Abstract
The long-term viability and plasmid retention of recombinant Escherichia coli strains were investigated by real-time testing of master cell banks (MCBs) stored at the Roche Molecular Systems Culture Collection (RMSCC). MCBs at the RMSCC were cryogenically frozen and stored at −80°C for long-term preservation. At regular intervals during a period of 5 to more than 10 years, representative cryovials of each MCB were tested for viability and plasmid retention. Plasmid retention and viability for all 30 MCBs were stable over time. Twenty-seven MCBs maintained high levels of plasmid retention (at or near 100%), while three MCBs showed lower plasmid retention rates (ranging from 13.9 to 96.5%) that were consistent over time. New MCBs with high plasmid retention were created from two of the MCBs with lower plasmid retention by selective pressure with high levels of antibiotics. These new MCBs have shown stable viability and high plasmid retention over the first 5 months of storage. In conclusion, this study shows that properly selected, frozen and stored MCBs retain viability and maintain plasmid retention over time. Moreover, it is possible to recover cultures with high plasmid retention from MCBs with low plasmid retention by selecting clones grown in the presence of high levels of antibiotics.
The Roche Molecular Systems Culture Collection (RMSCC) serves the Research, Development, Manufacturing and Patent departments of Roche Molecular Systems (RMS) with microbiological and cell culture support. In this capacity, the RMSCC acquires, preserves, authenticates, and distributes organisms and nucleic acids to support the projects and products of RMS. The RMSCC uses recombinant Escherichia coli strains to produce many of the enzymes and nucleic acid controls for RMS products. In order to reduce the amount of testing required and to ensure reproducibility, the RMSCC generally makes one master cell bank (MCB) for each recombinant E. coli strain that will last the life of the product. The MCB is grown from a single colony and provides the seed stock for any subsequent lots.
Given their central role as a source of materials for RMS products, it is important that the viability and plasmid retention of MCBs be maintained over time (4, 11). Cryogenically stored MCBs are maintained at the RMSCC for many years. At the RMSCC, we optimize the viability of MCBs by using cryopreservation, which has been shown to be a stable method for preservation of bacteria (2). To enhance plasmid retention, we select clones with high levels of antibiotic resistance (3, 20). Previously published studies of viability and plasmid retention have relied on short-term data (8). For example, recombinant E. coli showed no loss of viability or of plasmid retention after frozen storage for 10 months (12). Based on the experience of the American Type Culture Collection, the shelf life of frozen E. coli has been estimated to be greater than 30 years (7). However, the long-term plasmid retention of recombinant E. coli strains was not addressed in this study.
In order to better understand the long-term viability and plasmid retention of recombinant E. coli strains, I began a program of regular real-time testing of MCBs in the RMSCC. The prospective data that I have collected over the past 10 years demonstrates that cryogenically stored MCBs, when subjected to antibiotic selection followed by proper thawing, show no statistically significant loss of viability or plasmid retention. Furthermore, I found that new MCBs with high plasmid retention could be produced from recombinant E. coli strains with low plasmid retention by selecting resistant clones subjected to high levels of antibiotic.
MATERIALS AND METHODS
MCBs.
The enzyme production MCBs used in this study are shown in Table 1. These strains are used to produce enzymes for AMPLICOR, COBAS AMPLICOR, TaqMan, and research products and contain the protein coding region along with an inducible promoter (9, 10).
TABLE 1.
Enzyme production MCBs
| RMSCC IDa | Enzyme | Plasmid |
|---|---|---|
| 159 | DNA polymerase for AmpliTaq | pLSG5 |
| 657 | DNA polymerase for AmpliTaq, cycle sequencing | pRDA3-2 |
| 2843 | DNA polymerase for AmpliTaq, fluorescence sequencing | pLK103 |
| 160 | Uracil-DNA glycosylase | pFC101 |
| 395 | DNA polymerase for AmpliTaq, Stoffel fragment | pLSG8 |
| 410 | rTth DNA polymerase for AMPLICOR MONITOR assays | pLSG32 |
| 2925 | Pyrophosphatase | pAW143 |
| 3098 | Thermus species Z05 DNA polymerase | pAW151 |
ID, identification.
MCBs for the DNA and RNA controls used in this study are shown in Table 2. DNA controls for AMPLICOR and COBAS AMPLICOR tests contain the target sequence from the infectious organism, including the assay primer and probe binding regions which were cloned into a vector using standard methods and inserted into an E. coli K12-derived host strain (5, 16). The RNA controls for AMPLICOR and COBAS AMPLICOR tests contain the target sequence, a transcription promoter, and a poly(A) tail (19). The internal controls have the same primer binding sites as the positive controls, with a scrambled version of the positive control and an alternate primer (13, 15). The armored RNA controls for COBAS AmpliPrep kits contain the target sequence along with bacteriophage protein coding regions that provide the protective coating for the RNA molecules (14).
TABLE 2.
MCBs for production of RNA and DNA controls
| RMSCC IDa | Microorganism or virusb | Plasmid | Nucleic acid |
|---|---|---|---|
| Positive controls | |||
| 534 | Chlamydia trachomatis | pCHL-1 | DNA |
| 556 | Neisseria gonorrhoeae | pucGC2 | DNA |
| 741 | Mycobacterium tuberculosis | pKY5 | DNA |
| 742 | Mycobacterium intracellulare | pKY3 | DNA |
| 743 | Mycobacterium avium | pKY1 | DNA |
| 1686 | HCV | pHCVIIA | RNA |
| 1693 | HIV-1 | pRDW10 | DNA |
| 1854 | CMV | pCMA1 | DNA |
| 1968 | HIV-1 | pSYC35 | RNA |
| 3069 | HBV | pCABN | DNA |
| Internal controls | |||
| 1742 | M. tuberculosis | pSYC21 | DNA |
| 1785 | HIV-1 | pNAS-2 | RNA |
| 1795 | HCV | pHCVIIAS.2 | RNA |
| 1806 | C. trachomatis and N. gonorrhoeae | pSYC28 | DNA |
| 1860 | CMV | pSYC31 | DNA |
| 2451 | HCV | pSYC52 | RNA |
| 3068 | HBV | pTMN1 | DNA |
| 3223 | HIV-1 | pSDL150 | RNA |
| Armored RNA controls | |||
| 3502 | HCV | pAR-HCVW-1-1,2 | aRNAc |
| 3659 | HIV-1 | pCYK2-HIVPC | aRNA |
| 3732 | HCV | pARHCVqR2 | aRNA |
| 3661 | HIV-1 | pAR-HIVqR4 | aRNA |
ID, identification.
CMV, cytomegalovirus; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV-1, human immunodeficiency virus, type 1.
aRNA, armored RNA.
Recombinant E. coli strains from which MCBs were derived were originally created by scientists at RMS and deposited into the RMSCC. RMSCC staff grew clones of each strain on Luria agar (10 g of tryptone/liter, 5 g of yeast extract/liter, 10 g of NaCl/liter, 15 g of agar/liter) with the appropriate antibiotic, selected an isolated colony, and grew it in Luria broth (10 g of tryptone/liter, 5 g of yeast extract/liter, 10 g of NaCl/liter) with antibiotics at the appropriate temperature. Prior to 1995, 50 μg of ampicillin (Sigma, St. Louis, Mo.)/ml was the antibiotic selection used. After 1995, 100 μg of ampicillin/ml plus 100 μg of methicillin (Roche Molecular Biochemicals, Indianapolis, Ind.)/ml was used as the antibiotic selection for most of the MCBs. The addition of methicillin has been shown to inhibit satellite colony formation on solid medium (6). Strains for armored RNA controls were grown in the presence of 100 μg of carbenicillin (Sigma, St. Louis, Mo.)/ml as the antibiotic selection method.
For long-term preservation, cultures were grown for 12 to 16 h at 30°C for enzyme production MCBs and at 37°C for all other MCBs, adjusted to 10 to 15% glycerol (as a cryoprotectant), and then aliquoted into cryovials. The cryovials were placed into “Mr. Frosty” slow-freeze containers (Nalgene, Nunc International, Rochester, N.Y.) at room temperature and then placed in a −80°C freezer to cool the cells at approximately 1°C per min to minimize the cellular damage from freezing and to maintain maximum viability (17). After at least 4 h, the cryovials were moved into a liquid nitrogen tank for long-term storage. When a culture was needed, the cryovial was retrieved from the freezer and quickly thawed in a 37°C water bath to minimize cellular damage during the process (7). The culture was then quickly inoculated into growth medium to minimize any potential damage from the cryoprotectant. Approximately 1 week after freezing, cultures were subjected to restriction analysis and sequence confirmation before being deemed acceptable for manufacturing.
Viability and plasmid retention.
Viability and plasmid retention testing was performed on one representative cryovial of the MCB at regular intervals over a period of up to 11 years. The culture was thawed, serially diluted in 0.85% NaCl, plated on Luria agar without antibiotics, and incubated overnight. The lack of antibiotic at this stage of the test allows for detection of colonies that tend to lose their antibiotic resistance-encoding plasmids. After the colonies were counted for viability assessment, 200-well separated colonies were selected for plasmid retention testing (1).
The colonies were picked with a sterile toothpick and patched first onto an antibiotic-free plate and then onto an antibiotic-containing plate. The plates were marked so that the results from each individual colony on the two plates could be compared. After overnight incubation the numbers of colonies on the antibiotic-free and antibiotic-containing plates were determined. Plasmid retention was expressed as the percentage of colonies tested that grew on the antibiotic-containing plates divided by the total number that grew on the antibiotic-free plates.
Recovery of high plasmid retention strains.
Two of the recombinant E. coli strains in our study (pHCVIIA and pKY3) consistently demonstrated low plasmid retention. To produce new MCBs, these strains were streaked for isolation onto Luria-Bertani agar plates containing 50 μg/ml ampicillin. An isolated colony was added to 100 ml of Luria-Bertani broth containing freshly added 1,000-μg/ml ampicillin and 1,000-μg/ml methicillin in a 500-ml Erlenmeyer flask. The suspensions were incubated at 37°C for 12 h in a Brinkman Orbimix (Westbury, N.Y.) shaker.
Statistical analysis.
Sampling (or measurement) error is the primary source of statistical noise and is expected to be consistent across all MCBs. Therefore, while it is possible to test for decreased viability and retention for each strain separately, it is more efficient to examine strains together. This methodology improves the estimate of sampling error while allowing for simultaneous testing for overall and strain-specific decreases in viability and retention. As such, analysis of variance (ANOVA) was used to address the following two questions (18). First, is there evidence indicating a general trend of decreased viability or retention across all MCBs? Second, do any individual strains depart from the overall trend (decreasing or otherwise), and if so, which?
RESULTS
Long-term real-time analysis of MCBs.
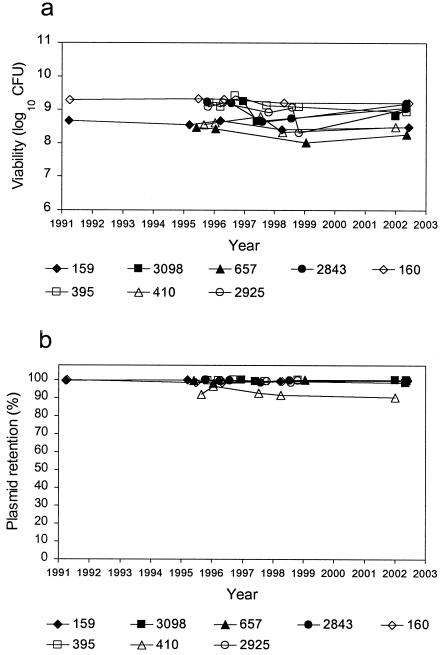
The viability and plasmid retention rate of the enzyme production MCBs are shown in Fig. 1a and b, respectively. All eight enzyme production MCBs showed stable viability, with CFU that remained within a 1-log10 range over time. The plasmid retention rate was maintained near 100% over time in all but one of the enzyme production MCBs. A slightly lower plasmid retention rate of 90.5 to 96.5% was observed for strain 410. The retention rate for this strain, however, appeared to be stable over time.
FIG. 1.
Viability (a) and plasmid retention (b) of enzyme production MCBs.
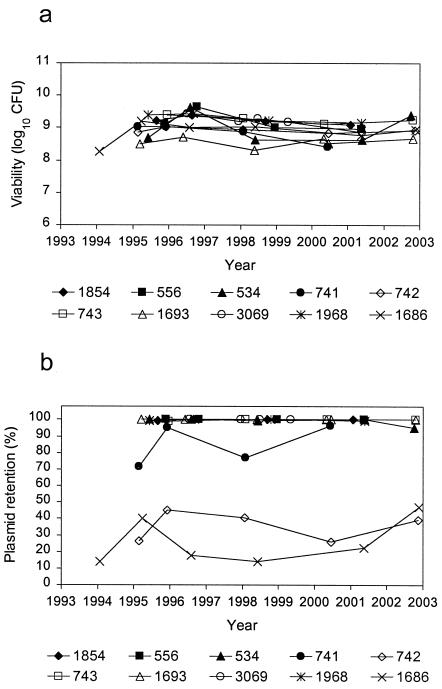
Figure 2 shows the viabilities and plasmid retention rates of MCBs for DNA and RNA positive controls. Although there were fluctuations in the CFU readings for some strains, CFU for all 10 positive control MCBs remained within a 1-log10 range over time. With regard to plasmid retention, all but three of the positive control MCBs maintained a plasmid retention rate at or near 100%. The retention rates for strain 741 ranged from 71.5 to 96.5%. Strains 742 and 1686 demonstrated even lower rates, ranging from 26.0 to 45.0% and from 13.9 to 47.0%, respectively. Despite these fluctuations, the plasmid retention rate appeared relatively stable in all three of these strains.
FIG. 2.
Viability (a) and plasmid retention (b) of positive control MCBs.
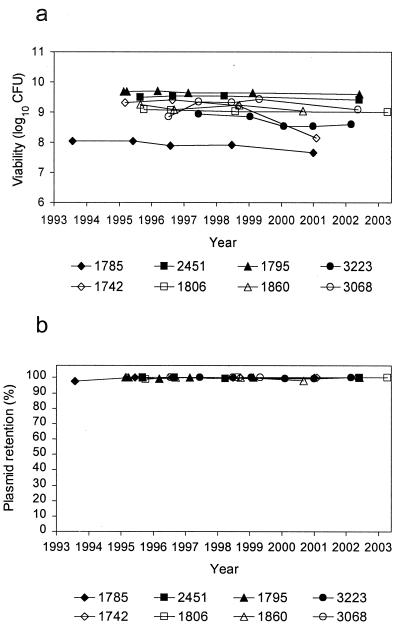
Figure 3 shows the viability and plasmid retention rates of MCBs for DNA and RNA internal controls. CFU readings remained within a 1-log10 range over time for all but one of the internal control MCBs. Strain 1742 showed CFU ranging from 8.14 to 9.32 log10. Strain 1785 showed a somewhat lower viability than the other MCBs; however, viability appeared to remain stable for this strain (7.67 to 8.04 log10 CFU). With regard to plasmid retention, all eight of the internal control MCBs maintained a plasmid retention rate at or near 100%.
FIG. 3.
Viability (a) and plasmid retention (b) of internal control MCBs.
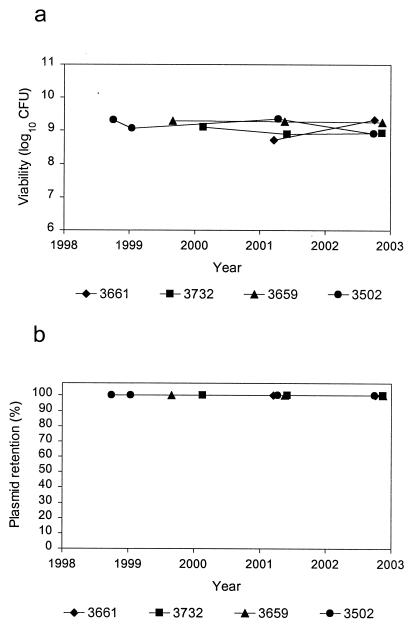
The viability and plasmid retention rate of the MCBs for the armored RNA controls are shown in Fig. 4a and b, respectively. Since these strains were not deposited in the RMSCC until 1998 or later, fewer data were available for these strains. Nonetheless, all of the armored RNA control MCBs appeared to be stable both for cell viability and plasmid retention.
FIG. 4.
Viability (a) and plasmid retention (b) of armored RNA control MCBs.
For all MCBs, ANOVA tests for a general trend of decreased viability or loss of plasmid retention across all lines were nonsignificant (P = 0.1047 or 0.5815, respectively). Likewise, tests for decreased viability or loss of plasmid retention in any line were similarly nonsignificant (P = 0.3461 and 0.9713, respectively). Such results indicate that changes observed within and across lines over the course of the experiment did not exceed expectation, given sampling error.
Creation of new MCBs with high plasmid retention.
During the course of this study, I noticed that two of the positive control MCBs, strain 1686 with plasmid pHCVIIA and strain 742 with plasmid pKY3, consistently had low plasmid retention rates. New MCBs for these two strains were created by selecting resistant clones derived from the original MCBs grown in the presence of high levels of antibiotic. These new MCBs were subjected to restriction analysis, functional analysis, and sequence analysis to ensure that they were equivalent to the original MCBs (data not shown).
Viability and plasmid retention testing of the new MCBs showed no significant change over time. The prefreeze viability was 1.8 × 108 CFU/ml for strain 742 and 6.8 × 108 CFU/ml for strain 1686. Plasmid retention tests performed the following day showed retention rates of 100% for both strains. The postfreeze viability for strain 742 was 1.2 × 108, 1.2 × 108, and 7.85 × 108 CFU/ml after storage for 1 week, 1 month, and 5 months, respectively. The postfreeze viability for strain 1686 was 3.8 × 108, 6.7 × 108, and 5.8 × 108 CFU/ml after storage for 1 week, 1 month, and 5 months, respectively. Plasmid retention rates remained 100% for both strains at all time points tested. My intention is to continue to monitor these new MCBs at regular intervals to ensure that they maintain high viability and plasmid retention over time.
DISCUSSION
Literature showing real-time survival data for preserved microorganisms is limited. Since the enzymes and nucleic acid controls are integral parts of RMS’s product lines, I wanted to be sure that we would not lose our production cultures over time. We therefore instituted a regular program of viability and plasmid retention testing.
To my knowledge, this is the first study to monitor viability and plasmid retention of cultures stored over a prolonged period of time. With few exceptions, the MCBs showed stable viability and also had consistently high plasmid retention rates over periods of up to 11 years. For the few MCBs that demonstrated lower viability and/or plasmid retention, these rates were usually apparent from the beginning and were likely due to the fact that the initial colonies were not properly selected. Hence, my results indicate that properly selected, frozen, and stored MCBs retain viability and plasmid retention over time. It is important to note that given the extended period of time the MCBs were monitored, the viability and plasmid retention testing was performed by different operators with various skill levels and years of experience. Although this may explain the fluctuation in viability and plasmid retention observed for some of the MCBs, the vast majority of MCBs were stable. This study did not address plasmid structural stability during storage, which may be more difficult to achieve than segregational stability. However, RMS’s manufacturing group has monitored the restriction patterns of the positive control strains over time and saw no change, indicating that there has been no significant gross structural instability (i.e., deletion, rearrangement, etc.) that could be detected at the population level.
The cultures stored in the RMSCC are used for the production of various enzymes and nucleic acid controls, and cultures have been chosen for generation of MCBs based on the features that are important for their various unique purposes. Prior to 1992, the protein production team knew that plasmid retention was important for optimizing protein yield during fermentation. Therefore, enzyme production clones were tested for antibiotic resistance in order to select those clones with high plasmid retention. It is therefore not surprising that all of the enzyme production MCBs initially showed high plasmid retention rates and have consistently maintained high rates over time. By comparison, RNA and DNA control clones were not routinely subjected to selection with high levels of antibiotics and plasmid retention testing prior to 1995. When the RMSCC instituted regular testing of control clones in 1995, some were found to have low or variable plasmid retention. Since that time, all new control clones have been subjected to the same rigorous selection and testing process as the enzymes, and MCBs made from those clones have maintained high plasmid retention.
The two internal control MCBs that exhibited low plasmid retention, strains 742 and 1686, were created prior to initiation of regular plasmid retention testing in the RMSCC. I was able to create new MCBs from these clones when they were grown under a high level of selective pressure with antibiotics. These new MCBs appear to have maintained high plasmid retention and viability over the 5-month period that they were monitored during this study. I will continue to monitor these newly created MCBs to confirm that they remain stable and am considering applying this rescue technique to two MCBs in the RMSCC that show low plasmid retention rates.
In summary, I have demonstrated that properly selected, frozen, and stored MCBs retain viability and plasmid retention over time. Moreover, it is possible to recover cultures with high plasmid retention from MCBs with low plasmid retention by selecting clones grown in the presence of high levels of antibiotics.
Acknowledgments
I gratefully acknowledge the contributions of Lawrence Elfbaum, Christina Kerksieck, Robert Kosecki, Katherine Mohr, Edith Peck, Huyen Phan, and Jill Williams for their technical expertise; Brian Rhees for statistical analysis; Michael Hagan for graphics; Linda Wuestehube for editorial assistance; and David Gelfand, Jonathan Raymond, and Gail Rodgers for critical analysis of the manuscript.
REFERENCES
- 1.Belt, A. 1996. Characterization of culture used for biotechnology and industry, p. 251-258. In J. C. Hunter-Cevera and A. Belt (ed.), Maintaining cultures for biotechnology and industry. Academic Press, New York, N.Y.
- 2.Dando, T., and I. Bousfield. 1991. Maintenance of industrial and marine bacteria and bacteriophages, p. 57-63. In B. Kirsop, and A. Doyle (ed.), Maintenance of industrial and marine bacteria and bacteriophages. Academic Press, New York, N.Y.
- 3.Dietz, A., and B. W. Churchill. 1985. Culture preservation and stability, p. 37-49. In A. T. Bull, and H. Dalton (ed.), Comprehensive biotechnology: the principles, applications and regulations of biotechnology in industry, agriculture and medicine, Pergamon Press, New York, N.Y.
- 4.Duncan, P. A. 2003. Characterization of microbial seeds used in the manufacture of biopharmaceuticals. BioProcess Int. 1:58-62. [Google Scholar]
- 5.Elion, E. 1997. Constructing recombinant DNA molecules by the polymerase chain reaction, p. 3.17. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 6.Fitzwater, T. 2002. Ligation and transformation protocol. U.S. Department of Commerce. [Online.] http://micro.nwfsc.noaa.gov/protocols/Lig_and_Trans.html.
- 7.Gherna, R. L. 1981. Preservation, p. 208-217. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 8.Heckly, R. J. 1978. Preservation of microorganisms, p. 1-53. In D. Perlman (ed.), Advances in applied microbiology. Academic Press, New York, N.Y. [DOI] [PubMed]
- 9.Lawyer, F. C., S. Stoffel, R. K. Saiki, S. Y. Chang, P. A. Landre, R. D. Abramson, and D. H. Gelfand. 1993. High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5′ to 3′ exonuclease activity. PCR Methods Appl. 2:275-287. [DOI] [PubMed] [Google Scholar]
- 10.Lawyer, F. C., S. Stoffel, R. K. Saiki, K. Myambo, R. Drummond, and D. H. Gelfand. 1989. Isolation, characterization, and expression in Escherichia coli of the DNA polymerase gene from Thermus aquaticus. J. Biol. Chem. 264:6427-6437. [PubMed] [Google Scholar]
- 11.Marquet, M., N. A. Horn, and J. A. Meek. 1997. Characterization of plasmid DNA vectors for use in human gene therapy, part 1. Biopharm 10:42-50. [Google Scholar]
- 12.Nierman, W., and T. Feldblyum. 1985. Cryopreservation of cultures that contain plasmids. Dev. Ind. Microbiol. 26:423-434. [Google Scholar]
- 13.Orle, K. A., C. A. Gates, D. H. Martin, B. A. Body, and J. B. Weiss. 1996. Simultaneous PCR detection of Haemophilus ducreyi, Treponema pallidum, and herpes simplex virus types 1 and 2 from genital ulcers. J. Clin. Microbiol. 34:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasloske, B. L., C. R. Walkerpeach, R. D. Obermoeller, M. Winkler, and D. B. DuBois. 1998. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. J. Clin. Microbiol. 36:3590-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenstraus, M., Z. Wang, S. Y. Chang, D. DeBonville, and J. P. Spadoro. 1998. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J. Clin. Microbiol. 36:191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., and D. W. Russell. 2001. Cloning PCR products by addition of restriction sites to the termini of amplified DNA, p. 8.37-8.41. In J. Argentine (ed.), Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Simione, F. P., Jr. 1992. Key issues relating to the genetic stability and preservation of cells and cell banks. J. Parenter. Sci. Technol. 46:226-232. [PubMed] [Google Scholar]
- 18.Steel, R. G. D., J. H. Torrie, and D. A. Dickey. 1997. Principles and procedures of statistics: a biometrical approach. McGraw-Hill Companies, Inc., New York, N.Y.
- 19.Wang, A. M., M. V. Doyle, and D. F. Mark. 1989. Quantitation of mRNA by the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 86:9717-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright, A. D., and T. J. Crease. 1996. Detection of “lost” plasmids from Escherichia coli using excess ampicillin. Anal. Biochem. 236:181-182. [DOI] [PubMed] [Google Scholar]