Abstract
Photoreceptors of the Xenopus laevis retina are the site of a circadian clock. As part of a differential display screen for rhythmic gene products in this system, we have identified a photoreceptor-specific mRNA expressed in peak abundance at night. cDNA cloning revealed an open reading frame encoding a putative 388 amino acid protein that we have named “nocturnin” (for night-factor). This protein has strong sequence similarity to the C-terminal domain of the yeast transcription factor, CCR4, as well as a leucine zipper-like dimerization motif. Nocturnin mRNA levels exhibit a high amplitude circadian rhythm and nuclear run-on analysis indicates that it is controlled by the retinal circadian clock at the level of transcription. Our observations suggest that nocturnin may function through protein–protein interaction either as a component of the circadian clock or as a downstream effector of clock function.
Circadian clocks are internal timing devices that assist the organism in its adaption to the external day-night cycle (1). Such clocks have a period near 24 hr and sustain rhythmic outputs including complex behavioral rhythms, hormone synthesis rhythms and rhythms of gene expression (2). Among vertebrates, circadian clocks have been localized to the suprachiasmatic nucleus (3), the pineal gland of nonmammalian forms (4) and the retina (5–8). Although the formal properties of such clocks have been well-studied and mutations leading to altered rhythmicity have been identified in both hamsters (9) and mice (10), the cellular and molecular mechanisms for generation of endogenous timing signals in vertebrates remains unknown.
A major clue to the understanding of vertebrate circadian clocks comes from molecular/genetic analyses of Drosophila and Neurospora. In these systems, rhythmic gene transcription appears to play a role in both the operation of the clock itself and in the output of the clock to control overt, downstream rhythms. Molecular components of the clock have been identified; these include products of the period (per) gene in Drosophila and the frequency (frq) gene in Neurospora (11–14). Current models for clock operation involve a transcription/translation loop in which the accumulation of either PER or FRQ protein results in feedback to negatively regulate transcription of the respective gene. The details of the mechanism are not defined, but in Drosophila, another gene timeless (tim) is also involved. The TIM protein interacts with PER (15) and in tim mutants, nuclear localization of PER is lost (16) and rhythmicity is abolished (17). In addition to rhythmic transcription of “clock genes” it also appears that one of the ways that the clock controls output rhythms is through regulation of the transcription of “clock-controlled genes” (2, 18, 19). Clock-controlled genes are downstream from the central clock mechanism and are important for expression of overt rhythms. Examples of clock-controlled genes are iodopsin (6) and tryptophan hydroxylase (20) in vertebrate photoreceptors.
Although rhythmic expression of mRNAs appears to be a central feature of both the clock mechanism and the downstream generation of overt circadian rhythms, clock components identified in Drosophila or Neurospora have not yet been found in vertebrates. Recently, we initiated a systematic screen (21) for rhythmic vertebrate mRNAs in Xenopus retina using mRNA differential display (22). Although such a screen for rhythmic gene products is ongoing in Neurospora (18), to our knowledge this is the first such screen in a vertebrate clock-containing tissue. Xenopus retina is particularly amenable to experimental analysis of circadian clock mechanisms because a circadian clock in the photoreceptor cells is functional and entrainable in culture (5, 23, 24). Our screen was designed to identify mRNAs that are expressed rhythmically in Xenopus retina in vitro in constant darkness. Messages identified from this screen should fall into one of two classes: clock-controlled genes (downstream of the central clock mechanism) or components of the clock. Identification of both classes of mRNA will be important in elucidation of vertebrate clock mechanisms.
One of the PCR products obtained from this screen (21), originally designated RM1 (rhythmic message 1), was particularly interesting due to its extremely high amplitude rhythmic expression. This message was present at high levels in early night and was undetectable during the day. Furthermore, it was retina-specific in Xenopus and within the retina was expressed only in photoreceptors (21), the site of a circadian clock (24). In this report, we present further characterization of the RM1 message that encodes a putative protein we have named “nocturnin.” An earlier version of this has been published in abstract form (25).
MATERIALS AND METHODS
Eyecup Culture.
Xenopus laevis (5–6.5 cm) were purchased from Nasco (Fort Atkinson, WI) and were maintained in 12-hr light/12-hr dark cyclic light. Animals used in these experiments were entrained in these lighting conditions at least 2 weeks before use. Care was in accordance with federal and institutional guidelines. Eyecups (including the retina, pigment epithelium, choroid, and sclera) were cultured in defined culture medium (23), in a humidified atmosphere of 95% O2/5% CO2. Incubations were carried out on a rotary shaker (60 rpm), in a constant temperature incubator at 21 ± 0.1°C under the indicated lighting conditions. During “dark” conditions, all manipulations were done under infrared lights. “Light” conditions indicate light intensity of 3.3 × 10−4 W·cm2·sec. All times are expressed as zeitgeber time (ZT) in hours, with respect to the original entraining light cycle, in which ZT 0 is defined as the time of normal light onset (dawn) and ZT 12 is defined as time of normal dark onset (dusk).
Preparation of Probes.
Random primed probes were prepared using the Random Primers DNA Labeling system (GIBCO/BRL). Antisense riboprobes were synthesized using the RNA Transcription kit (Stratagene). Both types of probes were purified with NucTrap gel filtration columns (Stratagene).
Cloning of the Nocturnin cDNA.
A mid-night Xenopus retinal cDNA library constructed in λZAPII (Stratagene) was screened as previously described (20) using a random prime-labeled probe (GIBCO/BRL) made from the original differential display PCR product (21). Seven positive clones were obtained. Partial sequence analysis was performed on all seven clones and two of the positive clones (RM1-2 and RM1-5) were characterized by complete sequence analysis of both strands using Sequenase (United States Biochemical).
Primer Extension.
Primer extension was performed on retinal mRNA using an oligonucleotide (5′-AGTCTGCTGGTGCTGTTTCC-3′) that is complementary to the nocturnin cDNA clone RM1-2 from bases 113–94 (26). Primer extensions on RNA from ZT 4 (nocturnin mRNA levels are low) and from ZT 14 (nocturnin mRNA levels are high) were performed at the same time and run side by side on the sequencing gel to verify authenticity of the extended product.
5′ Rapid Amplification of cDNA Ends (RACE).
5′-RACE was carried out according to kit protocols (GIBCO/BRL), with retinal RNA isolated from eyecups at ZT 14. The “gene-specific primer 1” (5′-AGAGGCTCAGACTGGTCATG-3′) used for first strand cDNA synthesis was complementary to nocturnin cDNA clone RM1-2, bases 203–184. The “gene-specific primer 2” (5′-ACAGCGGCACTACTGCTAAG-3′) used for PCR amplification was complementary to RM1-2, bases 155–136. The resulting PCR product was cloned into pCRII (Invitrogen) and sequenced.
Cloning of the Nocturnin Gene.
A 470-bp PstI fragment from the 5′-end of nocturnin cDNA clone RM1-2 was isolated, random prime labeled, and used to screen a partial Sau3A Xenopus genomic library (Stratagene). Four independent clones were obtained. One of these, RG1, containing a 17-kb insert was analyzed extensively by mapping and sequence analysis and used to determine the nocturnin gene structure.
Northern Blot Analysis.
At the appropriate times, retinas were removed from the cultured eyecups and were quickly frozen on dry ice for subsequent isolation of RNA. Isolation of RNA from single Xenopus retinas was done using a protocol based on the acid guanidinium thiocyanate-phenol-chloroform extraction method of Chomczynski and Sacchi (27) as described (28). Northern blot analyses were carried out exactly as described in (21) using antisense riboprobes made from nocturnin cDNA clone RM1-2. Filters were stripped by boiling twice for 10 min in 0.01× subsclerosing standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), 0.1% SDS, and rehybridized with antisense riboprobes made from β-actin clones (29) for normalization.
Quantitation of message levels was done directly from the radiolabeled filters using the PhosphorImager (Molecular Dynamics). Total counts per nocturnin band (minus background) were divided by the total counts per band of β-actin (minus background) to get numbers that were normalized for differences in lane loadings. Final results are expressed relative to the first ZT0 sample.
Nuclear Run on Analysis.
Nuclear run on analysis was carried out exactly as described in Green and Besharse (20), except 30 retinas were used for nuclear isolation for each time point. The resulting labeled transcripts were hybridized to Nytran filters containing 2 μg each of linearized plasmid DNA from nocturnin cDNA clone RM1-2, chicken β-actin, and the pBluescript vector (Stratagene) as described (20). The level of transcript initiation was quantitated by PhosphorImager as described above.
RESULTS
The original differential display PCR product, RM1 (21), was used as a probe to screen a night time Xenopus retinal cDNA library and cDNA clones were isolated and characterized by DNA sequencing. A single large open reading frame was identified that encodes a polypeptide of 388 amino acids (Fig. 1A). The sequence around the initiation methionine agrees well with consensus translation start sites (30) and a stop codon is present five codons upstream from this methionine, providing evidence that the cDNA clones contained the complete coding sequence.
Figure 1.
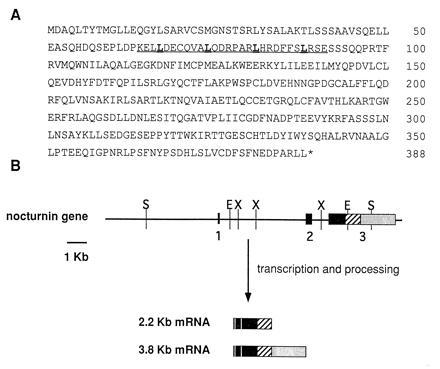
(A) Deduced amino acid sequence of nocturnin. Single letter amino acid abbreviations are used. The asterisk (∗) indicates the position of the stop codon. The leucine zipper motif is underlined, with leucines in bold type. (B) Nocturnin gene structure. The horizontal line represents the genomic DNA. The boxes indicate the relative size and position of the exons. The vertical lines represent restriction endonuclease cleavage sites: X, XbaI; E, EcoRI; S, SacI. ▪, the coding portions of the gene; ▨, the 3′-untranslated regions common to both mRNAs, and ░⃞, the 3′-untranslated sequence specific to the larger message. The compositions of the two resulting nocturnin mRNAs are diagrammed.
The transcription start site was identified using two independent methods: primer extension and 5′-RACE. Primer extension of retinal mRNA was done using an end-labeled oligonucleotide that is complementary to the nocturnin clone RM1-2, from bases 113–94. The resulting primer extended product was visualized on a sequencing gel and determined to be 127 bases in length (data not shown). 5′-RACE was performed on mRNA isolated from ZT 14 retinas and produced a PCR product that contained sequence matching the 5′ end of the cDNA clones, plus an additional 14 base pairs upstream of the 5′ end of the longest clone (RM1-2). The position of the 5′ end of the RACE product matched that predicted by the primer extension product exactly. The sequence of the RACE product was further verified by comparison to the nocturnin genomic clone (see below).
Previous northern blots using the differential display product as probe (21) had revealed two specific mRNA bands (3.8 kb and 2.2 kb; ref. 21). One of our cDNA clones contained sequence specific to the larger message. This clone contains additional 3′-untranslated sequence that is not present in the other clones. To further define the relationships of the two messages, we isolated four independent genomic clones from a Xenopus genomic library using a portion of one of the nocturnin cDNA clones as a probe. Characterization of these clones resulted in delineation of the nocturnin gene structure as shown in Fig. 1B. The gene contains three exons and two introns. The first two exons are small (96 and 267 bp, respectively). The third exon is large (3 kb) and contains the final 833 bp of the coding sequence, as well as the entire 3′-untranslated regions of both the small and the large message. The two messages result from the use of two different polyadenylylation sites within exon 3, as diagrammed (Fig. 1B).
The deduced nocturnin amino acid sequence contains a consensus leucine zipper-like motif near the amino terminal region of the putative protein, from amino acids 64–91. This motif contains four heptad repeats with leucines at every fourth position, and several charged amino acids contained within. These positively and negatively charged amino acids fit the prediction of alpha helix-stabilizing ion pairs that are part of the original leucine zipper model (31). Leucine zipper motifs, in which leucine residues are repeated at every seventh position, are postulated to form coiled coil structures in which the leucines promote dimerization through hydrophobic interactions. However, the zipper region of nocturnin does contain one proline (which can interrupt alpha-helical secondary structure) at the first position of the third heptad repeat.
Searches of GenBank with the nocturnin amino acid sequence, using the blast search algorithm, revealed significant sequence similarity to a transcription factor from Saccharomyces cerevisiae called CCR4 (Fig. 2). The region of similarity covers the C-terminal 291 amino acids of nocturnin and the 335 C-terminal amino acids of CCR4 (Fig. 2B). It begins at amino acid 98 of nocturnin, just downstream of the leucine-zipper like motif. Over this region, there is 31% identity and 49% similarity (conservative amino acid changes) between nocturnin and the analogous region of CCR4. CCR4 is a large, 837-amino acid protein (32), that is required for the transcription of a number of genes in yeast (33). CCR4 does not bind DNA itself, but is involved in protein–protein interactions through a “leucine-rich” interaction domain that lies upstream of the region similar to nocturnin. Both the “leucine-rich region” and the C-terminal region are necessary for transcriptional activation by CCR4 (33).
Figure 2.

Comparison of the nocturnin amino acid sequence with the yeast CCR4 sequence. (A) The last 291 amino acids of the nocturnin sequence (Top) are aligned with the last 335 amino acids of the CCR4 sequence (ref. 32; bottom and underlined) as determined by blast search algorithms. Positions of identical amino acids are designated by double dots (:), conservative amino acid changes are designated by single dots (.). Dashes within the sequence are gaps inserted for maximal alignment. The nocturnin sequence shown begins with amino acid number 98 and CCR4 sequence begins with amino acid number 606. (B) Diagrammatic representation of nocturnin and CCR4 alignment. Rectangular boxes represent the amino acid sequence with the N termini on the left and the C termini on the right. The regions of similarity between the two proteins are marked by the diagonal lines. The position of the other defined domains or motifs are marked (33).
The data base searches also revealed similarity of nocturnin to three human expressed sequence tags from human infant brain, human fetal liver/spleen, and human breast libraries. These tags were submitted to the database as partial cDNA sequences and the similarity to nocturnin spanned the entire region of these partial sequences. Nocturnin showed 41% identity and 51% similarity to the human breast sequence tag (H45114), 82% identity and 87% similarity to the fetal human liver/spleen tag (T87026), and 46% identity and 63% similarity to the infant human brain tag (H06019).
To analyze the expression pattern of the nocturnin mRNA, we performed northern blot analyses (Fig. 3A) on individual Xenopus retinas isolated at various times throughout the day and night from eyecups cultured in cyclic light, constant darkness and constant light. When measured at 6-hr time points over two days in culture, nocturnin mRNA levels were rhythmic with high amplitude in all three lighting conditions (Fig. 3B). However, in constant darkness and constant light, the amplitude of the rhythm decreases. The rhythm in constant darkness shows more animal to animal variability and a longer night-time expression time, while that in constant light is considerably damped on the second day. Finer resolution of message levels in cyclic light and constant dark (2-hr time points) from mid-day to mid-night, reveals a peak at ZT16 in both (Fig. 3C). In addition, the expression patterns and amplitudes of the two nocturnin mRNA species (3.8 and 2.2 kb) are the same.
Figure 3.
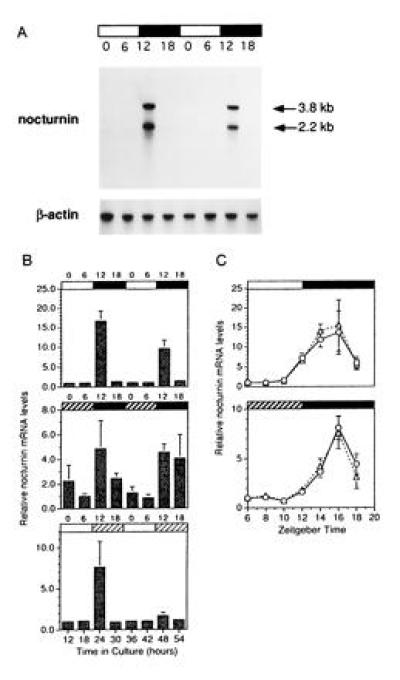
Expression patterns of nocturnin mRNA. Xenopus eyecups were cultured in cyclic light, constant darkness, or constant light. At the times indicated, retinas were isolated and RNA was prepared. Nocturnin mRNA levels were measured from individual retinas by Northern blot analyses, followed by quantitation by phosphorimaging. RNA levels are represented as “fold changes” over the first time point within each experiment (ZT 0 in B and ZT 6 in C). The absolute value of the “fold change” between different experiments cannot be directly compared. These quantitations are meant to illustrate the overall temporal pattern of expression of the message and the amount of variability between retinas, rather than to measure absolute levels of message. (A) Example of a Northern blot detection of nocturnin mRNA from retinas collected at 6-hr intervals over two days in cyclic light. Each lane contains about 1 μg of total RNA from a single retina. Bars above the blot indicate the lighting conditions: black bars indicate night (dark) and white bars indicate day (light). Numbers above the lanes denote ZT in hours, in which ZT 0 is defined as the time of normal light onset (dawn) and ZT 12 is defined as time of normal dark onset (dusk). The blot was stripped and reprobed with a chicken β-actin probe (29) for normalization of the lanes. (B) Quantitation of nocturnin mRNA levels at 6-hr intervals over two days in cyclic light (Top), constant dark (Middle), or constant light (Bottom). Numbers above the graphs indicate time of harvest (ZT). Bars above each graph denote lighting conditions at time of harvest. Bars: □, light in subjective day; ▪, dark in subjective night; , dark in subjective day; and ▨, light in subjective night. Each time point shown is an average of measurements of both the 3.8-kb and 2.2-kb messages from four or five individual retinas. Error bars = SEM. (C) Quantitation of nocturnin mRNA levels at two hr intervals from mid-day (ZT6) to mid-night (ZT18) in cyclic light (Upper) and constant dark (Lower). The two lineson the graphs represent the two different nocturnin mRNA species, which were quantitated separately; the triangles represent the 2.2-kb message, the circles represent the 3.8-kb message. The bars above the graphs indicate lighting conditions at the time of harvest as described in B. Each point represents measurements of nocturnin message levels from four to five individual retinas. Error bars = SEM.
The changes in nocturnin mRNA levels observed in our northern analyses could be due to changes in rates of synthesis, degradation, or both. To address the question of whether the rhythmic nocturnin message levels are due to changes in synthesis, we carried out nuclear run-on analyses to measure nocturnin transcript initiation at different times (Fig. 4). Because of the large number of pooled retinas required for these analyses, run-ons were conducted only at ZT 6 and 12 in cyclic light and ZT 4 and 13 in constant dark. Our results show that transcription initiation of the nocturnin gene is low early in the day and increases greater than 40-fold by early evening in cyclic light. In constant darkness, the amplitude of the rhythm of transcript initiation is 20-fold on two consecutive days in culture. Thus, the nocturnin gene is regulated transcriptionally by a circadian clock.
Figure 4.
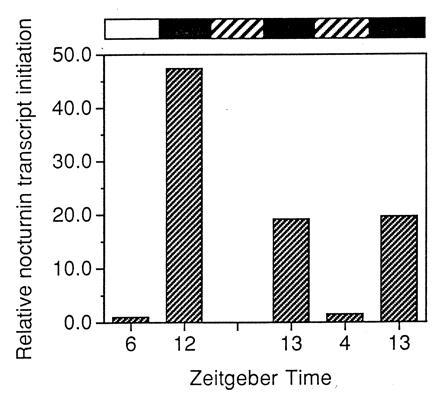
Nuclear run-on analysis of nocturnin transcript initiation. Eyecups were prepared in the light and cultured in cyclic light or constant darkness. At the times indicated, retinas were dissected from the eyecups in light (cyclic light samples) or under infrared light (constant dark samples). Thirty retinas were pooled for each time point and nuclei were isolated. Run-on transcriptions were performed with [α-32P]UTP, using 1 × 107 nuclei per reaction and the resulting labeled transcripts were purified and hybridized to slot-blotted cDNA of vector alone, β-actin, and nocturnin. Filters were exposed to x-ray film and were quantitated by PhosphorImager analysis. The rectangular bars above the graph represent the experimental conditions as described in Fig. 3. The first two samples are from cyclic light on the first day in culture and the last three samples are from constant dark conditions on the second and third day in culture.
DISCUSSION
We have described the identification of a novel, photoreceptor-specific mRNA that is transcriptionally regulated by the retinal circadian clock. This message contains an ORF that encodes a protein we have named nocturnin due to its high expression during the night. The similarity of nocturnin to the yeast transcription factor CCR4 and the presence of the leucine zipper-like domain suggest that it too may function to regulate transcription. A photoreceptor transcription factor that is transcriptionally regulated by the circadian clock and is expressed for only a few hours in the early night could be involved in either the regulation of clock-controlled rhythmic outputs or in the central mechanism generating circadian signals.
The region that exhibits similarity between nocturnin and CCR4 is necessary for transcriptional activation by CCR4 (33), but its exact role remains unknown. The CCR4 protein, like nocturnin, contains a protein interaction domain just N-terminal to this region (32). However, in CCR4 this is a “leucine-rich” domain, while in nocturnin, it is a putative leucine zipper. Neither protein contains an obvious DNA binding domain; CCR4 is thought to activate transcription via interaction with other DNA binding proteins (33–35). Defined N-terminal “activation” domains present in CCR4 are absent in nocturnin.
The structural features of nocturnin are reminiscent of the Drosophila clock component protein PER. The primary structure of PER also describes a novel protein with a domain (PAS domain) similar to known transcription factors, but contains no obvious DNA binding domain or transcription activation domain (36). The PAS domain is involved in protein–protein interactions and PER is thought to act as a transcription factor through binding to other proteins, although this has not been shown directly.
Proteins containing leucine zipper motifs fall into several groups. The majority identified to date are transcription factors and contain a basic DNA binding domain near the zipper motif (37–39). Recently, identification of shortsighted, a leucine zipper protein from Drosophila, was reported to lack the basic DNA binding region and to be localized to the cytoplasm (40), suggesting that it may interact with another leucine zipper-containing, DNA-binding protein to prevent its ability to activate transcription. There are also reports of other proteins containing leucine zipper-like motifs that are not transcription factors, such as a newly identified family of protein kinases (41, 42).
Although its precise function is undefined, nocturnin contains domains that appear to be highly conserved from yeast to humans. Three human expressed sequence tags exhibit high levels of similarity, in one case (T87026) exceeding 80% identity at the amino acid level. Evidence also exists for the presence of a CCR4-like protein in mouse. A yeast two-hybrid screen using yeast CCR4 as the bait, identified a mouse protein (mCAF) capable of functionally interacting with CCR4 (35). Therefore, a CCR4-like protein presumably exists in mouse.
The high amplitude, rhythmic expression of the nocturnin gene makes it valuable for identification and characterization of vertebrate clock-controlling promoter elements. This provides the opportunity to work “backwards” to identify components of the photoreceptor central clock mechanism. Additionally, because this gene is expressed at high levels specifically in photoreceptor cells, regulation of the nocturnin gene may provide insights into mechanisms of tissue-specific expression in photoreceptors and other clock cells.
Understanding the function and regulation of nocturnin will be important, both for studies of circadian clock mechanisms as well as for determining the role this protein is playing in the physiology of photoreceptor cells. If the leucine zipper is functional, identification of the protein or proteins with which nocturnin interacts will be critical. Finally, it remains to be determined where nocturnin fits into the makeup of the circadian system. Is it a downstream, clock-controlled gene or is it a clock component?
Acknowledgments
We gratefully acknowledge Sandra Parsons, Elizabeth Williams, and Chad Friel for technical assistance, and Dr. Donald Cleveland for the chicken β-actin cDNA clone. Additionally, we thank Dr. Greg Cahill, Dr. David Osterbur, and Brooke Sanger for many helpful discussions. This work was supported by National Institutes of Health Grant EY02414 (J.C.B.) and National Institutes of Health Postdoctoral Fellowship EY06489 (C.B.G.).
Footnotes
References
- 1.Pittendrigh C S. In: Handbook of Behavioral Neurobiology, Biological Rhythms. Aschoff J, editor. Vol. 4. New York: Plenum; 1981. pp. 57–80. [Google Scholar]
- 2.Takahashi J S. Curr Opin Genet Dev. 1993;3:301–309. doi: 10.1016/0959-437x(93)90038-q. [DOI] [PubMed] [Google Scholar]
- 3.Ralph M R, Foster R G, Davis F C, Menaker M. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 4.Deguchi T. Nature (London) 1979;282:94–96. doi: 10.1038/282094a0. [DOI] [PubMed] [Google Scholar]
- 5.Besharse J C, Iuvone P M. Nature (London) 1983;305:133–135. doi: 10.1038/305133a0. [DOI] [PubMed] [Google Scholar]
- 6.Pierce M E, Sheshberadaran H, Zhang Z, Fox L E, Applebury M L, Takahashi J S. Neuron. 1993;10:579–584. doi: 10.1016/0896-6273(93)90161-j. [DOI] [PubMed] [Google Scholar]
- 7.Cahill G M, Besharse J C. Prog Retinal Eye Res. 1995;14:267–291. [Google Scholar]
- 8.Tosini G, Menaker M. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 9.Ralph M R, Menaker M. Science. 1988;241:1225–1227. doi: 10.1126/science.3413487. [DOI] [PubMed] [Google Scholar]
- 10.Vitaterna M H, King D P, Chang A-M, Kornhauser J M, Lowrey P L, McDonald J D, Dove W F, Pinto L H, Turek F W, Takahashi J S. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunlap J C. Annu Rev Physiol. 1993;55:683–728. doi: 10.1146/annurev.ph.55.030193.003343. [DOI] [PubMed] [Google Scholar]
- 12.Hastings M. Curr Biol. 1994;4:720–723. doi: 10.1016/s0960-9822(00)00159-7. [DOI] [PubMed] [Google Scholar]
- 13.Hall J C. Trends Neurosci. 1995;18:230–240. doi: 10.1016/0166-2236(95)93908-g. [DOI] [PubMed] [Google Scholar]
- 14.Kay S A, Millar A J. Cell. 1995;83:361–364. doi: 10.1016/0092-8674(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 15.Gekakis N, Saez L, Delahaye-Brown A-M, Myers M, Sehgal A, Young M, Weitz C J. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 16.Vosshall L B, Price J L, Sehgal A, Saez L, Young M W. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- 17.Sehgal A, Price J L, Man B, Young M W. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 18.Loros J J, Denome S A, Dunlap J C. Science. 1989;243:385–388. doi: 10.1126/science.2563175. [DOI] [PubMed] [Google Scholar]
- 19.Florez J C, Takahashi J S. Ann Med. 1995;27:481–490. doi: 10.3109/07853899709002457. [DOI] [PubMed] [Google Scholar]
- 20.Green C B, Besharse J C. J Neurochem. 1994;62:2420–2428. doi: 10.1046/j.1471-4159.1994.62062420.x. [DOI] [PubMed] [Google Scholar]
- 21.Green C B, Besharse J C. Mol Brain Res. 1996;37:157–165. doi: 10.1016/0169-328x(95)00307-e. [DOI] [PubMed] [Google Scholar]
- 22.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 23.Cahill G M, Besharse J C. J Neurosci. 1991;11:2959–2971. doi: 10.1523/JNEUROSCI.11-10-02959.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cahill G M, Besharse J C. Neuron. 1993;10:573–577. doi: 10.1016/0896-6273(93)90160-s. [DOI] [PubMed] [Google Scholar]
- 25.Green C B, Besharse J C. Invest Ophthalmol Visual Sci. 1996;37:S638. [PubMed] [Google Scholar]
- 26.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 27.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 28.Green C B, Cahill G M, Besharse J C. Brain Res. 1995;677:283–290. doi: 10.1016/0006-8993(95)00166-n. [DOI] [PubMed] [Google Scholar]
- 29.Cleveland D W, Lopata M A, MacDonald R J, Cowan N J, Rutter W J, Kirschner M W. Cell. 1980;20:95–105. doi: 10.1016/0092-8674(80)90238-x. [DOI] [PubMed] [Google Scholar]
- 30.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landschulz W H, Johnson P F, McKnight S L. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 32.Malvar T, Biron R W, Kaback D B, Denis C L. Genetics. 1992;132:951–962. doi: 10.1093/genetics/132.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Draper M P, Liu H-Y, Nelsbach A H, Mosley S P, Denis C L. Mol Cell Biol. 1994;14:4522–4531. doi: 10.1128/mcb.14.7.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denis C L, Draper M P, Liu H-Y, Malvar T, Vallari R C, Cook W J. Genetics. 1994;138:1005–1013. doi: 10.1093/genetics/138.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Draper M P, Salvadore C, Denis C L. Mol Cell Biol. 1995;15:3487–3495. doi: 10.1128/mcb.15.7.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Z J, Edery I, Rosbash M. Nature (London) 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 37.Vinson C R, Sigler P B, McKnight S L. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 38.Harrison S C. Nature (London) 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- 39.Rabindran S K, Haroun R I, Clos J, Wisniewski J, Wu C. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- 40.Treisman J E, Lai Z-C, Rubin G M. Development (Cambridge, UK) 1995;121:2835–2845. doi: 10.1242/dev.121.9.2835. [DOI] [PubMed] [Google Scholar]
- 41.Dorow D S, Devereux L, Dietzsch E, De Kretser T. Eur J Biochem. 1993;213:701–710. doi: 10.1111/j.1432-1033.1993.tb17810.x. [DOI] [PubMed] [Google Scholar]
- 42.Mukai H, Ono Y. Biochem Biophys Res Commun. 1994;199:897–904. doi: 10.1006/bbrc.1994.1313. [DOI] [PubMed] [Google Scholar]


