Abstract
The diversity of bacterial floras in the ilea and ceca of chickens that were fed a vegetarian corn-soy broiler diet devoid of feed additives was examined by analysis of 1,230 partial 16S rRNA gene sequences. Nearly 70% of sequences from the ileum were related to those of Lactobacillus, with the majority of the rest being related to Clostridiaceae (11%), Streptococcus (6.5%), and Enterococcus (6.5%). In contrast, Clostridiaceae-related sequences (65%) were the most abundant group detected in the cecum, with the other most abundant sequences being related to Fusobacterium (14%), Lactobacillus (8%), and Bacteroides (5%). Statistical analysis comparing the compositions of the different 16S rRNA libraries revealed that population succession occurred during some sampling periods. The significant differences among cecal libraries at 3 and 7 days of age, at 14 to 28 days of age, and at 49 days of age indicated that successions occurred from a transient community to one of increasing complexity as the birds aged. Similarly, the ileum had a stable bacterial community structure for birds at 7 to 21 days of age and between 21 to 28 days of age, but there was a very unique community structure at 3 and 49 days of age. It was also revealed that the composition of the ileal and cecal libraries did not significantly differ when the birds were 3 days old, and in fact during the first 14 days of age, the cecal microflora was a subset of the ileal microflora. After this time, the ileum and cecum had significantly different library compositions, suggesting that each region developed its own unique bacterial community as the bird matured.
Intestinal bacteria play an important role in health through their effects on gut morphology, nutrition, pathogenesis of intestinal disease, and immune responses. The microbial flora is also believed to protect against colonization of the intestines by pathogens and to stimulate the immune response (35, 37). Intestinal bacteria are primarily responsible for degrading the copious amounts of mucus produced by goblet cells in the intestine (14, 36). However, many factors can affect the composition of the avian bacterial community, such as diet (27), age (27, 57, 62), antibiotic administration (27), and infection with pathogenic organisms (26).
Extensive studies of the culturable bacterial flora of chickens have been conducted (44). The predominant culturable bacteria in the chicken cecum are obligate anaerobes, at the level of 1011 per g of content (6, 7). At least 38 different types of anaerobic bacteria have been isolated from the chicken cecum (7, 8), and they comprise many different bacterial strains (34). Mead (34) found that gram-positive cocci such as Peptostreptococcus composed 28% of the total culturable bacteria, with other bacteria including Bacteroidaceae (20%), Eubacterium spp. (16%), Bifidobacterium spp. (9%), budding cocci (6%), Gemmiger formicilis (5%), and Clostridium spp. (5%). However, other authors have estimated that only 10 to 60% of the total bacteria in the cecum were detected by culture (6, 7, 8, 34, 46). Only recently have molecular approaches been used to investigate the bacterial ecology of the chicken intestine. Netherwood et al. (39) used hybridization to monitor the response of bacterial flora to probiotic administration, a common feed additive in meat animal production. Diet-related differences in the cecal intestinal microbial community were analyzed by Apajalahti et al. (4), using G+C% profiling of 16S ribosomal DNA (rDNA) sequences. They showed that most of the genes detected were not from well-known bacterial species. These findings were confirmed by Gong et al. (19) and Zhu et al. (62), who determined that many of the 16S rDNA sequences retrieved from a cecal library exhibited low sequence similarity to the genes of known bacterial genera. Several studies have also addressed the composition of the chicken small intestinal microflora and have revealed a surprisingly diverse bacterial community (27, 57).
The microbial community of the gastrointestinal tract ultimately reflects the coevolution of microorganisms with their animal host and the diet adopted by the host (13). Changes in the composition of the animal's microflora can have beneficial or detrimental effects on health, growth, and maturation of the animal host (23), as is evident from the beneficial effects of rearing food animals on feeds containing antibiotics (38, 52). With concerns over agricultural use of these growth-promoting antibiotics and the emergence of antibiotic resistance in human or zoonotic pathogens, there is increasing pressure to eliminate this practice from animal husbandry. Unfortunately, it is unknown how these antibiotics influence the intestinal microflora and ultimately affect feed conversion, growth, and health of the food animal.
Previous chicken microflora studies have focused on animals reared on a feed containing an anticoccidial agent and protein supplementation (19, 62). Additional protein sources used to supplement poultry feed are generally derived from rendered food animals and, although initially sterile, the animal by-product is contaminated with microorganisms in the final production steps for poultry feed (24). If we are to identify the bacterial species that promote or interfere with good gastrointestinal health as a consequence of the use of feed antibiotics, it is important to first define the gastrointestinal microflora of chickens reared on a simple, vegetable diet devoid of other dietary supplements that may influence the microflora's composition.
In this study, we describe bacterial community succession in the ileal and cecal ecosystems of broiler chickens that were fed a corn-soy diet devoid of animal protein, antibiotics, or anticoccidials during their maturation to market weight. An understanding of the development of the normal bacterial community will allow us to detect disruption in the flora and analyze the effects of food animal management changes. This information may allow us to manipulate the intestinal flora with the intention of enhancing intestinal health and feed conversion.
MATERIALS AND METHODS
Animal sampling.
Sixty 1-day-old commercial Ross-hybrid broiler chicks, placed on sawdust bedding, were used as the source of bacteria for DNA extractions. The most popular breed of meat chickens, Ross-hybrids, has been bred for rapid growth and efficient feed conversion. Under standard management practices, this breed reaches market weight in 35 to 55 days (grow-out). Chicks were fed ad libitum a commercial corn-soy diet that did not contain animal protein, growth-promoting antibiotics, or coccidiostats. A starter feed (23% protein) was provided until the birds were 14 days of age and then was replaced by a grower feed (20% protein) for the remainder of the grow-out. The major difference between the two feeds is the percentage of crude protein (soy); fat is constant at 6% and the balance is made up by carbohydrates from corn and soy. Ten chicks were sacrificed at 3 and 7 days of age, and the intestinal contents were removed and pooled. At 14, 21, 28, and 49 days of age, five chicks were sacrificed and their intestinal contents were pooled. Intestinal contents were collected in the morning and pooled to reduce individual variation. One cecum was removed from each bird and the ileum was removed between the duodenum and Merkel's diverticulum, they were cut open aseptically, and the contents were placed into sterile tubes containing brain heart infusion broth. The tubes were kept on ice until the bacteria were collected.
Bacterial DNA isolation.
The bacterial fraction was recovered from the intestinal contents by density gradient centrifugation as described by Apajalahti et al. (3). Bacterial cells were lysed with the beads, solution 1, and IRS of the Mo Bio kit (Mo Bio Laboratories Inc., Solana Beach, Calif.) by beating at 6,000 rpm for 20 min on a vortex shaker. Genomic DNA was extracted as follows. Lysed cells were treated with sodium dodecyl sulfate (final concentration, 0.5%) and proteinase K (final concentration, 0.1 mg/ml) and incubated at 37°C for 30 min. The sample was extracted twice with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1). DNA was concentrated with a 0.6 volume of isopropanol and resuspended in sterile water. DNA concentration was measured with a Beckman DU640 spectrophotometer (Beckman Instruments, Inc., Fullerton, Calif.).
PCR for construction of 16S rDNA clone libraries.
For construction of the 16S rRNA gene clone libraries, three sets of primers which target the domain Bacteria were used (31). These were 8F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-TAC GG(C/T) TAC CTT GTT ACG ACT T-3′) (set A), 8F and 1522R (5′-AAG GAG GTG ATC CAN CCR CA-3′) (set B), and 8F and 926R (5′-ACC GCT TGT GCG GGC CC-3′) (set C). Primer 1492R was synthesized as a mixture of oligonucleotides with either T or C at position 1497 (Escherichia coli numbering). Primer sets A and B are frequently used for molecular diversity studies because they result in a nearly full-length 16S rDNA product and are considered universal for the domain Bacteria and for the prokaryotes, respectively (32). Primer set C was used to minimize the effect of template concentration on PCR bias (11). Final reaction mixtures included 25 or 100 ng of template DNA with primer set C and 25 ng with other primer sets, 1× PCR buffer, 2.0 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 1 μM (each) primers, and 0.05 U of Taq DNA polymerase (Roche Molecular Biochemicals) in a final reaction volume of 25 μl. Initial DNA denaturation was performed at 94°C for 2 min in a PTC200 thermocycler (MJ Research, Inc., Watertown, Mass.), followed by 15 cycles of denaturation at 94°C for 1 min, annealing at 54°C (primer set A), 48°C (primer set B), or 58°C (primer set C) for 30 s, and elongation at 72°C for 1 min and then a final elongation step at 72°C for 10 min. Fifteen cycles of PCR were performed rather than 30 cycles to minimize PCR bias (11) associated with preferential amplification of certain 16S rDNA types (54). Three separate PCR amplifications were performed to minimize potential bias and to increase the DNA yield for subsequent cloning. The nine PCRs generated with the three PCR primer sets were pooled together for cloning. The amplified PCR products were purified with the Wizard PCR product purification kit (Promega, Madison, Wis.). The purified products were ligated into pGEM-T Easy (Promega). Ligation was done at 4°C overnight, followed by transformation into competent E. coli JM109 cells by heat shock (45 s at 42°C). The clones were screened for α-complementation of β-galactosidase by using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside) (5).
Plasmid extraction and sequencing.
DNA preparations for sequencing were made with the QIAprep spin plasmid kit (Qiagen, Valencia, Calif.) as specified by the manufacturer. Plasmids were eluted with 50 μl of water and stored at −70°C. Sequencing reactions were performed by use of a PE-ABI Big Dye terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) as described by the manufacturer, and electrophoresis and readout were performed with an ABI PRISM 3700 DNA analyzer (Applied Biosystems). Primers T7 and SP6 were used to sequence both strands of each PCR product.
Analysis of DNA sequences.
Resulting DNA sequences were edited to exclude primer binding sites and ambiguous bases and were assembled into contiguous sequences (570 to 650 bp) by use of the Sequencher program, version 4.10 (Gene Code Corp., Ann Arbor, Mich.). The resulting DNA sequence information was analyzed by use of the programs FASTA (41) and BLAST (2). Chimeric sequences were detected as described by Suau et al. (53).
The estimate of library coverage was calculated as described by Good (20) and was applied to quantitative comparisons of 16S rRNA gene sequence libraries as described by Singleton et al. (49). The same definitions for the variables in the formula Cx = 1 − (Nx/n) were used as those by Singleton et al. (49), i.e., Cx is the homologous coverage of sample X, Nx is the number of unique sequences, and n is the total number of sequences in the sample. Coverage was determined at 98% 16S rDNA sequence similarity (32, 53). Nx was analyzed by using the Sequencher program (v 4.1). Differences in the composition of the floras were estimated by comparisons of the libraries as described by Singleton et al. (49), using the LIBSHUFF utility (www.arches.uga.edu/∼whitman/libshuff.html). If the P values were <0.05, the two libraries were considered significantly different from each other, indicating that the microbial compositions were different.
Nucleotide sequence accession numbers.
Representative DNA sequences were submitted to GenBank under accession numbers AY235577 to AY235675.
RESULTS
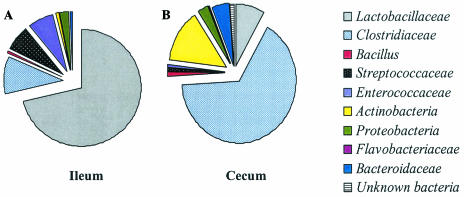
Total 16S rRNA partial gene sequences (350 to 410 bp) (1,230) were compared to sequences present in GenBank. This analysis showed that four major groups dominated the chicken intestinal bacterial community: low- and high-G+C gram-positive bacteria, proteobacteria, and the Cytophaga/Flexibacter/Bacteroides group (Fig. 1). The low-G+C gram-positive group, which included Lactobacillus, clostridia, Bacillus, and streptococci, was the most abundant group of organisms in both the ileum and the cecum. In a previous study, we also found a community of low-G+C gram-positive bacteria in broiler litter (manure), which is a mix of feces and composted bedding material (31). Thirteen genera were found to be common to both the ileum and cecum; however, the abundances of these were quite different (Table 1). In the ileum, Lactobacillus species accounted for 67% of the total 16S rDNA sequences, but the Clostridiaceae (68%) dominated the bacterial community of the cecum. Streptococcus and Enterococcus 16S rDNA sequences were also more abundant in the ileum than in the cecum, but Fusobacterium sequences were detected at higher levels in the cecum (14.3%) than in the ileum (0.9%). Streptococcus and Enterococcus were more abundant in the ileum (13.2%) than in the cecum (1.7%). Only a very small percentage of proteobacteria-related sequences were represented in the libraries (2.5% in the ileal library and 2.8% in the cecal library). Statistical comparisons of the libraries revealed that the composition of the ileal and cecal bacterial floras did not significantly differ when the birds were 3 days of age (Table 2). However, during the period of 7 to 14 days of age, the cecal microflora was a subset of the ileal microflora. After this time, the ileum and cecum had significantly different library compositions, suggesting that each region developed its own unique bacterial community.
FIG. 1.
Composition of the bacterial floras of the ilea and ceca of broiler chickens as determined by sequencing of 1,230 clones from a 16S rDNA community DNA library. (A) Bacterial composition of the ileum. (B) Bacterial composition of the cecum.
TABLE 1.
Bacterial genera detected in both the ileal and cecal 16S rDNA libraries
| Group (% of total) | Genus | % of genus in:a
|
|
|---|---|---|---|
| Ileum | Cecum | ||
| Low G+C, gram positive | Lactobacillus | 67.59 | 7.75 |
| (ileum, 94.18; cecum, 76.9) | Weisella | 1.05 | 0.48 |
| Clostridium | 9.69 | 39.26 | |
| Ruminococcus | 0.44 | 16.48 | |
| Eubacterium | 0.73 | 9.85 | |
| Bacillus | 0.67 | 1.45 | |
| Staphylococcus | 0.95 | 0 | |
| Streptococcus | 6.63 | 0.65 | |
| Enterococcus | 6.43 | 0.97 | |
| High G+C gram positive | Fusobacterium | 0.73 | 13.89 |
| (ileum, 0.92; cecum, 13.89) | Bifidobacterium | 0.19 | 0 |
| Proteobacteria (gram negative) | Ochrobacterium | 0.18 | 0.81 |
| (ileum, 2.28; cecum, 2.75) | Alcaligenes | 0.88 | 0.65 |
| Escherichia | 0.35 | 1.29 | |
| Campylobacter | 0.88 | 0 | |
| Cytophaga/Flexibacter/Bacteroides | Flavobacterium | 0 | 0.16 |
| (ileum, 0.6; cecum, 5.19) | Bacteroides | 0.60 | 5.01 |
A total of 614 and 616 sequences were analyzed from the ileum and cecum, respectively.
TABLE 2.
P value distribution of 16S rDNA gene sequence libraries compared among samples from ileum and cecuma
| Age of chicken (days) | Pair-wise comparison | P value |
|---|---|---|
| 3 | Cecum-ileum | 0.567 |
| Ileum-cecum | 0.095 | |
| 7 | Cecum-ileum | 0.092 |
| Ileum-cecum | 0.001 | |
| 14 | Cecum-ileum | 0.083 |
| Ileum-cecum | 0.001 | |
| 21 | Cecum-ileum | 0.001 |
| Ileum-cecum | 0.001 | |
| 28 | Cecum-ileum | 0.001 |
| Ileum-cecum | 0.001 | |
| 49 | Cecum-ileum | 0.001 |
| Ileum-cecum | 0.001 |
Differences were determined by pair-wise comparisons of the cecal clone libraries with the ileal libraries and vice versa. Except for a similar composition at 3 days of age (P > 0.05), the ileal composition was significantly different from that of the cecum at all ages. However, at 7 and 14 days, the composition of the cecal library appeared to be a subset of the ileal library.
Ileal (614) and cecal (616) sequences were analyzed to calculate library coverage. There were 78 and 142 unique 16S rDNA sequences identified from the ileum and cecum, respectively, at a similarity level of 98%. Thus, the coverage was 88 and 77% for the ileum and cecum, respectively, suggesting that these unique ribotypes represent the major genera and species present in the chicken intestine. We identified several 16S sequences demonstrating homology to bacteria that are potentially pathogenic for chickens, such as Staphylococcus aureus (50), Clostridium perfringens (17), E. coli (9), and Campylobacter coli (47).
Microbial composition of the cecum.
Of the 619 clones, the most abundant sequences (65.6%) were homologous to Clostridiaceae (low-G+C, gram positive), while Fusobacterium (high-G+C, gram positive) strains accounted for 13.9% (Fig. 1). Less than 10% of the cecal 16S rDNA sequences were related to gram-negative bacteria, and of these, 2.8% were proteobacteria, and Bacteroidaceae comprised 5.1% of the total. The diversity and composition of the microfloras at different ages are shown in Table 3. At 3 days of age, 15% of the total 16S rDNA sequences from the broiler chicken cecum were identified as belonging to Proteobacteria (α, β, and γ). Lactobacillus represented one-fourth of the total sequences at 3 days of age, with Lactobacillus delbrueckii and Lactobacillus acidophilus as the dominant species. At later times, the lactobacilli were reduced in abundance (1 to 11% of total), but no single species of Lactobacillus was clearly dominant.
TABLE 3.
Abundance of bacterial 16S rDNA sequences (n = 619) isolated from the cecal floras of broiler chickens
| Group | Class | Genus or species | Abundance of sequence (no. of sequence [%]) at day:
|
|||||
|---|---|---|---|---|---|---|---|---|
| 3 | 7 | 14 | 21 | 28 | 49 | |||
| Low G+C, gram positive | Lactobacillaceae | Lactobacillus spp. | 3 (2.86) | 1 (1.01) | 1 (0.88) | |||
| L. acidophilus | 8 (7.62) | 1 (1.08) | 4 (3.60) | 4 (4.12) | ||||
| L. crispatus | 1 (0.95) | 3 (3.23) | 3 (2.70) | |||||
| L. delbrueckii | 14 (13.33) | 2 (1.80) | ||||||
| L. reuteri | 1 (0.90) | 1 (1.03) | ||||||
| L. aviarius | 1 (1.03) | |||||||
| Weissella spp. | 1 (0.95) | 1 (0.90) | 1 (1.03) | |||||
| Clostridiaceae | Clostridium spp. | 33 (31.43) | 56 (60.22) | 42 (37.84) | 25 (25.25) | 34 (29.82) | 33 (34.02) | |
| C. perfringens | 13 (12.38) | |||||||
| C. lactofermentum | 2 (1.75) | 1 (1.03) | ||||||
| C. orbiscindens | 4 (4.04) | |||||||
| Ruminococcus | 7 (6.67) | 16 (17.20) | 29 (26.13) | 16 (16.16) | 18 (15.79) | 16 (16.49) | ||
| Eubacterium spp. | 1 (0.95) | 7 (7.53) | 10 (9.00) | 11 (11.11) | 10 (8.77) | 22 (22.68) | ||
| Bacillaceae | Bacillus | 3 (2.70) | 4 (4.04) | 2 (1.75) | ||||
| Streptococcaceae | Streptococcus | 3 (2.86) | 1 (1.03) | |||||
| Enterococcaceae | Enterococcus faecium | 2 (1.90) | 2 (2.15) | 1 (0.90) | 1 (1.03) | |||
| Enterococcus durans | 1 (1.03) | |||||||
| High G+C, gram positive | Fusobacteriaceae | Fusobacterium spp. | 2 (2.15) | 10 (9.00) | 27 (27.27) | 40 (35.09) | 7 (7.22) | |
| Proteobacteria (gram negative) | α | Ochrobacter anthropi | 5 (4.76) | |||||
| β | Achromobacter xylosoxidans | 4 (3.81) | ||||||
| Alcaligenes sp. | ||||||||
| γ | Escherichia coli | 7 (6.67) | 1 (0.9) | |||||
| Cytophaga/Flexibacter/Bacteroides | Flavobacteriaceae | Flavobacterium ferrugineum | 1 (0.95) | |||||
| Bacteroidaceae | Bacteroides | 1 (0.95) | 5 (5.38) | 4 (3.60) | 10 (10.10) | 6 (5.26) | 5 (5.15) | |
| Unknown bacteria | 1 (0.95) | 1 (1.08) | 1 (1.01) | 1 (0.88) | 3 (3.09) | |||
| Total | 105 | 93 | 111 | 99 | 114 | 97 | ||
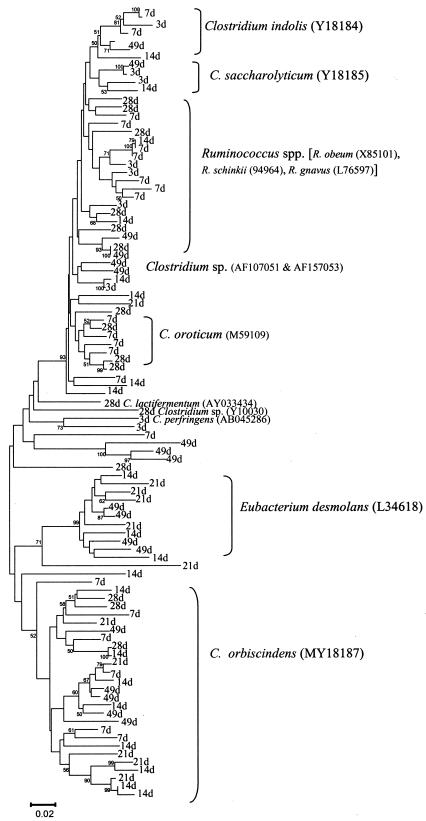
The Clostridiaceae were the major group in all sample ages, representing 51 to 85% of the total cecal 16S rDNA sequences. Many of these cecal 16S rDNA sequences had relatively low percent identity with clostridial sequences present in the GenBank 16S rDNA database at the time of this analysis. Except for a few sequences with 98 to 99% identity to C. perfringens, Clostridium lactifermentum, segmented filamentous bacteria, and Clostridium orbiscindens, most of the sequence similarities ranged from 89 to 95%. Clostridial cecal 16S rDNA sequences were grouped in several clusters. Figure 2 shows the relatedness of these clostridial sequences; the cluster was labeled if percent identity values were ≥95%. The Ruminococcus species (R. schinkii, R. obeum, and R. gnavus) represented the most abundant population of clostridia in the cecum (15.6% of the total sequences). While sequences related to Clostridium indolis accounted for 8.5%, C. orbiscindens 4.4%, and Eubacterium 10.0%, sequences with low homology to known Clostridium spp. made up 12.3% of the total cecal sequences.
FIG. 2.
Phylogenetic tree showing 16S rDNA sequences related to the Clostridiaceae. The majority of the sequences were isolated from the cecal library. The tree was constructed by neighbor-joining analysis of a distance matrix obtained from a multiple-sequence alignment. Bootstrap values (expressed as percentages of 100 replications) are shown at branch points; values of <50 were not considered significant. The cluster was labeled if some sequences exhibited percent identity values of at least 95%.
At 7 days of age, sequences related to the Clostridium saccharolyticum (24.7%), Clostridium oroticum (15.1%), and C. orbiscindens (12.9%) clusters dominated the cecal bacterial community, suggesting that these bacterial strains replaced the initially diverse population of clostridia. Similarly, at 14 to 28 days of age, 20 to 36% of the sequences were related to R. schinkii and 7 to 15% were related to C. indolis, demonstrating that the community had again undergone population succession. The sequences at 49 days of age had the highest Eubacterium (22.7%) abundance, but 27% of the Clostridium sequences had low similarity to known species. Fusobacterium sequences were recovered at 7 to 49 days of age but were in very high abundance at 21 (27.3%) and 28 (35.1%) days of age. At all ages, Clostridium, Eubacterium, and Ruminococcus dominated the bacterial community of the cecum.
The composition of chicken litter is believed to reflect the composition of cecal droppings of chickens. The most abundant genera in litter are Facklamia, Salinococcus, and Corynebacterium (31), which we did not detect in the 16S libraries of the cecum. A significant percentage of the total litter bacterial community was also composed of β-proteobacteria, but we detected few of these species in the cecal flora. These differences suggest that the microbial community of litter is substantially modified by the modest amount of composting that occurs with aging of the litter in the flock house.
In order to study the changes of the bacterial community as the birds matured, we compared the libraries at different ages in a pair-wise manner (49). Table 4 shows the results of the statistical evaluation; at some ages the composition of the cecal microflora varied significantly (P < 0.05). For example, the composition of the cecal flora was significantly different (P = 0.001) at 3 and 7 days of age compared to the other ages, suggesting that the composition was transient in young chicks. However, the flora at 14 to 28 days of age was not significantly different, except that the 14-day library appeared to be a subset of the 28-day library, suggesting increasing complexity of the flora as the birds aged. The 49-day library was significantly different from those of the other ages, indicating further maturation of the bacterial flora.
TABLE 4.
P-value distribution of 16S rDNA gene sequence libraries among samples from birds of different agesa
| Source of library | Age (days) |
P value at day:
|
|||||
|---|---|---|---|---|---|---|---|
| 3 | 7 | 14 | 21 | 28 | 49 | ||
| Cecum | 3 | 1 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| 7 | 0.001 | 1 | 0.008 | 0.001 | 0.134 | 0.002 | |
| 14 | 0.001 | 0.001 | 1 | 0.231 | 0.743 | 0.293 | |
| 21 | 0.001 | 0.001 | 0.10 | 1 | 0.669 | 0.003 | |
| 28 | 0.001 | 0.001 | 0.015 | 0.100 | 1 | 0.014 | |
| 49 | 0.001 | 0.001 | 0.003 | 0.001 | 0.020 | 1 | |
| Ileum | 3 | 1 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| 7 | 0.001 | 1 | 0.048 | 0.041 | 0.001 | 0.001 | |
| 14 | 0.001 | 0.937 | 1 | 0.172 | 0.436 | 0.001 | |
| 21 | 0.044 | 0.997 | 0.740 | 1 | 0.567 | 0.001 | |
| 28 | 0.001 | 0.001 | 0.001 | 0.249 | 1 | 0.028 | |
| 49 | 0.001 | 0.001 | 0.001 | 0.001 | 0.124 | 1 | |
Differences were determined by pair-wise comparisons of the heterologous coverage of each clone library with that of the other libraries and with itself, using LIBSHUFF. If the P values were <5% (P < 0.05), significant differences existed between the two clone libraries. If x compared to y is not significantly different, but y compared to x produces P of <0.05, then x is considered a subset of y.
Microbial composition of the ileum.
Of the 614 16S rDNA clones, Lactobacillus species were most abundant, at 68.5% of the total sequences. Most of the sequences were related to those of cultivated species of bacteria, such as Enterococcus cecorum and Enterococcus faecium (identity, 98 to 100%), Streptococcus alactolyticus and Streptococcus intestinalis (identity, 97 to 99%), and C. perfringens (identity, 99%). Table 5 shows the composition of the ileal libraries. Only a few (2.5%) Proteobacteria-related sequences were detected; these were related to Achromobacter xylosoxidans, Alcaligenes faecalis, Campylobacter coli, and E. coli. Other sequences related to Clostridiaceae (9.7%) were represented mainly by Clostridium lituseburense relatives (identity, 94 to 98%; abundance, 6.4%), C. perfringens (identity, 98 to 99%), and R. gnavus (identity, 93 to 94%; abundance, 0.5%).
TABLE 5.
Abundance of bacterial 16S rDNA sequences (n = 610) isolated from the ileal floras of broiler chickens
| Group | Class | Genus or species | Abundance of sequence (no. of sequences [%]) at day:
|
|||||
|---|---|---|---|---|---|---|---|---|
| 3 | 7 | 14 | 21 | 28 | 49 | |||
| Low G+C, gram positive | Lactobacillaceae | Lactobacillus spp. | 1 (1.05) | 2 (2.22) | 1 (0.88) | 4 (4.04) | ||
| L. acidophilus | 7 (7.37) | 54 (58.9) | 54 (52.9) | 57 (50) | 3 (2.72) | |||
| L. crispatus | 4 (4.21) | 1 (1.11) | 8 (7.84) | 3 (2.63) | 82 (74.5) | 36 (36.4) | ||
| L. reuteri | 3 (2.94) | 5 (4.39) | 8 (7.27) | 1 (1.01) | ||||
| L. delbrueckii | 40 (42.1) | 1 (1.11) | ||||||
| Weisella spp. | 6 (6.32) | |||||||
| L. salivarius | 6 (5.26) | 2 (1.82) | 28 (28.3) | |||||
| L. gasseri | 3 (2.63) | |||||||
| Clostridiaceae | Clostridium spp. | 1 (1.05) | 1 (1.11) | 7 (6.86) | 9 (7.89) | 7 (6.36) | 19 (19.2) | |
| C. perfringens | 15 (15.8) | |||||||
| Ruminococcus | 3 (2.63) | |||||||
| Eubacterium spp. | 5 (4.39) | |||||||
| Bacillus spp. | 4 (4.04) | |||||||
| Staphylococcaceae | Staphylococcus | 2 (2.11) | 3 (2.63) | |||||
| Streptococcaceae | Streptococcus | 2 (2.11) | 16 (17.78) | 17 (16.67) | 3 (2.63) | 1 (0.91) | ||
| Enterococcaceae | Enterococcus | 3 (3.16) | 14 (15.56) | 13 (12.75) | 3 (2.63) | 3 (2.72) | 2 (2.02) | |
| High G+C, gram positive | Fusobacteriaceae | Fusobacterium prausnitzii | 5 (4.39) | |||||
| Bifidobacteriaceae | Bifidobacterium spp. | 1 (1.11) | ||||||
| Proteobacteria (gram negative) | α | Ochrobacterium | 1 (1.05) | |||||
| β | Alcaligenes | 4 (4.21) | ||||||
| A. faecalis | 1 (1.05) | |||||||
| ɛ | Campylobacter | 5 (5.26) | ||||||
| γ | E. coli | 2 (2.11) | ||||||
| Cytophaga/Flexibacter/Bacteroides | Bacteroidaceae | Bacteroides spp. | 3 (2.63) | 1 (1.01) | ||||
| Unknown bacteria | 2 (2.11) | 5 (4.39) | 4 (3.60) | 4 (4.04) | ||||
| Total | 95 | 90 | 102 | 114 | 110 | 99 | ||
L. acidophilus, Clostridium, Streptococcus, and Enterococcus were dominant 16S rDNA sequences in the ileal libraries for 7 to 21 days of age. Similar to in the cecum, 13% of clones from the ileal 16S rDNA library at 3 days of age had similarity to α, β, ɛ, and γ proteobacterial sequences, but these sequences were not identified among numerous clones sequenced from subsequent 16S rDNA ileal libraries. The ileal library at 3 days of age was statistically significantly different (P < 0.05) from those at all other ages, suggesting that the composition of the ileum of young chicks is transient (Table 4). The analysis indicated that most sequences from the clone libraries of days 3, 7, and 49 had a high dissimilarity to the sequences from the libraries of the other ages. However, the analysis showed that the 14-, 21-, and 28-day clone libraries were not significantly different (P > 0.10), indicating that the ileal flora was very similar during this time. This is somewhat surprising since the diet composition changed from starter to grower feed during this time. For birds at 7 to 21 days of age and between 21 and 28 days of age, the ileum had a stable bacterial community structure, but there was a very unique community structure at 3 and 49 days of age.
The dominant sequences homologous to Lactobacillus varied from L. delbrueckii at 3 days to L. acidophilus from days 7 to 21 and Lactobacillus crispatus from days 28 to 49. At 49 days of age, the community exhibited another shift in the Lactobacillus population, with an increase in Lactobacillus salivarius. The abundance of sequences with homology to clostridia tended to increase from 3 to 49 days of age, with the exception of an abundance of C. perfringens at 3 days of age. About 15% of the cecal 16S rDNA total sequences at 3 days of age had homology to C. perfringens, which is an important cause of necrotic enteritis in broilers and is generally controlled by use of growth-promoting antibiotics (18, 30). However, we detected sequences in the ileal 16S rDNA library at 14 days of age that were homologous to the segmented filamentous bacteria commonly associated with a healthy gastrointestinal tract in animals (51).
DISCUSSION
The chicken intestinal microflora has been investigated extensively by use of culture-based methods (6, 7, 8, 33, 46). These studies have shown that gram-positive bacteria were usually cultured, and the abundance of gram-positive bacteria has been confirmed by microscopic examination (19). Our data, generated by molecular detection, corroborated the culture-based results in that lactobacilli predominated in the small intestine (with smaller numbers of streptococci and enterobacteria), whereas the cecal flora is composed mainly of anaerobes and fewer numbers of facultative bacteria (6). Salanitro et al. (46) enumerated anaerobic bacteria from the ilea and ceca of 14-day-old chicks and showed that the predominant cultured flora of the ileum included Lactobacillus (33.8 to 59%), Streptococcus (8.9 to 16.8%), E. coli (14.7 to 33%), and eubacteria (9 to 24.3%), while eubacteria (60.6%) and Bacteroides (12.8) dominated the flora of the cecum. Our findings were similar, with Lactobacillus and Streptococcus accounting for 62.7 and 12.2% of the microflora, respectively. However, we detected many species of Clostridiaceae in both the ileum and cecum, while the abundance of proteobacteria was fairly low. Barnes et al. (6) and Salanitro et al. (46) also observed that the microbial community structure varies with age. Our studies not only confirmed this microbial succession in young chicks, but it also indicated that the microbial community structure was fairly stable during the period of rapid skeletal growth (14 to 28 days of age) and then had a significant change during the period of significant weight gain at the end of the grow-out (49 days). The unique microbial community at 3 days of age suggests that the early bacterial community is relatively transient and is replaced by a stable bacterial community later in life. However, broiler chickens are harvested as juvenile birds, as puberty begins at about 210 days (30 weeks) of age; therefore, the bacterial community of a 49-day-old broiler reflects that of a relatively young bird.
Although there are limitations of molecular methods such as PCR in providing accurate quantitative measurements of the actual composition of a microbial community (15, 54), recent studies of the chicken intestine have produced remarkably similar results. Gong et al. (19) used a combination of culture, 16S rDNA libraries, and 16S rDNA terminal restriction fragment length polymorphism analysis to characterize the bacterial community of the ceca of 6-week-old (42 days) broiler chickens that were fed a corn-soy diet without growth promoters. The most abundant groups comprising the libraries were the low-G+C gram-positive bacteria (Clostridium and Ruminococcus) and Fusobacterium praunitzii (19). Zhu et al. (62) reported similar results, using 16S rDNA libraries and temperature gradient gel electrophoresis to study the floras of broiler chickens fed a corn-soy diet that contained animal protein and an anticoccidial compound. Clostridia were the dominant group; however, they reported that 40% of their library sequences were related to Sporomusa or enteric bacteria related to the γ-proteobacteria, such as E. coli. Similar studies have focused on the small intestinal microflora and illustrate that lactobacilli are abundant (27, 57).
Recent molecular approaches have revealed that the composition of the microbiota varies with different diets, feed additives (4, 27, 62), and probiotics (39). The microfloras of our birds, which were fed a vegetarian diet lacking growth-promoting antibiotics and antiparasitic agents, contained more clostridia and Bacillus-Lactobacillus-Streptococcus than those reported by Zhu et al. (62). However, we did not detect sequences related to the Sporomusa group, Actinomyces, or Atopobium, and our libraries contained few sequences related to the γ-proteobacteria that were so abundant in the Zhu study. It is possible that PCR bias contributed to the differences in the findings; differences in PCR conditions have been shown to inaccurately reflect the abundance of some genera (11). Differences in universal bacterial primer sequences can also bias the abundance (11), but Gong et al. (19) used the same primer sets that were used by us and Zhu et al. (62), suggesting that diet may have been the main contributor to the differences in microbial community among the investigations. Unfortunately, we are unable to provide relevant comparisons, because the study by Zhu et al. (62) contains limited information relating to the composition of the chickens' diets, while the study by Gong et al. (19) contains limited information relating to the abundance of the bacterial flora.
Numerous studies of the intestinal microbiota of humans (53, 60), pigs (43), mice (12), termites (40), and chickens (62) have contributed novel 16S rRNA sequences to GenBank. Regardless of animal species, there is a clear demarcation in microbial composition within the gastrointestinal tract (28). In chickens, there are clear differences in the carbohydrate composition of glycoproteins and glycolipids that adorn the epithelial cells lining the gastrointestinal tract (1). The microorganisms that occupy these niches have evolved the ability to utilize various unique polysaccharides present in mucin and the glycolipids and glycoproteins that adorn the epithelial cells lining the gastrointestinal tract. Microbial composition may therefore reflect the host's unique carbohydrate composition of mucins, glycolipids, and glycoproteins and the coevolution of lectins and glycosidases that allow the bacteria to adhere to these substrates and to liberate sugar for metabolism (29, 36, 45, 48). This demarcation within the gastrointestinal tract also reflects the different physiological functions within this organ system and coevolution of the microorganisms with their animal host, producing microbial processes that serve to promote a healthy gastrointestinal tract (25, 42, 55, 56, 61). It is therefore not surprising to find the same genera or species occupying the same niche in the gastrointestinal tract, regardless of animal host, especially when the physiological function is similar among animal species. We identified several 16S rDNA sequences from chickens with identity to gastrointestinal microflora sequences from other animal species, including humans (22, 43, 58). However, we also identified several clones from our 16S rDNA libraries that exhibited the highest identity to sequences from bacteria isolated from chickens or chicken litter. These results suggest that host animals may have coevolved with unique strains of bacteria that have adapted to utilizing polysaccharides unique to the animal host (13). Studies have shown that fecal bacteria from animals can be differentiated on the basis of ribosomal operon sequencing, ribotyping, or amplified fragment length polymorphism analysis (10, 16, 21, 59). Additional studies using molecular typing techniques should reveal a host of relevant data concerning source point contamination, microbial evolution, microbial symbiosis, and effect of diet and other host factors on microbial community structure.
Acknowledgments
We thank William Whitman for critically reviewing the manuscript.
This work was funded by the United States Department of Agriculture (USDA NRICGP 99-35212-8680 to J. J. Maurer, USDA NRICGP 01-35212-10877 to C. H. Hofacre) and USDA Formula Funds (M. D. Lee and B. G. Harmon).
REFERENCES
- 1.Alroy, J., V. Goyal, N. W. Lukacs, R. L. Taylor, R. G. Strout, H. D. Ward, and M. E. A. Pereira. 1989. Glycoconjugates of the intestinal epithelium of the domestic fowl (Gallus domesticus): a lectin histochemistry study. Histochem. J. 21:187-193. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Apajalahti, J. H. A., L. K. Sarkilahti, B. R. E. Maki, J. P. Heikkinen, P. H. Nurminen, and W. E. Holben. 1998. Effective recovery of bacterial DNA and percent guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl. Environ. Microbiol. 64:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apajalahti, J. H. A., A. Kettunen, M. R. Bedford, and W. E. Holben. 2001. Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens. Appl. Environ. Microbiol. 67:5656-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struh. 1993. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 6.Barnes, E. M., G. C. Mead, D. A. Barnum, and E. G. Harry. 1972. The intestinal flora of the chicken in the period 2 to 6 weeks of age, with particular reference to the anaerobic bacteria. Br. Poult. Sci. 13:311-326. [DOI] [PubMed] [Google Scholar]
- 7.Barnes, E. M., and C. S. Impey. 1972. Some properties of the nonsporing anaerobes from poultry caeca. J. Appl. Bacteriol. 35:241-251. [DOI] [PubMed] [Google Scholar]
- 8.Barnes, E. M. 1979. The intestinal microflora of poultry and game birds during life and after storage. J. Appl. Bacteriol. 46:407-419. [DOI] [PubMed] [Google Scholar]
- 9.Barnes, H. J., and W. B. Gross. 1997. Colibacillosis, p. 131-142. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. R. McDougald, and Y. M. Saif (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames, Iowa.
- 10.Buchan, A., M. Alber, and R. E. Hodson. 2001. Strain-specific differentiation of environmental Escherichia coli isolates via denaturing gradient gel electrophoresis (DGGE) analysis of the 16S-23S intergenic spacer region. FEMS Microbiol. Ecol. 35:313-321. [DOI] [PubMed] [Google Scholar]
- 11.Chandler, D. P., J. K. Fredrickson, and F. J. Brockman. 1997. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol. Ecol. 6:475-482. [DOI] [PubMed] [Google Scholar]
- 12.Depalncke, B., K. R. Hristova, H. A. Oakley, V. J. McCracken, R. Aminov, R. I. Mackie, and H. R. Gaskins. 2000. Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 66:2166-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drasar, B. S., and P. A. Barrow. 1985. Intestinal microbiology, p. 19-42. American Society for Microbiology, Washington, D.C.
- 14.Falk, P. G., L. V. Hooper, T. Midtvedt, and J. I. Gordon. 2000. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field, K. G., A. E. Bernhard, and T. J. Brodeur. 2003. Molecular approaches to microbiological monitoring: fecal source detection. Environ. Monit. Assess. 81:313-326. [PubMed] [Google Scholar]
- 17.Finken, M. D., and D. P. Wages. 1997. Necrotic enteritis, p. 261-267. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. R. McDougald, and Y. M. Saif (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames, Iowa.
- 18.George, B. A., C. L. Quarles, and D. J. Fagerberg. 1982. Virginiamycin effects on controlling necrotic enteritis infection in chickens. Poult. Sci. 61:447-450. [DOI] [PubMed] [Google Scholar]
- 19.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, P. M. Sabour, R. Wheatcroft, and S. Chen. 2002. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 208:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 21.Guan, S., R. Xu, S. Chen, J. Odumeru, and C. Gyles. 2002. Development of a procedure for discriminating among Escherichia coli isolates from animal and human sources. Appl. Environ. Microbiol. 68:2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heilig, H. G., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill, M. J. 1982. Gut flora associated diseases in man. J. Vet. Med. 33(Suppl.):32-36. [Google Scholar]
- 24.Hofacre, C. L., D. G. White, J. J. Maurer, C. Morales, C. Lobsinger, C. R. Hudson, and S. G. Thayer. 2001. Antibiotic resistant bacteria in rendered animal products. Avian Dis. 45:953-961. [PubMed] [Google Scholar]
- 25.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 26.Kimura, N., F. Mimura, S. Nishida, and A. Kobayashi. 1976. Studies on the relationship between intestinal flora and cecal coccidiosis in chicken. Poult. Sci. 55:1375-1383. [DOI] [PubMed] [Google Scholar]
- 27.Knarreborg, A., M. A. Simon, R. M. Engberg, B. B. Jensen, and G. W. Tannock. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Environ. Microbiol. 68:5918-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koopman, J. P., H. M. Kennis, A. M. Stadhouders, and H. De Boer. 1983. Some aspects of the gastrointestinal microflora of germfree mice associated with cultured microfloras. Lab. Anim. 17:188-195. [DOI] [PubMed] [Google Scholar]
- 29.Leblond, F., H. Boureau, and P. Bourliquix. 1997. The cloning of sialidase genes from Clostridium indolis and Clostridium coleatum. Clin. Infect. Dis. 25(Suppl. 2):S156-S157. [DOI] [PubMed] [Google Scholar]
- 30.Long, J. R. 1973. Necrotic enteritis in broiler chickens. I. A review of the literature and prevalence of the disease in Ontario. Can. J. Comp. Med. 37:302-308. [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, J., S. Sanchez, C. Hofacre, J. J. Maurer, B. G. Harmon, and M. D. Lee. 2003. Evaluation of broiler litter with reference to the microbial composition as assessed by using 16S rDNA and functional gene markers. Appl. Environ. Microbiol. 69:901-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mead, G. C., and B. W. Adams. 1975. Some observations on the caecal microflora of the chick during the first two weeks of life. Br. Poult. Sci. 16:169-176. [DOI] [PubMed] [Google Scholar]
- 34.Mead, G. C. 1989. Microbes of the avian cecum: types present and substrates utilized. J. Exp. Zool. 3(Suppl.):48-54. [DOI] [PubMed] [Google Scholar]
- 35.Mead, G. C. 2000. Prospects for competitive exclusion treatment to control salmonellas and other foodborne pathogens in poultry. Vet. J. 159:111-123. [DOI] [PubMed] [Google Scholar]
- 36.Meslin, J. C., N. Fontaine, and C. Andrieux. 1999. Variation of mucin distribution in the rat intestine, caecum and colon: effect of the bacterial flora. Comp. Biochem. Physiol. A 123:235-239. [DOI] [PubMed] [Google Scholar]
- 37.Metchnikoff, E. 1903. Prolongation of life. G. Putnam's Sons, New York, N.Y.
- 38.Moore, P. A., A. Evenson, T. D. Luckey, E. McCoy, C. A. Elvehjem, and E. B. Hart. 1946. Use of sulfasuxidine, streptothricin, and streptomycin in nutritional studies with the chick. J. Biol. Chem. 165:437-441. [PubMed] [Google Scholar]
- 39.Netherwood, T., H. J. Gilbert, D. S. Parker, and A. G. O'Donnell. 1999. Probiotics shown to change bacterial community structure in the avian gastrointestinal tract. Appl. Environ. Microbiol. 65:5134-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohkuma, M., and T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 42.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133-139. [DOI] [PubMed] [Google Scholar]
- 43.Pryde, S. E., A. J. Richardson, C. S. Stewart, and H. J. Flint. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 65:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rolfe, R. D. 2000. The role of probiotic cultures in the control of gastrointestinal health. J. Nutr. 130(Suppl.):396S-402S. [DOI] [PubMed] [Google Scholar]
- 45.Roos, S., and H. Jonsson. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433-442. [DOI] [PubMed] [Google Scholar]
- 46.Salanitro, J. P., I. G. Fairchilds, and Y. D. Zgornicki. 1974. Isolation, culture characteristics, and identification of anaerobic bacteria from the chicken cecum. Appl. Microbiol. 27:678-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shane, S. M. 1997. Campylobacteriosis, p. 235-246. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. R. McDougald, and Y. M. Saif (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames, Iowa.
- 48.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:99-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skeeles, J. K. 1997. Staphylococcosis, p. 247-254. In B. W. Calnek, H. J. Barnes, C. W. Beard, L. R. McDougald, and Y. M. Saif (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames, Iowa.
- 51.Snel, J., P. P. Heinen, H. J. Blok, R. J. Carman, A. J. Duncan, P. C. Allen, and M. D. Collins. 1995. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus arthromitus.” Int. J. Syst. Bacteriol. 45:780-782. [DOI] [PubMed] [Google Scholar]
- 52.Stokstad, E. L. R., J. H. Jukes, J. Pierce, A. C. Page, and A. L. Franklin. 1949. The multiple nature of the animal protein factor. J. Biol. Chem. 180:647-654. [PubMed] [Google Scholar]
- 53.Suau, A., R. Bonnet, M. Sutren, J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of gene encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torrallardona, D., C. I. Harris, and M. F. Fuller. 2003. Pigs' gastrointestinal microflora provide them with essential amino acids. J. Nutr. 133:1127-1131. [DOI] [PubMed] [Google Scholar]
- 56.Umesaki, Y., H. Setoyama, S. Matsumoto, A. Imaoka, and K. Itoh. 1999. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 67:3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Wielen, P. W., D. A. Keuzenkamp, L. J. Lipman, F. van Knapen, and S. Biesterveld. 2002. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 44:286-293. [DOI] [PubMed] [Google Scholar]
- 58.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1996. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 62:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wheeler, A. L., P. G. Hartel, D. G. Godfrey, J. L. Hill, and W. I. Segars. 2002. Potential of Enterococcus faecalis as a human fecal indicator for microbial source tracking. J. Environ. Qual. 31:1286-1293. [DOI] [PubMed] [Google Scholar]
- 60.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye, H. Q., D. H. Mallonee, J. E. Wells, I. Bjorkhem, and P. B. Hylemon. 1999. The bile acid-inducible baiF gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A hydrolase. J. Lipid Res. 40:17-23. [PubMed] [Google Scholar]
- 62.Zhu, X. Y., T. Zhong, Y. Pandya, and R. D. Joerger. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124-137. [DOI] [PMC free article] [PubMed] [Google Scholar]