL | 1,678.7 | 1,481.8 | 957.4 | 279.8 |
| Octanoyl-HSL | 1,905.9 | 1,772.9 | 760.7 | 275.1 |
| Decanoyl-HSL | 1,781.6 | 37.1 | 467.3 | 2.2 |
| Dodecanoyl-HSL | 501.2 | 8 | 43.6 | 3 |
| 3-Hydroxynonanoyl-HSL | 1,796.3 | 1,683 | 1,487.5 | 437.6 |
| 3-Hydroxydodecanoyl-HSL | 1,602.6 | 197.9 | 216.4 | 6.9 |
| 2-Butenoyl-HSL | 73.8 | 8 | 10.4 | 2.3 |
| 2-Pentenoyl-HSL | 42.6 | 7.5 | 12.1 | 2.2 |
| 2-Hexenoyl-HSL | 1,672.1 | 346.3 | 766.6 | 5.2 |
| 2-Octenoyl-HSL | 1,653.1 | 1,303.8 | 1,114.7 | 58.9 |
| 2-Nonenoyl-HSL | 1,733.1 | 1,304.2 | 1,261.3 | 61.1 |
| 2-Decenoyl-HSL | 1,536.9 | 924.1 | 1,319.4 | 9.5 |
| 2-Butynoyl-HSL | 1,744.1 | 54.2 | 669.3 | 3 |
| 2-Hexynoyl-HSL | 1,576.1 | 18.4 | 285.6 | 1.7 |
| 5-Hexynoyl-HSL | 657.8 | 9.4 | 56.3 | 1.6 |
| 2-Octynoyl-HSL | 1,448.8 | 36.3 | 813.5 | 1.9 |
| 3-Oxo-7-octynoyl-HSL | 2,115.1 | 1,854.9 | 1,862.9 | 646.9 |
| 3-Oxo-11-octadecenoyl-HSL | 813.4 | 25 | 67.3 | 3.3 |
| Di-HSL-decandioate | 967 | 16.5 | 49.2 | 1.8 |
| Di-HSL-3,12-dioxotetradecandioate | 1,208.8 | 210.3 | 529.6 | 8.3 |
| p-Propylbenzoyl-HSL | 136 | 11.5 | 14.2 | 1.7 |
| O-Hexyl-N-HSL | 1,963.4 | 717.5 | 1,974.4 | 8.3 |
| HSL | 19.7 | 4.1 | 11.1 | 2.9 |
AHL analogs were provided at the indicated concentrations to cultures of KYC55(pJZ372)(pJZ384)(pJZ410) (left two columns of values) and WCF47(pCF218)(pCF372) (right two columns of values) in AT broth. Bacteria were cultured for 12 h and assayed for β-galactosidase activity. β-Galactosidase expression in the absence of autoinducers was less than 5 Miller units for both strains.
We used this bioassay strain to test an isolate of Mesorhizobium huakuii for AHL production. This bacterium is a moderately fast-growing rhizobium that fixes nitrogen in symbiotic association with the legume Astragalus sinicus (Chinese milk vetch), an economically important green manure grown during the winter in eastern Asia (5, 19). There are no previous reports of cell-cell communication in the Mesorhizobium genus, although the genome sequence of M. loti contains 10 predicted LuxR homologs and six predicted AHL synthases (20). Several species of Rhizobium and Sinorhizobium produce AHLs that control various functions, such as exopolysaccharide production (22), rhizosphere-related gene expression (31), nodulation (9, 23), and the conjugal transfer of pSym plasmids (17).
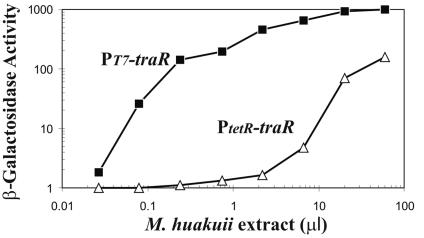
M. huakuii strain 93 was cultured in 100 ml of TY medium (41) at 28°C to stationary phase, and the cells were removed from the broth by centrifugation. The supernatant was extracted with ethyl acetate, and the extract was dried and resuspended in 1 ml of distilled H2O (1% of the original culture volume). This sample was serially diluted into 1 ml of AT medium (14) containing each of the two bioassay strains. The older bioassay strain, WCF47(pCF218)(pCF372), was induced only when the AHLs were added at the highest concentrations tested (Fig. 3). In contrast, the newer, more sensitive strain, KYC55(pJZ372)(pJZ384)(pJZ410), was able to detect 200-fold smaller amounts of this extract.
FIG. 3.
PT7-traR sensitively detects acyl-HSLs in M. huakuii spent culture supernatant. A 100-ml volume of supernatant from stationary-phase M. huakuii 93 growing in TY medium was extracted with ethyl acetate, dried, and resuspended in 1 ml of distilled H2O. The indicated volumes were added to 1 ml of AT medium containing approximately 107 cells of either strain KYC55(pJZ372)(pJZ384)(pJZ410) (squares) or WCF47(pCF218)(pCF372) (diamonds). The cultures were incubated with aeration for 12 h and assayed for β-galactosidase specific activity.
Reverse-phase thin-layer chromatography (TLC) was used to visualize the AHLs produced by this strain (35). Synthetic AHLs and a supernatant from A. tumefaciens R10(pCF218) (45) were used as controls. This A. tumefaciens strain was previously shown to produce primarily OOHL and smaller amounts of other AHLs, including 3-oxohexanoyl-HSL and octanoyl-HSL (3, 35, 45). When 1 μl of supernatant was spotted on the TLC plate, the new bioassay strain detected all three compounds (Fig. 4, left panel), while the older, less sensitive strain detected only OOHL (Fig. 4, right panel).
FIG. 4.
TLC of autoinducers synthesized by M. huakuii. Lanes: 1, standard mixture of 3-oxo derivatives with acyl chain lengths of C6 (2.5 pmol), C8 (0.25 pmol), and C12 (0.5 nmol); 2, 3-hydroxyl derivatives with acyl chain lengths of C6 (20 pmol), C8 (20 pmol), and C10 (20 pmol); 3, HSLs with fully reduced acyl chain lengths of C6 (100 pmol), C8 (30 pmol), and C10 (40 pmol); 4, activities of autoinducers obtained from 1 ml of M. huakuii growing in TY medium to stationary phase; 5, activities of autoinducers obtained from 1 μl of A. tumefaciens R10(pCF218). TLC plates were chromatographed as described previously and overlaid with 100 ml of agar containing AT medium, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and approximately 107 bacteria per ml [strains KYC55(pJZ372)(pJZ384)(pJZ410) (left panel) and WCF47(pCF218)(pCF372) (right panel)]. TLC plates were incubated overnight at 28°C and examined for X-Gal hydrolysis.
The more sensitive bioassay strain detected one major autoinducer from the M. huakuii 93 supernatant, while the less sensitive strain did not detect any AHL unless considerably more extract was spotted onto the TLC plate. The Rf of this compound is the same as that of octanoyl-HSL. This strain therefore synthesizes a compound active in the A. tumefaciens bioassay, almost certainly an AHL, and possibly octanoyl-HSL. The chemical structure of this autoinducer is currently under investigation.
The new bioassay strain described here has the greatest sensitivity and the broadest range of AHL detection yet reported. This strain will be a useful tool for detecting extremely low concentrations of AHLs or possibly other bioactive signal molecules. It could have applications in AHL detection in lakes, streams, soils, and other dilute environmental samples. This strain could also be useful in the detection of AHL synthase genes in cosmid libraries, even those that are expressed very weakly in E. coli. We are currently using it to attempt to detect AHL synthase genes in environmental DNA samples. The extreme sensitivity of this strain may also be useful in biochemical studies of AHL synthase genes.
ADDENDUM IN PROOF
We have recently tested the bioassay strain for detection of AHLs containing 14, 16, or 18 carbon acyl chains. All compounds were readily detected when provided at a 5 μM concentration, resulting in 300 to 1,600 U of β-galactosidase activity. AHLs containing 3-oxo substituents were also detectable at a concentration of 5 nM, while AHLs with unsubstituted acyl chains were not detected at this lower concentration.
Acknowledgments
Jun Zhu and Yunrong Chai contributed equally to this work.
We thank Anatol Eberhard for the generous supply of AHLs used in this study.
This study was supported by grant GM42893 from the National Institute of General Medical Sciences to S.CW., by the NJAU Initiative Fund to J.Z., and by a 973 project grant (001CB1089) from China to Z.Z.
REFERENCES
- 1.Brelles-Marino, G., and E. J. Bedmar. 2001. Detection, purification and characterisation of quorum-sensing signal molecules in plant-associated bacteria. J. Biotechnol. 91:197-209. [DOI] [PubMed]
- 2.Brunschwig, E., and A. Darzins. 1992. A two-component T7 system for the overexpression of genes in Pseudomonas aeruginosa. Gene 111:35-41. [DOI] [PubMed] [Google Scholar]
- 3.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. Y., and S. C. Winans. 1991. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J. Bacteriol. 173:1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, W. X., G. S. Li, Y. L. Qi, E. T. Wang, H. L. Yuan, and J. L. Li. 1991. Rhizobium huakuii sp. nov. isolated from the root nodules of Astragalus sinicus. Int. J. Syst. Bacteriol. 41:275-280. [Google Scholar]
- 6.Cho, K., C. Fuqua, and S. C. Winans. 1997. Transcriptional regulation and locations of Agrobacterium tumefaciens genes required for complete catabolism of octopine. J. Bacteriol. 179:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, S. H., and E. P. Greenberg. 1991. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc. Natl. Acad. Sci. USA 88:11115-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, S. H., and E. P. Greenberg. 1992. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J. Bacteriol. 174:4064-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels, R., D. E. De Vos, J. Desair, G. Raedschelders, E. Luyten, V. Rosemeyer, C. Verreth, E. Schoeters, J. Vanderleyden, and J. Michiels. 2002. The cin quorum sensing locus of Rhizobium etli CNPAF512 affects growth and symbiotic nitrogen fixation. J. Biol. Chem. 277:462-468. [DOI] [PubMed] [Google Scholar]
- 10.Dunny, G. M., and S. C. Winans. 1999. Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 11.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua, C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 14.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hajdukiewicz, P., Z. Svab, and P. Maliga. 1994. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25:989-994. [DOI] [PubMed] [Google Scholar]
- 17.He, X., W. Chang, D. L. Pierce, L. O. Seib, J. Wagner, and C. Fuqua. 2003. Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 185:809-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holden, M. T., S. Ram Chhabra, R. de Nys, P. Stead, N. J. Bainton, P. J. Hill, M. Manefield, N. Kumar, M. Labatte, D. England, S. Rice, M. Givskov, G. P. Salmond, G. S. Stewart, B. W. Bycroft, S. Kjelleberg, and P. Williams. 1999. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol Microbiol. 33:1254-1266. [DOI] [PubMed] [Google Scholar]
- 19.Jarvis, B. D. W., P. Van Berkum, W. X. Chen, S. M. Nour, M. P. Fernandez, J. C. Cleyet-Marel, and M. Gillis. 1997. Phylogenetic and genetic relationships of Mesorhizobium tianshanense and related rhizobia. Int. J. Syst. Bacteriol. 47:895-898. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7(Suppl.):381-406. [DOI] [PubMed] [Google Scholar]
- 21.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 22.Marketon, M. M., S. A. Glenn, A. Eberhard, and J. E. Gonzalez. 2003. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 185:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marketon, M. M., M. R. Gronquist, A. Eberhard, and J. E. Gonzalez. 2002. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J. Bacteriol. 184:5686-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 25.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 26.More, M. I., L. D. Finger, J. L. Stryker, C. Fuqua, A. Eberhard, and S. C. Winans. 1996. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science 272:1655-1658. [DOI] [PubMed] [Google Scholar]
- 27.Pappas, T., and S. C. Winans. 2003. A LuxR-type regulator from Agrobacterium tumefaciens elevates Ti plasmid copy number by activating transcription of plasmid replication genes. Mol. Microbiol. 48:1059-1073. [DOI] [PubMed] [Google Scholar]
- 28.Parsek, M. R., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 96:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 30.Qin, Y., Z. Q. Luo, A. J. Smyth, P. Gao, S. Beck von Bodman, and S. K. Farrand. 2000. Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO J. 19:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodelas, B., J. K. Lithgow, F. Wisniewski-Dye, A. Hardman, A. Wilkinson, A. Economou, P. Williams, and J. A. Downie. 1999. Analysis of quorum-sensing-dependent control of rhizosphere-expressed (rhi) genes in Rhizobium leguminosarum bv. viciae. J. Bacteriol. 181:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer, A. L., E. P. Greenberg, and M. R. Parsek. 2001. Acylated homoserine lactone detection in Pseudomonas aeruginosa biofilms by radiolabel assay. Methods Enzymol. 336:41-47. [DOI] [PubMed] [Google Scholar]
- 33.Schaefer, A. L., T. A. Taylor, J. T. Beatty, and E. P. Greenberg. 2002. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J. Bacteriol. 184:6515-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer, A. L., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1996. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc. Natl. Acad. Sci. USA 93:9505-9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, M. Stoffels, K. Riedel, M. Givskov, A. Hartmann, C. Langebartels, and L. Eberl. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steidler, L., J. M. Wells, A. Raeymaekers, J. Vandekerckhove, W. Fiers, and E. Remaut. 1995. Secretion of biologically active murine interleukin-2 by Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 61:1627-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens, A. M., and E. P. Greenberg. 1999. Transcriptional activation by luxR, p. 231-242. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 39.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vannini, A., C. Volpari, C. Gargioli, E. Muraglia, R. Cortese, R. De Francesco, P. Neddermann, and S. D. Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vincent, J. M. 1970. A manual for the practical study of root nodule bacteria. Blackwell Scientific Publications, Oxford, United Kingdom.
- 42.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 43.Winson, M. K., M. Camara, A. Latifi, M. Foglino, S. R. Chhabra, M. Daykin, M. Bally, V. Chapon, G. P. Salmond, B. W. Bycroft, A. Lazdunski, G. S. A. B. Stewart, and P. Williams. 1995. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:9427-9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, R. G., T. Pappas, J. L. Brace, P. C. Miller, T. Oulmassov, J. M. Molyneaux, J. C. Anderson, J. K. Bashkin, S. C. Winans, and A. Joachimiak. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971-974. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, J., J. W. Beaber, M. I. More, C. Fuqua, A. Eberhard, and S. C. Winans. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180:5398-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 96:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, J., and S. C. Winans. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA 98:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]