The anaerobic way of life comprises a great metabolic diversity and, correspondingly, diversity of energy-generating mechanisms (52). The facultative anaerobes have a respiratory system that uses preferentially oxygen, but in the absence of oxygen and presence of alternative electron acceptors, such as NO3−, NO2−, Fe(III), or others, alternative electron transport pathways are induced. The situation is completely different in strict anaerobes which can derive energy only by fermentation and/or by ion gradient-driven phosphorylation. In general, the energy yield in strict anaerobes is only a small fraction of the one gained by aerobes (43). Obligately fermenting organisms rely mostly on substrate level phosphorylation (SLP), and the energy yield ranges from 1 to 4 mol of ATP per mol of hexose fermented. However, a number of organisms have evolved additional mechanisms to increase their ATP yield. Some employ electrogenic end product efflux (51) and, in addition, some of the fermentation pathways comprise one or more membrane-bound reactions which result in the generation of a transmembrane ion gradient across the membrane (43). Typical examples for the latter are the fumarate reductase system and the Na+-motive decarboxylases (9, 36). Since the latter do not catalyze oxidation/reduction reactions, the term electron transport does not fit these mechanisms and, therefore, the more general term ion-gradient-driven phosphorylation is favored. At the end of this line are the chemolithoautotrophic anaerobes. Methanogenic archaea reduce CO2 to CH4 and do not employ SLP but have membrane-bound enzymes that couple methyltransfer reactions and electron transfer reactions to the export of Na+ and H+, respectively (7, 21). The Na+ and H+ gradients established are used to drive the synthesis of ATP by membrane-bound A1A0 ATPases (48). Although the analyses of the structure and function of A1A0 ATPases has made some progress in recent years, the ion specificity of these enzymes needs to be elucidated, and the question how both ion gradients are used to drive ATP synthesis has not been settled (47). Acetogens use a similar pathway for CO2 reduction as methanogens, the Wood-Ljungdahl pathway, but with respect to the energy conserving mechanisms they can be divided into two groups, the Na+ and the H+ organisms, which apparently differ also in some aspects of the pathway (38, 44). In recent years it became evident that CO2 is neither the only nor the preferred electron sink in acetogens but acetogens can use alternative electron acceptors (12). This review will focus on the energetics of CO2 reduction (homoacetogenesis) and reduction of other electron acceptors in acetogens.
PATHWAYS OF ENERGY CONSERVATION IN ACETOGENS
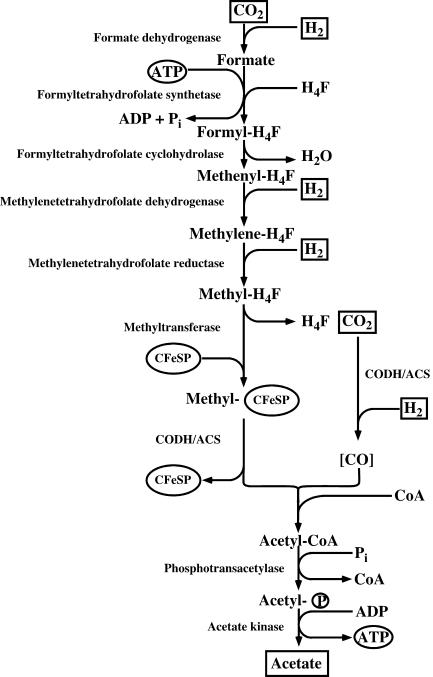
Organisms that are able to reduce CO2 to acetate via the acetyl coenzyme A (acetyl-CoA) or Wood-Ljungdahl pathway (Fig. 1) are termed acetogens. This metabolic capability differentiates acetogens from organisms that synthesize acetate by other metabolic pathways. Acetogens are strictly anaerobic bacteria that can grow by the conversion of C1 compounds such as H2-CO2, CO, or formate to acetate (8). Phylogenetically, acetogens are rather diverse, and 19 bacterial genera have been described to date. Acetogens can inhabit very diverse ecosystems ranging from different soils to termite hindgut, and also extremophiles with respect to temperature, pH, and salinity have been found. It is estimated that acetogenesis yields billions of tons of acetate globally each year and, therefore, acetogens play an important role in the global carbon cycle (15).
FIG. 1.
Wood-Ljungdahl pathway. Reductants are shown as hydrogen, but electrons can also be derived from oxidation of organic substrates.
Acetogens can grow on a variety of different substrates such as, for example, hexoses, C2 and C1 compounds. Hexoses are converted exclusively to acetate:
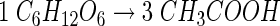 |
(1) |
and, therefore, this fermentation is also referred to as homoacetogenesis. The pathway of hexose consumption starts with their oxidation via the Embden-Meyerhof-Parnas pathway to pyruvate, which is then oxidized by pyruvate:ferredoxin oxidoreductase to acetyl-CoA, reduced ferredoxin, and CO2. The acetyl-CoA is then converted to acetate via acetyl phosphate. The oxidative branch of the pathway (equation 2) is coupled to the synthesis of 4 mol of ATP by SLP:
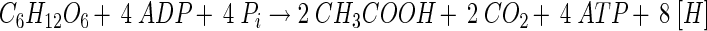 |
(2) |
Second, the reducing equivalents gained during glycolysis and pyruvate:ferredoxin-oxidoreductase are reoxidized by reducing the two mol of CO2 to another mol of acetate via the Wood-Ljungdahl pathway:
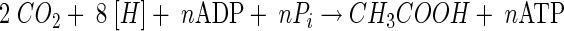 |
(3) |
During growth on sugars or other organic substrates, CO2 can be formally regarded as an electron sink and, per se, there is no need that the Wood-Ljungdahl pathway is coupled to energy conservation. Energy is gained during glycolysis, and the redox balance is maintained by operation of the Wood-Ljungdahl pathway. However, it is important to note that the Wood-Ljungdahl pathway also enables growth on H2-CO2 according to:
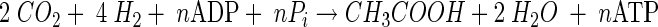 |
(4) |
and, therefore, it must be coupled to net ATP synthesis. The overall free energy change of the reaction (ΔGo′ = −95 kJ/mol) could allow for the synthesis of 1 to 2 mol of ATP. Indeed, it is now well established that CO2 reduction via the Wood-Ljungdahl pathway according to equations 3 and 4 is coupled to ion gradient-driven phosphorylation.
OVERVIEW OF THE WOOD-LJUNGDAHL PATHWAY: BIOCHEMISTRY AND ENERGETICS
The Wood-Ljungdahl pathway is widespread in anaerobic bacteria and archaea and is used for both anabolism and catabolism (19, 39, 55, 77). First, CO2 is reduced to formate by action of formate dehydrogenase, and then formate is activated and bound to tetrahydrofolate (H4F), yielding formyl-H4F (Fig. 1). Water is split off, and the resulting methenyl group is reduced via methylene-H4F to methyl-H4F. The methyl group is then transferred to a protein containing a corrinoid cofactor and iron sulfur clusters, the corrinoid iron-sulfur protein (CFeSP). From there, the methyl group is transferred to the bifunctional CO dehydrogenase/acetyl-CoA synthase (CODH/ACS) that plays a central role in the pathway (11, 56). The methyl group is condensed on CODH/ACS with carbon monoxide, derived from another mol of CO2 oxidized by the CODH activity, to acetyl-CoA. Acetate is produced from acetyl-CoA by action of phosphotransacetylase and acetate kinase. The reducing equivalents required are gained by oxidation of molecular hydrogen (during autotrophic growth) or NADH and reduced ferredoxin (during heterotrophic growth).
The energetics of this pathway is rather interesting. One mole of ATP is produced by SLP in the acetate kinase reaction, but one mole of ATP is consumed in the formyl-H4F synthetase reaction (see Fig. 1). Therefore, the net ATP gain by SLP is zero, and ion gradient-driven phosphorylation must occur as well (for the organisms can grow chemolithoautotrophically). In recent years, experimental evidence was presented that ion gradient-driven phosphorylation indeed occurs in acetogens, but the sites of energy conservation and the mechanisms employed are different. From a bioenergetic point of view, acetogens can be divided into two groups: the Na+-dependent acetogens with Acetobacterium woodii (46) and the H+-dependent acetogens with Moorella thermoacetica (formerly Clostridium thermoaceticum) as model organisms (38). The latter group contains cytochromes and a membrane-bound, H+ motive electron transport chain. The Na+-dependent acetogens lack cytochromes but have membrane-bound corrinoids and couple the Wood-Ljungdahl pathway to primary and electrogenic translocation of Na+. The ion gradient established is taken advantage of by H+ and Na+ translocating ATP synthases, respectively.
ENERGY CONSERVATION IN H+-DEPENDENT ACETOGENS
In the pioneering work of the group of L. G. Ljungdahl, several membrane-integral electron carriers were identified in Moorella thermoacetica and the closely related Moorella thermoautotrophica (formerly Clostridium thermoautotrophicum). Menaquinone MK-7 (2-methyl-3-heptaprenyl-1.4-naphthoquinone; Eo′ = −74 mV) and two b-type cytochromes (cytochrome b559, Eo′ = −215 mV; cytochrome b554, Eo′ = −57 mV) were already identified in 1975, and later on it was shown that a flavoprotein copurified with cytochrome b559 (3, 22). These components are suggested to be involved in electron transport processes (6, 38). The nature of the electron donor and acceptor systems is less clear. It should be mentioned that all of the enyzmes of the Wood-Ljungdahl pathway were isolated so far from the cytosolic compartment after French press disrupture of cells and, therefore, their involvement in energy conservation seems questionable. However, this harsh procedure might have led to a desintegration of membrane-bound enzymes and liberation of the enzymes or catalytically active subcomplexes thereof into the cytosolic compartment. Therefore, it is a necessity to prepare inside-out vesicles under mild conditions that prevent or reduce the loss of membrane-bound enzymes. Using such vesicle preparations, it was indeed shown that certain enzymes may attach to the cytoplasmic membrane: the potential electron donors hydrogenase, CODH, and NADH dehydrogenase, as well as the potential electron acceptor methylene-H4F reductase (30).
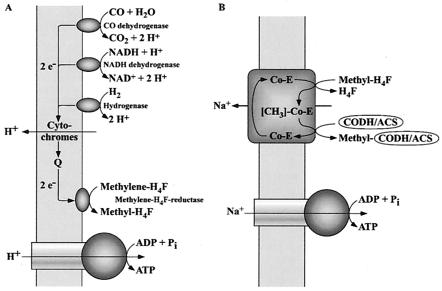
Thauer et al. (73) suggested in 1977 that the methylene-H4F reductase may be the last limb of an electron transport chain that could lead from an electron donor such as hydrogenase, CODH, and NADH dehydrogenase via the membrane-bound electron carriers to methylene-H4F. This electron transport could result in the generation of a transmembrane proton potential that then drives ATP synthesis (Fig. 2A). Although this is a reasonable scenario, experimental evidence for either membrane-bound electron transport to methylene-H4F or the generation of a transmembrane electrochemical potential (ΔμH+) during this reaction is still missing. However, a ΔμH+ was established in membrane vesicles during oxidation of CO (by CODH) coupled to the reduction of ferrycyanide (28). The ΔμH+ was used to drive the uptake of amino acids (29). This clearly proved CO-dependent formation of ΔμH+ but did not reveal the components and mechanisms involved.
FIG. 2.
Hypothetical model of energy conservation in acetogens. (A) H+ organisms; (B) Na+ organisms. For explanations, see the text.
The ΔμH+ established is taken advantage of by a H+ F1F0 ATPase to synthesize ATP. The enzyme and subcomplexes thereof have been isolated from membranes of Moorella thermoacetica and Moorella thermoautotrophica (6, 32). The enzyme contained, like the H+ F1F0 ATPase from Escherichia coli, subunits α, β, γ, δ, ɛ, and c, but was devoid of the essential subunits a and b. Therefore, it was speculated that the architecture of the clostridial F1F0 ATPase is simpler than that from other bacteria (4). However, the genes encoding the F1F0 ATPase are organized in an operon that also contains the genes coding for subunits a and b (5). Therefore, it can be assumed that the enzyme from Moorella thermoacetica and Moorella thermoautotrophica, like that from other bacteria, has an α3β3γδɛab2c9-14 architecture. The discrepancy between the biochemical and molecular data are most likely due to the loss of subunits during purification of the ATPase. Indeed, loss of subunits during purification of F1F0 ATPases is encountered very often and also observed in A. woodii in which the same subunits were missing in the purified enzyme but later on shown to be present in the in vivo complex (see below).
ENERGY CONSERVATION IN NA+-DEPENDENT ACETOGENS
With the discovery that methanogenic archaea that also use in part the Wood-Ljungdahl pathway are strictly dependend on Na+ for CO2 reduction (53, 54) and that they couple the methyltetrahydromethanopterin:coenzyme M methyltransferase to vectorial Na+ transport across the cytoplasmic membrane (37, 45, 49), it was obvious to look for Na+ dependence of acetogenesis. These studies revealed a Na+ dependence for growth and acetate formation in A. woodii (26), Thermoanaerobacter kivui (formerly Acetogenium kivui) (78), and Ruminococcus productus (formerly Peptostreptococcus productus) (20). A. woodii was used as a model system to unravel the molecular basis of the Na+ dependence.
(i) Generation of a sodium motive force in A. woodii.
From studies with cell suspensions of A. woodii, it was apparent that the Wood-Ljungdahl pathway is Na+ dependent and accompanied by the generation of a sodium motive force across the cytoplasmic membrane by a primary, electrogenic pump involved in acetate formation from CO2. Further studies with substrates that are fed into the Wood-Ljungdahl pathway at different levels identified the reaction sequence leading from methylene-H4F to a methylated intermediate as the Na+-dependent reaction sequence of the pathway (26). This reaction sequence contains the methylene-H4F reductase, and one (or two) methyltransferase reactions as presented below:
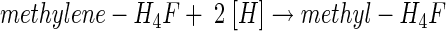 |
(5) |
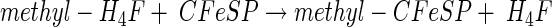 |
(6) |
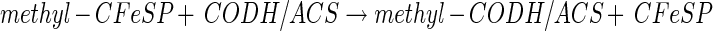 |
(7) |
Equation 5 is catalyzed by the methylene-H4F reductase. Because the enzyme was found to be localized to 100% in the cytoplasm (R. Heise, V. Müller, and G. Gottschalk, unpublished data), we do not consider this reaction sequence to be sodium motive. Furthermore, studies with the Na+-dependent acetogen R. productus also made an involvement of the methylene-H4F reductase in Na+ transport unlikely (76). This leaves the reactions shown in equations 6 and 7 as the most likely candidates for Na+ transport. This reaction sequence has never been studied in Na+-dependent acetogens; all we know results from studies with proton-dependent organisms in which the entire sequence seems to be catalyzed by soluble, cytoplasmic enzymes (55, 69). However, in Na+-dependent acetogens, the situation might be different. The analogy of the pathways in acetogens and methanogens, as well as the finding of membrane-bound corrinoids in Na+-dependent acetogens (2), led to the proposal that the methyl transfer from methyl-H4F to CODH/ACS is the site of Na+ extrusion in Na+-dependent acetogens (46). In analogy to the membrane-bound, Na+-pumping methyltetrahydromethanopterin:coenzyme M methyltransferase of methanogens, the corrinoid could be a cofactor of a multisubunit, membrane-bound, Na+-translocating methyltransferase (Fig. 2B). However, this still has to be verified by experimental analyses.
(ii) A. woodii synthesizes ATP by means of an Na+ F1F0 ATP synthase.
Upon addition of H2-CO2, A. woodii produced acetate and Na+ was extruded from the cells giving rise to a chemical Na+ potential (ΔpNa+) of −90 mV (26). Experiments with cell suspensions revealed that the Na+ gradient drives phosphorylation of ADP. ATP synthesis was not inhibited by protonophores but by sodium ionophores, indicating a direct coupling of ATP synthesis to the Wood-Ljungdahl pathway by ΔμNa+ (24). Studies with cell suspensions of A. woodii clearly demonstrated the presence of an Na+-translocating ATPase/ATP synthase (25, 27).
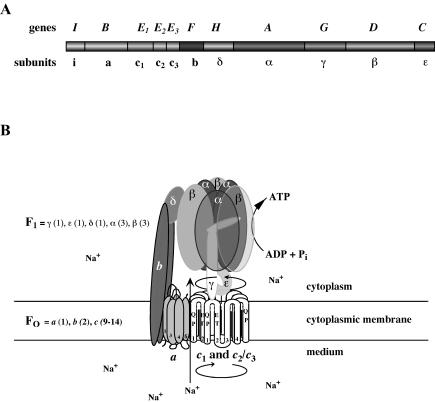
The ATPase was purified to apparent homogeneity after solubilization of membranes with Triton X-100 by (poly)ethyleneglycol precipitation and gel filtration. This preparation contained (only) six subunits (α, β, γ, δ, ɛ, and c2/3) as the clostridial enzyme (62). However, later studies using separation of membrane protein complexes by blue native-polyacrylamide gel electrophoresis and subsequent sodium dodecyl sulfate-polyacrylamide gel electrophoresis in the second dimension revealed the presence of three additional subunits (b, a, and c1) (1). Biochemical, immunological, and molecular studies identified the ATPase of A. woodii as a member of the family of Na+ F1F0 ATP synthases (17, 58, 61, 62). In bacterial F1F0 ATPases, three copies of each subunit α and β alternate around a central stalk built by subunit γ. Subunit ɛ is part of the central stalk and connects it with the membrane domain via subunit c, the proteolipid. A ring of subunit c (9 to 14 copies), subunit a (1 copy), and subunit b (2 copies) are localized in the membrane and build the motor of the rotary device (47). Ion flow through the membrane along the a-c interface is coupled to a rotation of the ring of proteolipids (66), and this rotation drives rotation of the central stalk (subunit γ) (79). Rotation of subunit γ within the a3b3 headpiece results in the liberation of ATP from the β subunits. Such a mechanism requires a stator, which is most likely build by subunits b and δ (Fig. 3). It is noteworthy that the same architecture and mechanisms apply to Na+ and H+ F1F0 ATPases. The ion specificity is altered by only a few amino acid changes in the motor component (see below).
FIG. 3.
The Na+ F1F0 ATPase of A. woodii. (A) Genomic organization of the atp genes; (B) hypothetical model of the enzyme. Please note that the stoichiometry of subunits c1 and c2/3 is unknown. Residues involved in Na+ binding are indicated.
(iii) Na+ F1F0 ATP synthase of A. woodii: the first enzyme with a heterooligomeric motor comprising “bacterial-like” and “eukaryal-like” proteolipids.
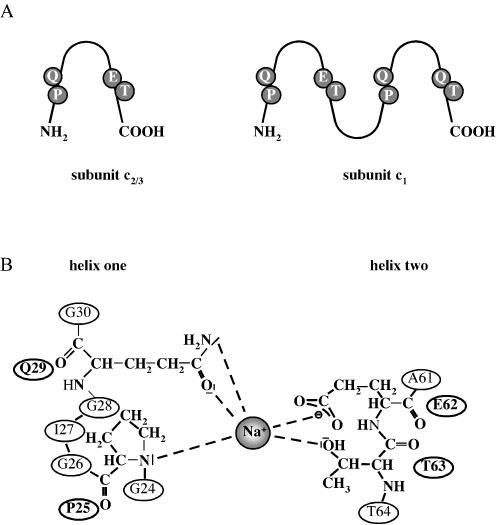
The atp operon of A. woodii that encodes the Na+ F1F0 ATP synthase contains homologues of the nine genes present in the E. coli atp operon (17, 58) (Fig. 3). The order of the genes is atpIBE1E2E3FHAGDC. Northern blot analyses revealed that the genes form one polycistronic message. In contrast to any other known F1F0 ATPase operon, the atp operon from A. woodii contains three tandemly organized genes (atpE1, atpE2, and atpE3) encoding subunit c, the proteolipid. AtpE2 (subunit c2), and AtpE3 (subunit c3) are 100% identical at the amino acid level, and only 18 substitutions occurred on the DNA level (58). This is strong evidence for a duplication of an ancestral gene. The deduced molecular mass of the polypeptides is 8.18 kDa. As their bacterial homologues, they are supposed to be organized in the membrane like a hairpin with two transmembrane helices connected by a polar loop. Most interestingly, atpE1 with 546 bp is more than double the size of atpE2/3. The first and second halves are 66% identical on the DNA level, indicating a duplication of a precursor and subsequent fusion of the two gene copies. The deduced molecular mass of AtpE1 (subunit c1) is 18.37 kDa, with four predicted transmembrane helices arranged in two hairpins. However, the membrane-buried ion-binding residue (Glu62 in AtpE2/3; Glu79 in hairpin one of AtpE1), which is also conserved and involved in H+ transport in H+ F1F0 ATPases, is substituted by a glutamine residue in hairpin two (Fig. 4). It is important to note that homologues of AtpE1 were previously found only in eukaryal V1V0 ATPases (40, 50). These enzymes are not able to synthesize ATP in vivo, and their inability to synthesize ATP results from the fact that the ion-translocating group is missing in one of the two hairpins of subunit c. Therefore, it was important to demonstrate the presence of a eukaryal-like proteolipid in an ATP-synthesizing ATPase. Biochemical studies verified that the proteolipid oligomer of the F1F0 ATPase from A. woodii indeed comprises a mixture of “bacterial-like” 8- and “eukaryal-like” 16-kDa polypeptides, the first found in nature (1). The stoichiometry of the different polypeptides in the c-oligomer has not yet been determined, but it may vary with the substrate and may be used to change the enzymatic reaction from preferentially hydrolysis to synthesis of ATP and vice versa.
FIG. 4.
Na+-binding site in the Na+ F1F0 ATPase of A. woodii.(A) Na+-binding site in subunits c1 and hairpin one of subunit c2/3. Please note that hairpin two of subunit c1 does not contain the conserved glutamate and, therefore, is not capable of binding Na+. (B) Closer look on the Na+-binding site.
(iv) Sodium ion binding site in FO.
The Na+ binding site resides in the motor component of the ATPase (34). The Na+ binding site in subunit c of F1F0 ATPases was elucidated by various techniques and groups. Glu62, Thr63, and Gln29 (numbering of A. woodii subunit c2/3) were identified as part of the binding site by site-directed mutagenesis in E. coli and Propionigenium modestum (35, 80). Furthermore, Pro25 (A. woodii numbering) of subunit c is only conserved in the two Na+ F1F0 ATPases (60). This might indicate the involvement of Pro25 in Na+ binding, but this has to be verified by site-directed mutagenesis. The sodium ion binding site is depicted in Fig. 4.
Apart from subunit c, a second subunit, subunit a, is required for ion transport; both are envisaged to make the ion channel of the ATPase (10, 16). Only one residue of subunit a from the H+ F1F0 ATPase, Arg210, was indicated by mutant studies to be directly involved in H+ transport (75). The residues involved in Na+ binding of subunit a from Na+ F1F0 ATPases are unknown, but a comparison of the sequences of subunit a from the Na+ F1F0 ATPase from P. modestum and A. woodii to those from H+ F1F0 ATPases revealed determinants likely to be important for Na+ binding (59).
(v) Why multiplication and duplication of the proteolipid encoding gene?
One of the most striking and unique features of the atp operon of A. woodii is the presence of multiple copies of proteolipid encoding genes. Multiplication of proteolipid encoding genes has been found before only in V1V0 ATPases from eukarya and A1A0 ATPases from archaea (47, 48). What could be the selective pressure for multiplication of proteolipid-encoding genes? One has to keep in mind that the subunits of the ATPase are present in different stoichiometries (a1b2c9-14δα3γβ3ɛ), and the proteolipid has by far the highest copy number (9 to 14 copies of the bacterial 8-kDa proteolipid, depending on the species) in the complex. Most of our knowledge concerning the regulation of the synthesis of the proteolipid derived from the paradigm E. coli. There, the proteolipid encoding gene is part of a polycistronic message, and enhanced synthesis of the proteolipid is achieved by enhancement of translation (42). In addition, but to a lesser extent, regulation by differential mRNA stability contributes to differential gene expression (41). Apparently, multiplication of the atpE gene and embedding the copies into the operon is another way to increase the concentration of subunit c. This strategy is apparently realized by A. woodii, but this does not exclude additional mechanisms.
Another interesting finding is that the proteolipid of the A1A0 ATPase of the hyperthermophilic archaeon Methanopyrus kandleri is deduced from the genome sequence to have a proteolipid with 13 times the size of bacterial 8-kDa proteolipids that is encoded by only a single gene (70). The rotor is apparently monomeric and built by only one copy of a proteolipid with 13 hairpin domains. Methanopyrus kandleri grows optimally at 98°C, with a maximum at 110°C. Because the rotor is located in the cytoplasmic membrane, it “feels” the heat more intensely than heat-protected enzymes from the cytoplasm; this extremely high temperature might favor a monomeric rotor. However, the existence of a monomeric rotor is only deduced from the genomic sequence but not yet proven by biochemical data. Methanocaldococcus jannaschii and Methanothermobacter thermoautotrophicus grow optimally at 85 and 65°C, and they have proteolipid monomers with three- and two-hairpin domains (63, 64), whereas Methanosarcina mazei and other mesophiles have 8-kDa proteolipids with only one hairpin (65). Therefore, larger proteolipids could be of evolutionary origin favored by high temperature environments.
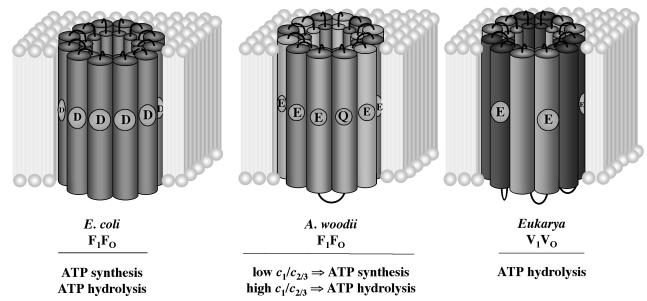
The duplication of the proteolipid encoding gene per se has no consequence for the function of the ATPase since it was shown before that genetically engineered duplicated proteolipids from E. coli are functional in H+ transport and ATP synthesis (33). The striking feature, however, of subunit c1 of A. woodii is not its size but rather the fact that the ion-translocating residue is not conserved in helix two. A loss of one ion-translocating residue is also encountered in the eukaryal “16-kDa proteolipids” from V1V0 ATPases, and this loss has the dramatic consequence that the V1V0 ATPases are not able to synthesize ATP in vivo (47). The ability to synthesize ATP is directly dependent on the number of ions translocated per ATP synthesized. According to the equation ΔGp = −nFΔp, a phosphorylation potential (ΔGp) of ∼50 to 70 kJ/mol is sustained by the use of n = 3 to 4 ions/ATP at a physiological electrochemical ion potential of −180 mV (Δp). However, if the number of ions is lower, then ATP can no longer be synthesized. Although it was demonstrated that the number of monomers in the proteolipid ring may vary from 9 to 14 (67, 71, 72), for the following calculation it is assumed that, as in the case of the bacterial and archaeal “8-kDa proteolipids” with two transmembrane helices, 12 monomers and 12 ion-translocating carboxyl groups are present per oligomer. Once we take into account three ATP-synthesizing or hydrolyzing centers, this gives a stoichiometry of four ions/ATP. In contrast, six copies of the “16-kDa proteolipid” with four transmembrane helices constitute the proteolipid oligomer of V1V0 ATPases. Since the ion-translocating group is lost in the first pair of transmembrane helices, the stoichiometry is only two ions/ATP, which is too low to allow ATP synthesis. On the other hand, if the number of ions is low, the same ΔGp can account for a much higher Δp, making the enzyme a better ion pump, a function required by the physiology of the eukaryotic cell. In general, the lower the number of carboxylates per ring, the worse the coupling efficiency. Taking this into account, it is now reasonable to assume that a cell could, depending on the cellular needs, alter the function of the ATPase between ATP synthesis and ATP hydrolysis by varying the number of ion translocating residues. This is a very attractive idea for A. woodii, which can grow by fermentation of hexoses or by anaerobic respiration or by using pathways which neither involve SLP nor respiration but most likely methyl transfer-driven Na+ extrusion during the operation of the Wood-Ljungdahl pathway. During fermentation, the F1F0 ATPase has to work as an ion pump to generate the membrane potential, whereas during autotrophic growth on H2-CO2 it has to work as a synthase. The switch from pump to synthase could be performed by changing the ratio of c1/c2/3 (Fig. 5). To test this hypothesis is a challenging task for future experiments.
FIG. 5.
Rotors of different ATPases. E. coli F1F0 is assumed to have 12 copies of the 8-kDa proteolipid, V1V0 ATPase to have six copies of a duplicated 16-kDa proteolipid in which the H+-translocating glutamate is only conserved in hairpin two. The rotor of A. woodii comprises a mixture of “bacterial-like” 8- and “eukaryal-like” 16-kDa proteolipids with unknown stoichiometry.
UTILIZATION OF ALTERNATIVE ELECTRON ACCEPTORS BY ACETOGENS
As outlined above, the fermentation of hexoses (but also other substrates) yields reducing equivalents that are used to reduce 2 mol of CO2 by the action of the Wood-Ljungdahl pathway, thereby allowing the synthesis of 3 mol of acetate per mol of hexose fermented. In addition to CO2, alternative electron acceptors, such as aromatic acrylate groups, fumarate, dimethyl sulfoxide, nitrate, and nitrite can be used by some acetogens. Under these conditions, the Wood-Ljungdahl pathway may be turned off completely and only 2 mol of acetate are formed. Furthermore, acetate synthesis may be blocked completely (14). A typical substrate combination that does not yield acetate is the oxidation of a methyl group coupled to the reduction of a phenylacrylate (13). The use of alternative electron acceptors was, in some instances, shown to be coupled to energy conservation. A. woodii, for example, is known to reduce the carbon-carbon double bond of phenylacrylate ethers such as caffeate as shown in Fig. 6.
FIG. 6.
Reduction of the carbon-carbon double bond of phenylacrylate ethers such as caffeate by A. woodii.
The electrons can be derived from various donors such as fructose, methanol, or hydrogen. Cell yield measurements with cells grown on fructose or methyl group-containing substrates showed not only that is caffeate used as an electron sink but also that caffeate reduction is coupled to energy conservation (74). Very clear evidence for ATP synthesis coupled with caffeate reduction was obtained with resting cells of A. woodii in which hydrogen-dependent caffeate reduction was accompanied by the synthesis of ATP (23). Recently, it was shown that ATP synthesis occurred by a chemiosmotic mechanism (31). Most interestingly, like CO2 reduction, both hydrogen-dependent caffeate reduction and ATP synthesis coupled to caffeate reduction were strictly Na+ dependent, and the latter was dependent on a transmembrane Na+ gradient. These studies were fully compatible with the following sequence of events: caffeate reduction → generation of a transmembrane Na+ gradient → generation of ATP by the Na+ F1F0 ATP synthase (31).
It is likely that the electrons are channeled from hydrogen to caffeate via a membrane-bound electron transport chain. Oxidation of hydrogen is catalyzed by a hydrogenase and, in earlier studies, a hydrogenase was purified from A. woodii (57). Because more than 99% of the activity was found in the cytoplasm, the enzyme was described as a soluble, cytoplasmic enzyme. This suggests that an additional electron carrier such as NAD+ or ferredoxin transports the electrons to the membrane and would require a membrane-bound NADH dehydrogenase or reduced ferredoxin dehydrogenase. On the other hand, it cannot be excluded that a membrane-bound hydrogenase might be present as well. After oxidation of the electron donors, the electrons are transferred to the acceptor, caffeate. Cytochromes or quinones were not detected in caffeate-grown cultures (74) and, therefore, the components involved in the electron transport from hydrogen to caffeate are still obscure.
The utilization of caffeate is induced by caffeate, and in the presence of both caffeate and CO2, acetate is no longer produced from CO2. The site of repression of the Wood-Ljungdahl pathway in A. woodii is unknown. On the other hand, it was demonstrated the H+ organism Moorella thermoacetica uses preferentially nitrate as an electron acceptor (68). In the presence of nitrate it does not contain cytochrome b and, therefore, acetate can no longer be produced by the Wood-Ljungdahl pathway (18).
CONCLUSIONS
Although the elucidation of the mechanisms of energy conservation in acetogens is still in its infancy, it turned out that A. woodii is one of the rare cases in which the entire energetics (substrate accumulation, flagellar rotation, and ATP synthesis) is based on an Na+ current across the cytoplasmic membrane. However, the ΔμNa+-generating enzyme is still unknown, and future experiments should address the nature of this Na+ pump. The Na+ F1F0 ATPase of A. woodii is of special interest since it represents the first ATPase that combines features of bacterial and eukaryal enzymes. Whether this might enable the organism to regulate its cellular energy metabolism depending on the growth conditions remains to be seen. The electron transport and the ΔμH+-generating mechanisms in H+-dependent acetogens still needs to be solved. Furthermore, elucidation of the bioenergetics, enzymology, and regulatory processes involved in the utilization of alternative electron acceptors presents challenging tasks for future studies. Genome analyses will certainly pave the way toward a better understanding of the energetics, biochemistry, and physiology of acetogens.
Acknowledgments
I am indebted to my coworkers for their excellent work.
I thank the Deutsche Forschungsgemeinschaft for continuous and generous support.
REFERENCES
- 1.Aufurth, S., H. Schägger, and V. Müller. 2000. Identification of subunits a, b, and c1 from Acetobacterium woodii Na+-F1F0-ATPase: subunits c1, c2, and c3 constitute a mixed c-oligomer. J. Biol. Chem. 275:33297-33301. [DOI] [PubMed] [Google Scholar]
- 2.Dangel, W., H. Schulz, G. Diekert, H. König, and G. Fuchs. 1987. Occurence of corrinoid-containing membrane proteins in anaerobic bacteria. Arch. Microbiol. 148:52-56. [Google Scholar]
- 3.Das, A., J. Hugenholtz, H. van Halbeek, and L. G. Ljungdahl. 1989. Structure and function of a menaquinone involved in electron transport in membranes of Clostridium thermoautotrophicum and Clostridium thermoaceticum. J. Bacteriol. 171:5823-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das, A., D. M. Ivey, and L. G. Ljungdahl. 1997. Purification and reconstitution into proteoliposomes of the F1F0 ATP synthase from the obligately anaerobic gram-positive bacterium Clostridium thermoautotrophicum. J. Bacteriol. 179:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das, A., and L. G. Ljungdahl. 1997. Composition and primary structure of the F1F0 ATP synthase from the obligately anaerobic bacterium Clostridium thermoaceticum. J. Bacteriol. 179:3746-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das, A., and L. G. Ljungdahl. 2003. Electron-transport systems in acetogens, p. 191-204. In L. G. Ljungdahl, M. W. Adams, L. L. Barton, J. G. Ferry, and M. K. Johnson (ed.), Biochemistry and physiology of anaerobic bacteria. Springer, New York, N.Y.
- 7.Deppenmeier, U., V. Müller, and G. Gottschalk. 1996. Pathways of energy conservation in methanogenic archaea. Arch. Microbiol. 165:149-163. [Google Scholar]
- 8.Diekert, G., and G. Wohlfarth. 1994. Metabolism of homoacetogens. Antonie Leeuwenhoek Int. J. Gen. Microbiol. 66:209-221. [DOI] [PubMed] [Google Scholar]
- 9.Dimroth, P. 1997. Primary sodium ion translocating enzymes. Biochim. Biophys. Acta 1318:11-51. [DOI] [PubMed] [Google Scholar]
- 10.Dimroth, P., U. Matthey, and G. Kaim. 2000. Critical evaluation of the one- versus the two-channel model for the operation of the ATP synthase's motor. Biochim. Biophys. Acta 1459:506-513. [DOI] [PubMed] [Google Scholar]
- 11.Doukov, T. I., T. M. Iverson, J. Seravalli, S. W. Ragsdale, and C. L. Drennan. 2002. A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl CoA synthase. Science 298:567-572. [DOI] [PubMed] [Google Scholar]
- 12.Drake, H. L., S. Daniel, K. Küsel, C. Matthies, C. Kuhner, and S. Braus-Strohmeyer. 1997. Acetogenic bacteria: what are the in situ consequences of their diverse metabolic diversities? Biofactors 1:13-24. [DOI] [PubMed] [Google Scholar]
- 13.Drake, H. L., S. L. Daniel, C. Matthies, and K. Küsel. 1994. Acetogenesis: reality in the laboratory, uncertainty elsewhere, p. 273-302. In H. L. Drake (ed.), Acetogenesis. Chapman & Hall, New York, N.Y.
- 14.Drake, H. L., and K. Küsel. 2003. How the diverse physiologic potentials of acetogens determine their in situ realities, p. 171-190. In L. G. Ljungdahl, M. W. Adams, L. L. Barton, J. G. Ferry, and M. K. Johnson (ed.), Biochemistry and physiology of anaerobic bacteria. Springer, New York, N.Y.
- 15.Drake, H. L., K. Küsel, and C. Matthies. Acetogenic prokaryotes. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., in press. Springer-Verlag, New York, N.Y.
- 16.Fillingame, R. H., W. Jiang, and O. Y. Dmitriev. 2000. Coupling H+ transport to rotary catalysis in F-type ATP synthases: structure and organization of the transmembrane rotary motor. J. Exp. Biol. 203:9-17. [DOI] [PubMed] [Google Scholar]
- 17.Forster, A., R. Daniel, and V. Müller. 1995. The Na+-translocating ATPase of Acetobacterium woodii is a F1F0-type enzyme as deduced from the primary structure of its β, γ, and ɛ subunits. Biochim. Biophys. Acta 1229:393-397. [DOI] [PubMed] [Google Scholar]
- 18.Fröstl, J. M., C. Seifritz, and H. L. Drake. 1996. Effect of nitrate on the autotrophic metabolism of the acetogens Clostridium thermoautotrophicum and Clostridium thermoaceticum. J. Bacteriol. 178:4597-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs, G. 1986. CO2 fixation in acetogenic bacteria: variations on a theme. FEMS Microbiol. Rev. 39:181-213. [Google Scholar]
- 20.Geerligs, G., P. Schönheit, and G. Diekert. 1989. Sodium-dependent acetate formation from CO2 in Peptostreptococcus productus (strain Marburg). FEMS Microbiol. Lett. 57:253-258. [DOI] [PubMed] [Google Scholar]
- 21.Gottschalk, G., and R. K. Thauer. 2001. The Na+-translocating methyltransferase complex from methanogenic archaea. Biochim. Biophys. Acta 1505:28-36. [DOI] [PubMed] [Google Scholar]
- 22.Gottwald, M., J. R. Andreesen, J. LeGall, and L. G. Ljungdahl. 1975. Presence of cytochrome and menaquinone in Clostridium formicoaceticum and Clostridium thermoaceticum. J. Bacteriol. 122:325-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, B., M. Bokranz, P. Schönheit, and A. Kröger. 1988. ATP formation coupled to caffeate reduction by H2 in Acetobacterium woodii NZva16. Arch. Microbiol. 150:447-451. [Google Scholar]
- 24.Heise, R., V. Müller, and G. Gottschalk. 1993. Acetogenesis and ATP synthesis in Acetobacterium woodii are coupled via a transmembrane primary sodium ion gradient. FEMS Microbiol. Lett. 112:261-268. [Google Scholar]
- 25.Heise, R., V. Müller, and G. Gottschalk. 1992. Presence of a sodium-translocating ATPase in membrane vesicles of the homoacetogenic bacterium Acetobacterium woodii. Eur. J. Biochem. 206:553-557. [DOI] [PubMed] [Google Scholar]
- 26.Heise, R., V. Müller, and G. Gottschalk. 1989. Sodium dependence of acetate formation by the acetogenic bacterium Acetobacterium woodii. J. Bacteriol. 171:5473-5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heise, R., J. Reidlinger, V. Müller, and G. Gottschalk. 1991. A sodium-stimulated ATP synthase in the acetogenic bacterium Acetobacterium woodii. FEBS Lett. 295:119-122. [DOI] [PubMed] [Google Scholar]
- 28.Hugenholtz, J., D. M. Ivey, and L. G. Ljungdahl. 1987. Carbon monoxide-driven electron transport in Clostridium thermoautotrophicum membranes. J. Bacteriol. 169:5845-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hugenholtz, J., and L. G. Ljungdahl. 1990. Amino acid transport in membrane vesicles of Clostridium thermoautotrophicum. FEMS Microbiol. Lett. 69:117-122. [DOI] [PubMed] [Google Scholar]
- 30.Hugenholtz, J., and L. G. Ljungdahl. 1989. Electron transport and electrochemical proton gradient in membrane vesicles of Clostridium thermoaceticum. J. Bacteriol. 171:2873-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imkamp, F., and V. Müller. 2002. Chemiosmotic energy conservation with Na+ as the coupling ion during hydrogen-dependent caffeate reduction by Acetobacterium woodii. J. Bacteriol. 184:1947-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivey, D. M., and L. G. Ljungdahl. 1986. Purification and characterization of the F1-ATPase from Clostridium thermoaceticum. J. Bacteriol. 165:252-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, P. C., and R. H. Fillingame. 1998. Genetic fusions of subunit c in the F0 sector of H+-transporting ATP synthase. Functional dimers and trimers and determination of stoichiometry by cross-linking analysis. J. Biol. Chem. 273:29701-29705. [DOI] [PubMed] [Google Scholar]
- 34.Kaim, G., and P. Dimroth. 1993. Formation of a functionally active sodium-translocating hybrid F1F0 ATPase in Escherichia coli by homologous recombination. Eur. J. Biochem. 218:937-944. [DOI] [PubMed] [Google Scholar]
- 35.Kaim, G., F. Wehrle, U. Gerike, and P. Dimroth. 1997. Molecular basis for the coupling ion selectivity of F1F0 ATP synthases: probing the liganding groups for Na+ and Li+ in the c subunit of the ATP synthase from Propionigenium modestum. Biochemistry 36:9185-9194. [DOI] [PubMed] [Google Scholar]
- 36.Kröger, A., V. Geisler, E. Lemma, F. Theis, and R. Lenger. 1992. Bacterial fumarate respiration. Arch. Microbiol. 158:311-314. [Google Scholar]
- 37.Lienard, T., B. Becher, M. Marschall, S. Bowien, and G. Gottschalk. 1996. Sodium ion translocation by N5-methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanosarcina mazei Gö1 reconstituted in ether lipid liposomes. Eur. J. Biochem. 239:857-864. [DOI] [PubMed] [Google Scholar]
- 38.Ljungdahl, L. G. 1994. The acetyl-CoA pathway and the chemiosmotic generation of ATP during acetogenesis, p. 63-87. In H. L. Drake (ed.), Acetogenesis. Chapman & Hall, New York, N.Y.
- 39.Ljungdahl, L. G. 1986. The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu. Rev. Microbiol. 40:415-450. [DOI] [PubMed] [Google Scholar]
- 40.Mandel, M., Y. Moriyama, J. D. Hulmes, Y.-C. E. Pan, H. Nelson, and N. Nelson. 1988. cDNA sequence encoding the 16-kDa proteolipid of chromaffin granules implies gene duplication in the evolution of H+-ATPases. Proc. Natl. Acad. Sci. USA 85:5521-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy, J. E., B. Gerstel, B. Surin, U. Wiedemann, and P. Ziemke. 1991. Differential gene expression from the Escherichia coli atp operon mediated by segmental differences in mRNA stability. Mol. Microbiol. 10:2447-2458. [DOI] [PubMed] [Google Scholar]
- 42.McCarthy, J. E., H. U. Schairer, and W. Sebald. 1985. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 4:519-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller, V. 2001. Bacterial fermentation. In Encyclopedia of life sciences. [Online.] Macmillan, London, United Kingdom. http://www.els.net.
- 44.Müller, V., S. Aufurth, and S. Rahlfs. 2001. The Na+ cycle in Acetobacterium woodii: identification and characterization of a Na+ translocating F1F0-ATPase with a mixed oligomer of 8 and 16 kDa proteolipids. Biochim. Biophys. Acta 1505:108-120. [DOI] [PubMed] [Google Scholar]
- 45.Müller, V., M. Blaut, and G. Gottschalk. 1993. Bioenergetics of methanogenesis, p. 360-406. In J. G. Ferry (ed.), Methanogenesis. Chapman & Hall, New York, N.Y.
- 46.Müller, V., and G. Gottschalk. 1994. The sodium ion cycle in acetogenic and methanogenic bacteria: generation and utilization of a primary electrochemical sodium ion gradient, p. 127-156. In H. L. Drake (ed.), Acetogenesis. Chapman & Hall, New York, N.Y.
- 47.Müller, V., and G. Grüber. 2003. ATP synthases: structure, function and evolution of unique energy converters. Cell. Mol. Life Sci. 60:474-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Müller, V., C. Ruppert, and T. Lemker. 1999. Structure and function of the A1A0 ATPases from methanogenic archaea. J. Bioenerg. Biomembr. 31:15-28. [DOI] [PubMed] [Google Scholar]
- 49.Müller, V., C. Winner, and G. Gottschalk. 1988. Electron transport-driven sodium extrusion during methanogenesis from formaldehyde + H2 by Methanosarcina barkeri. Eur. J. Biochem. 178:519-525. [DOI] [PubMed] [Google Scholar]
- 50.Nelson, N. 1992. Structural conservation and functional diversity of V-ATPases. J. Bioenerg. Biomembr. 24:407-414. [DOI] [PubMed] [Google Scholar]
- 51.Otto, R., A. S. M. Sonnenberg, H. Veldkamp, and W. N. Konings. 1980. Generation of an electrochemical proton gradient in Streptococcus cremoris by lactate efflux. Proc. Natl. Acad. Sci. USA 77:5502-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peinemann, S., and G. Gottschalk. 1992. The anaerobic way of life, p. 300-311. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer Verlag, Berlin, Germany.
- 53.Perski, H. J., J. Moll, and R. K. Thauer. 1981. Sodium dependence of growth and methane formation in Methanobacterium thermoautotrophicum. Arch. Microbiol. 130:319-321. [Google Scholar]
- 54.Perski, H. J., P. Schönheit, and R. K. Thauer. 1982. Sodium dependence of methane formation in methanogenic bacteria. FEBS Lett. 143:323-326. [Google Scholar]
- 55.Ragsdale, S. W. 1992. Enzymology of the acetyl-CoA pathway of autotrophic CO2 fixation. Crit. Rev. Biochem. Mol. Biol. 26:261-300. [DOI] [PubMed] [Google Scholar]
- 56.Ragsdale, S. W., and M. Kumar. 1996. Ni containing carbon monoxide dehydrogenase/acetyl-CoA synthase. Chem. Rev. 96:2515-2539. [DOI] [PubMed] [Google Scholar]
- 57.Ragsdale, S. W., and L. G. Ljungdahl. 1984. Hydrogenase from Acetobacterium woodii. Arch. Microbiol. 139:361-365. [DOI] [PubMed] [Google Scholar]
- 58.Rahlfs, S., S. Aufurth, and V. Müller. 1999. The Na+-F1F0-ATPase operon from Acetobacterium woodii. Operon structure and presence of multiple copies of atpE which encode proteolipids of 8- and 18-kDa. J. Biol. Chem. 274:33999-34004. [DOI] [PubMed] [Google Scholar]
- 59.Rahlfs, S., and V. Müller. 1999. Sequence of subunit a of the Na+-translocating F1F0-ATPase of Acetobacterium woodii: proposal for residues involved in Na+ binding. FEBS Lett. 453:35-40. [DOI] [PubMed] [Google Scholar]
- 60.Rahlfs, S., and V. Müller. 1997. Sequence of subunit c of the Na+-translocating F1F0 ATPase of Acetobacterium woodii: proposal for determinants of Na+ specificity as revealed by sequence comparisons. FEBS Lett. 404:269-271. [DOI] [PubMed] [Google Scholar]
- 61.Reidlinger, J., F. Mayer, and V. Müller. 1994. The molecular structure of the Na+-translocating F1F0-ATPase of Acetobacterium woodii, as revealed by electron microscopy, resembles that of H+-translocating ATPases. FEBS Lett. 356:17-20. [DOI] [PubMed] [Google Scholar]
- 62.Reidlinger, J., and V. Müller. 1994. Purification of ATP synthase from Acetobacterium woodii and identification as a Na+-translocating F1F0-type enzyme. Eur. J. Biochem. 223:275-283. [DOI] [PubMed] [Google Scholar]
- 63.Ruppert, C., H. Kavermann, S. Wimmers, R. Schmid, J. Kellermann, F. Lottspeich, H. Huber, K. O. Stetter, and V. Müller. 1999. The proteolipid of the A1A0 ATP synthase from Methanococcus jannaschii has six predicted transmembrane helices but only two proton-translocating carboxyl groups. J. Biol. Chem. 274:25281-25284. [DOI] [PubMed] [Google Scholar]
- 64.Ruppert, C., R. Schmid, R. Hedderich, and V. Müller. 2001. Selective extraction of subunit D of the Na+-translocating methyltransferase and subunit c of the A1A0 ATPase from the cytoplasmic membrane of methanogenic archaea by chloroform/methanol and characterization of subunit c of Methanothermobacter thermoautotrophicus as a 16-kDa proteolipid. FEMS Microbiol. Lett. 195:47-51. [DOI] [PubMed] [Google Scholar]
- 65.Ruppert, C., S. Wimmers, T. Lemker, and V. Müller. 1998. The A1A0 ATPase from Methanosarcina mazei: cloning of the 5′ end of the aha operon encoding the membrane domain and expression of the proteolipid in a membrane-bound form in Escherichia coli. J. Bacteriol. 180:3448-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sambongi, Y., Y. Iko, M. Tanabe, H. Omote, A. Iwamoto-Kihara, I. Ueda, T. Yanagida, Y. Wada, and M. Futai. 1999. Mechanical rotation of the c subunit oligomer in ATP synthase (F1F0): direct observation. Science 286:1722-1724. [DOI] [PubMed] [Google Scholar]
- 67.Seelert, H., A. Poetsch, N. A. Dencher, A. Engel, H. Stahlberg, and D. J. Müller. 2000. Structural biology: proton-powered turbine of a plant motor. Nature 405:418-419. [DOI] [PubMed] [Google Scholar]
- 68.Seifritz, C., S. L. Daniel, A. Gossner, and H. L. Drake. 1993. Nitrate as a preferred electron sink for the acetogen Clostridium thermoaceticum. J. Bacteriol. 175:8008-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seravalli, J., S. Zhao, and S. W. Ragsdale. 1999. Mechanism of transfer of the methyl group from (6S)-methyltetrahydrofolate to the corrinoid/iron-sulfur protein catalyzed by the methyltransferase from Clostridium thermoaceticum: a key step in the Wood-Ljungdahl pathway of acetyl-CoA synthesis. Biochemistry 38:5728-5735. [DOI] [PubMed] [Google Scholar]
- 70.Slesarev, A. I., K. V. Mezhevaya, K. S. Makarova, N. N. Polushin, O. V. Shcherbinina, V. V. Shakhova, G. I. Belova, L. Aravind, D. A. Natale, I. B. Rogozin, R. L. Tatusov, Y. I. Wolf, K. O. Stetter, A. G. Malykh, E. V. Koonin, and S. A. Kozyavkin. 2002. The complete genome of the hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA 99:4644-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stahlberg, H., D. J. Müller, K. Suda, D. Fotiadis, A. Engel, T. Meier, U. Matthey, and P. Dimroth. 2001. Bacterial Na+-ATP synthase has an undecameric rotor. EMBO Rep. 2:229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stock, D., A. G. Leslie, and J. E. Walker. 1999. Molecular architecture of the rotary motor in ATP synthase. Science 286:1700-1705. [DOI] [PubMed] [Google Scholar]
- 73.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tschech, A., and N. Pfennig. 1984. Growth yield increase linked to caffeate reduction in Acetobacterium woodii. Arch. Microbiol. 137:163-167. [Google Scholar]
- 75.Valiyaveetil, F. I., and R. H. Fillingame. 1997. On the role of Arg-210 and Glu-219 of subunit a in proton translocation by the Escherichia coli FOF1-ATP synthase. J. Biol. Chem. 272:32635-32641. [DOI] [PubMed] [Google Scholar]
- 76.Wohlfarth, G., and G. Diekert. 1991. Thermodynamics of methylenetetrahydrofolate reduction to methyltetrahydrofolate and its implications for the energy metabolism of homoacetogenic bacteria. Arch. Microbiol. 155:378-381. [Google Scholar]
- 77.Wood, H. G., S. W. Ragsdale, and E. Pezacka. 1986. The acetyl-CoA pathway of autotrophic growth. FEMS Microbiol. Rev. 39:345-362. [Google Scholar]
- 78.Yang, H., and H. L. Drake. 1990. Differential effects of sodium on hydrogen- and glucose-dependent growth of the acetogenic bacterium Acetogenium kivui. Appl. Environ. Microbiol. 56:81-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoshida, M., E. Muneyuki, and T. Hisabori. 2001. ATP synthase-a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell. Biol. 2:669-677. [DOI] [PubMed] [Google Scholar]
- 80.Zhang, Y., and R. H. Fillingame. 1995. Changing the ion binding specificity of the Escherichia coli H+-transporting ATP synthase by directed mutagenesis of subunit c. J. Biol. Chem. 270:87-93. [DOI] [PubMed] [Google Scholar]