Abstract
We have previously reported that l-proline has cryoprotective activity in Saccharomyces cerevisiae. A freeze-tolerant mutant with l-proline accumulation was recently shown to carry an allele of the PRO1 gene encoding γ-glutamyl kinase, which resulted in a single amino acid substitution (Asp154Asn). Interestingly, this mutation enhanced the activities of γ-glutamyl kinase and γ-glutamyl phosphate reductase, both of which catalyze the first two steps of l-proline synthesis and which together may form a complex in vivo. Here, we found that the Asp154Asn mutant γ-glutamyl kinase was more thermostable than the wild-type enzyme, which suggests that this mutation elevated the apparent activities of two enzymes through a stabilization of the complex. We next examined the gene dosage effect of three l-proline biosynthetic enzymes, including Δ1-pyrroline-5-carboxylate reductase, which converts Δ1-pyrroline-5-carboxylate into l-proline, on l-proline accumulation and freeze tolerance in a non-l-proline-utilizing strain. Overexpression of the wild-type enzymes has no influence on l-proline accumulation, which suggests that the complex is very unstable in nature. However, co-overexpression of the mutant γ-glutamyl kinase and the wild-type γ-glutamyl phosphate reductase was effective for l-proline accumulation, probably due to a stabilization of the complex. These results indicate that both enzymes, not Δ1-pyrroline-5-carboxylate reductase, are rate-limiting enzymes in yeast cells. A high tolerance for freezing clearly correlated with higher levels of l-proline in yeast cells. Our findings also suggest that, in addition to its cryoprotective activity, intracellular l-proline could protect yeast cells from damage by oxidative stress. The approach described here provides a valuable method for breeding novel yeast strains that are tolerant of both freezing and oxidative stresses.
Frozen-dough technology has recently been used in the baking industry to supply oven-fresh bakery products to consumers. Many freeze-tolerant yeasts have been isolated from natural sources and have also been constructed by conventional mutation techniques (9, 10, 15, 19, 20). However, the mechanism of freeze tolerance is not well understood, and a baker's yeast that provides good leavening qualities for both sweet- and lean-thawed doughs after frozen storage has not yet been developed.
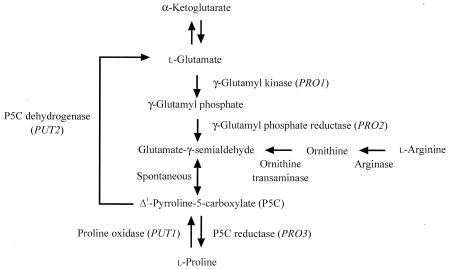
We previously investigated the cryoprotective effects of amino acids on freezing stress in the yeast Saccharomyces cerevisiae and found that l-proline, known as an osmoprotectant (4, 7), has cryoprotective activity that is nearly equal to that of glycerol or trehalose (17, 31). S. cerevisiae synthesizes l-proline from l-glutamate catalyzed by three enzymes, γ-glutamyl kinase (γ-GK; the PRO1 gene product), γ-glutamyl phosphate reductase (γ-GPR; the PRO2 gene product), and Δ1-pyrroline-5-carboxylate reductase (P5CR; the PRO3 gene product), although the rate-limiting step has not yet been determined (Fig. 1). On the other hand, l-proline is converted to l-glutamate within mitochondria by the action of two enzymes, proline oxidase (the PUT1 gene product) and P5C dehydrogenase (the PUT2 gene product) (Fig. 1). We also showed that there is a positive correlation between intracellular l-proline levels and resistance to these stresses in S. cerevisiae, although there is strain-to-strain variability in freezing tolerance (17, 31, 32).
FIG. 1.
Biosynthesis and metabolism of l-proline in Saccharomyces cerevisiae. Genes encoding particular enzymes are shown in parentheses.
We therefore isolated a mutant, derived from l-proline analogue-resistant mutants of S. cerevisiae, which exhibited both l-proline accumulation and freeze tolerance (31). The mutant was recently found to carry an allele of the PRO1 gene encoding γ-GK, to have a single amino acid replacement of Asp by Asn at position 154, and to show a prominent increase in both γ-GK and γ-GPR activities (18). In Escherichia coli, the γ-GK and γ-GPR enzymes are believed to form a heterodimer to channel the unstable intermediate, γ-glutamyl phosphate, in vivo (1). As reported by Smith et al. (29), γ-GK activity was undetected unless purified γ-GPR had been added to the in vitro assay system. In a moth bean P5C synthetase, which exhibited both γ-GK and γ-GPR activities, a leucine zipper sequence is present in each of the enzymatic domains. Leucine zippers may function intramolecularly to maintain the structure of the two domains of the Vigna P5C synthetase, and homodimer or heterodimer formation may occur through the zippers to allow close association between originally separate domains (12). Tomenchok and Brandriss (33) reported that the S. cerevisiae PRO1 gene complemented the E. coli proB-deleted strain, which lacks γ-GK activity, and suggested that S. cerevisiae γ-GK can complex with E. coli γ-GPR. Our results also suggested that yeast γ-GK and γ-GPR together form a complex to function with each other in vivo (18).
Thus, we report here the gene dosage effect of PRO1, PRO2, and PRO3 in the pathway of l-proline biosynthesis on the intracellular l-proline level and freeze tolerance. In addition, a possible mechanism for l-proline accumulation is discussed.
MATERIALS AND METHODS
Yeast and bacterial strains.
The S. cerevisiae strains used in this study are described in Table 1. Strain MB329-17C was derived from a cross between S288C and Σ1278b (34). An l-azetidine-2-carboxylic acid (AZC)-resistant mutant strain, FH515, with higher levels of intracellular l-proline was isolated from strain MB329-17C after ethyl methanesulfonate mutagenesis (31). In this study, put1 gene disruptant strain INVDput1 was constructed from strain INVSc1 (Invitrogen, Carlsbad, Calif.), which is the wild-type strain with an S288C background. E. coli strain DH5α [F− λ− Φ80lacZΔM15 Δ(lacZYA argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) supE44 thi-1 gyrA96] was used to construct the expression plasmids for the yeast genes.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype or strain (plasmids) | Background and/or description |
|---|---|---|
| MB329-17C | α trp1 ura3-52 put1-54 PRO1 | S288C and ∑1278b |
| FH515 | α trp1 ura3-52 put1-54 mutated PRO1 | MB329-17C, l-Proline-accumulated mutant |
| INVSc1 | ahis3-Δ1 leu2 trp1-289 ura3-52 | S288C, His− Leu− Trp− Ura− |
| INVDput1 | ahis3-Δ1 leu2 trp1-289 ura3-52 put1::CgHIS3 | INVSc1, put1 disruptant, Leu− Trp− Ura− |
| INV-WT | INVSc1 (pAD4, pTV3, pUV2, pHV1) | INVSc1, wild type |
| INVDput1-WT | INVDput1 (pAD4, pTV3, pUV2) | INVSc1, put1 disruptant |
| INVDput1-W1 | INVDput1 (pAD-WTPRO1, pTV3, pUV2) | High-copy PRO1 |
| INVDput1-M1 | INVDput1 (pAD-D154NPRO1, pTV3, pUV2) | High-copy mutated PRO1 |
| INVDput1-W12 | INVDput1 (pAD-WTPRO1, pTV-PRO2, pUV2) | High-copy PRO1, PRO2 |
| INVDput1-M12 | INVDput1 (pAD-D154NPRO1, pTV-PRO2, pUV2) | High-copy mutated PRO1, PRO2 |
| INVDput1-W123 | INVDput1 (pAD-WTPRO1, pTV-PRO2, pUV-PRO3) | High-copy PRO1, PRO2, PRO3 |
| INVDput1-M123 | INVDput1 (pAD-D154NPRO1, pTV-PRO2, pUV-PRO3) | High-copy mutated PRO1, PRO2, PRO3 |
| INVDput1-W2 | INVDput1 (pAD4, pTV-PRO2, pUV2) | High-copy PRO2 |
| INVDput1-W3 | INVDput1 (pAD4, pTV3, pUV-PRO3) | High-copy PRO3 |
Plasmids.
Yeast episomal plasmids pAD4, pTV3, and pUV2 (supplied by J. Nikawa) (24), all of which are S. cerevisiae-E. coli shuttle vectors containing the bacterial ampicillin resistance gene and the LEU2, TRP1, and URA3 genes, respectively, were used for complementing the auxotrophic markers and for expressing the PRO1, PRO2, and PRO3 genes, respectively, in S. cerevisiae. Plasmid pAD4 contains the S. cerevisiae ADH1 promoter and terminator regions. Plasmid pCgHIS3 (supplied by S. Harashima) was used for disruption of the PUT1 gene.
Culture media.
The media used for growth of S. cerevisiae were SD (2% glucose, 0.67% Bacto Yeast Nitrogen Base without amino acids; Difco Laboratories, Detroit, Mich.) and YPD (2% glucose, 1% Bacto Yeast Extract, 2% Bacto Peptone). The SD medium contains ammonium sulfate (0.1%) as the nitrogen source. When appropriate, required supplements were added to the media for auxotrophic strains. Yeast strains were also cultured on SD agar plates containing an l-proline analogue, AZC (Sigma Chemical Co., St. Louis, Mo.). The E. coli recombinant strains were grown in Luria-Bertani medium (26) containing ampicillin (50 μg/ml). When necessary, 2% agar was added to solidify the medium.
Disruption of the PUT1 genes.
The enzymes used for DNA manipulation were obtained from Takara Shuzo (Kyoto, Japan) and were used under the conditions recommended by the supplier. Conventional techniques were used for DNA manipulation and transformation as described previously (24). The DNA fragment containing the Candida glabrata HIS3 gene was amplified by PCR with plasmid pCgHIS3 and oligonucleotide primers 5′-TTG GAA TTT CCT TTC GGC AAT GGC TTT CCG GTT ACC ACG CGT TGT AAA ACG ACG GCC AGT-3′ and 5′-TAA GCC TGA CGA CGA CAA GCC ACT TTA CTA CCG ATT TAG GCA CAG GAA ACA GCT ATG ACC-3′ (the underlining indicates the sequences 322 bp upstream of the ATG initiation codon and 287 bp downstream of the TGA termination codon of the PUT1 gene, respectively). The unique amplified band of 2.0 kb containing the C. glabrata HIS3 gene was purified and then integrated into the PUT1 locus in strain INVSc1 by transformation. The resultant put1 disruptant, INVDput1, was selected from among several His+ transformants, and the correct disruption was verified by chromosomal PCR analysis.
Construction of plasmids for expression of the PRO1, PRO2, and PRO3 genes in S. cerevisiae.
To overexpress the wild-type and mutated PRO1 genes, a DNA fragment of the open reading frame was prepared by PCR with genomic DNA from strains MB329-17C and FH515 and oligonucleotide primers 5′-ACC CAA GCT TTG GTC AGT GGC ACA G-3′ and 5′-ACC CGA GCT CGA AGG ATT TTA ACG GAT CAC A-3′ (the underlining indicates the positions of HindIII and SacI, respectively). The unique band of 1.9 kb amplified from genomic DNA of MB329-17C and FH515 was digested with HindIII and SacI and then ligated to the large fragment of pAD4 digested with HindIII and SacI to construct pAD-WTPRO1 and pAD-D154NPRO1, respectively. The nucleotide sequences of the wild-type and mutated PRO1 genes were confirmed by DNA sequencing.
To overexpress the PRO2 and PRO3 genes, a DNA fragment of the open reading frame was prepared by PCR with genomic DNA from strain INVSc1 and oligonucleotide primers 5′-CCA ACT GCA GTT GTG GCG TTG GGT C-3′ and 5′-ACC CGA GCT CCT TGA AGC TTC CGC C-3′ for the PRO2 gene and 5′-CCA ACT GCA GCG TAC AAA AGG ACA AGA TC-3′ and 5′-ACC CGA GCT CTT ATC GGA CCG ACG G-3′ for the PRO3 gene (the underlining indicates the positions of PstI and SacI, respectively). The unique amplified bands corresponding to 1.7 and 1.1 kb, respectively, were digested with PstI and SacI and then ligated to the PstI and SacI sites of pAD4 to construct pAD-PRO2 and pAD-PRO3, respectively. The nucleotide sequences of the PRO2 and PRO3 genes were confirmed by DNA sequencing. The 4.0- and 3.4-kb BamHI fragments from pAD-PRO2 and pAD-PRO3 were then ligated into the BamHI site of pTV3 and pUV2 to construct pTV-PRO2 and pUV-PRO3, respectively.
These genes were placed under the control of the S. cerevisiae ADH1 promoter and terminator in the resultant plasmids. These plasmids were then introduced into strain INVDput1.
Enzyme assay.
To determine the activities of γ-GK (EC 2.7.2.11), strains MB329-17C and FH515 were grown in 50 ml of SD medium at 30°C for 24 h with shaking. The whole-cell extracts were prepared by vortexing the cells with glass beads. An ammonium sulfate precipitate (70% saturation) of the extracts was then desalted with a PD-10 column (Amersham Pharmacia Biotech, Buckinghamshire, England) and used as the enzyme source. Protein concentrations were determined using a Bio-Rad (Hercules, Calif.) protein assay kit with bovine serum albumin as the standard protein.
The γ-GK activity was assayed by the procedure of Smith et al. (29), and the reaction mixture contained the following in a final volume of 0.25 ml at pH 7.0: 50 mM l-glutamate, 10 mM ATP, 20 mM MgCl2, 100 mM hydroxylamine-HCl, 50 mM Tris base, and the enzyme plus water. The reaction was carried out at 37°C for 30 min to 2 h and then terminated by the addition of 1 ml of stop solution (55 g of FeCl3-6H2O, 20 g of trichloroacetic acid, 21 ml of 12 N HCl per liter). Precipitated proteins were removed by centrifugation, and the absorbance at 535 nm was recorded against a blank identical to the one mentioned above but lacking ATP. The amount of γ-glutamyl hydroxamate was measured from the absorbance at 535 nm by comparison with a standard curve of γ-glutamyl hydroxamate (Sigma). One unit of activity was defined as the amount of enzyme required to produce 1 μmol of γ-glutamyl hydroxamate per h.
Intracellular contents of l-proline and freeze-tolerance test.
In a 500-ml flask, yeast cells were grown to the stationary phase in 50 ml of SD medium at 30°C for 48 h with shaking. For the determination of intracellular l-proline, 5 ml of cell suspension (approximately 5 × 108 cells) was removed and the cells were washed twice with 0.9% NaCl and suspended in 0.5 ml of distilled water. The 1.5-ml microcentrifuge tube containing cells was transferred to a boiling-water bath, and intracellular amino acids were extracted by boiling for 10 min. After centrifugation (5 min at 15,000 × g), each supernatant was subsequently quantitated with an amino acid analyzer (L-8500A; Hitachi Co., Tokyo, Japan). l-Proline content was expressed as a percentage of dry weight.
For the freeze-tolerance test, 0.1 ml of cell suspension (approximately 107 cells) was stored at −20°C. Under these conditions, it took about 1 h until the cells were frozen, assuming that the cooling rate was low (approximately 0.5 to 1.0°C/min). Samples of the frozen cells were thawed at room temperature for 15 min, serial dilutions in distilled water were prepared, and aliquots were plated on YPD plates. After incubation at 30°C for 2 days, the survival rates were expressed as percentages, which were calculated as follows: [(number of colonies after freezing at −20°C)/(number of colonies before freezing)] × 100.
H2O2 tolerance test.
Yeast cells were cultured to the exponential growth phase (optical density at 600 nm of 1.0) in 5 ml of SD medium at 30°C with shaking and were exposed to 3 mM H2O2 for various times. Before addition of H2O2 and at intervals thereafter, 0.1 ml of the culture was removed and diluted in distilled water and aliquots were plated on YPD plates. After incubation at 30°C for 2 days, the survival rates were expressed as percentages, which were calculated as follows: [(number of colonies after addition of H2O2)/(number of colonies before addition of H2O2)] × 100.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the PRO1, PRO2, PRO3, PUT1, and C. glabrata HIS3 genes are M85293, U43565, M57886, M18107, and U31470, respectively.
RESULTS
Asp154Asn mutant γ-GK had increased stability.
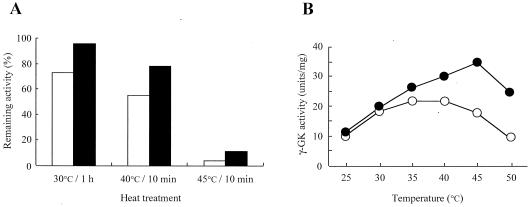
We have previously shown that the allele of PRO1 enhanced the activities of both γ-GK and γ-GPR, which together may form a complex in vivo (18). It has been suggested that the Asp154Asn substitution in the γ-GK protein causes a stabilization of the complex, which leads to an elevation in the apparent activities of the two enzymes. Therefore, we first examined the thermostabilities and optimum temperatures of the wild-type and Asp154Asn mutant γ-GK enzymes in the crude extracts prepared from strains MB329-17C and FH515 (Fig. 2). The mutant γ-GK had a residual activity that was 1.3- to 1.4-fold greater than that of the wild-type enzyme under all of the conditions tested (Fig. 2A). The optimum temperature of the mutant γ-GK also increased to 45°C, whereas the wild-type enzyme had maximal activity at 35°C and a remarkable drop in activity was observed when the enzyme was assayed at 45°C (Fig. 2B). These results indicate that the Asp154Asn mutant γ-GK becomes more stable than the wild-type enzyme.
FIG. 2.
The Asp154Asn mutant γ-GK is highly thermostable compared with the wild-type enzyme. (A) Thermostability of the wild-type (open bars) and Asp154Asn mutant (filled bars) γ-GK. γ-GK activity remaining after incubation at the indicated conditions was determined at 37°C and was expressed as a percentage of the original activity. The variations in the values were less than 5%. (B) Temperature dependence of the relative enzymatic activity of the wild-type (open circles) and Asp154Asn mutant (filled circles) γ-GK. The values are means of results from three independent experiments. The variations in the values were less than 5%.
However, these characteristics of γ-GPR enzymes were not analyzed at temperatures above 30°C, because some unexpected contaminants in the crude extracts perturbed the reverse reaction of γ-GPR by a phosphate-dependent reduction of NADP+ with glutamate-γ-semialdehyde (derived from equilibrium with P5C) as the substrate.
Overexpression of mutant γ-GK and wild-type γ-GPR causes l-proline accumulation.
Enhancement of the apparent γ-GK and γ-GPR activities due to substitution of one amino acid in the γ-GK protein leads to oversynthesis of l-proline in yeast cells. Therefore, because the rate-limiting step has not been determined yet in S. cerevisiae, one might expect that the increase of enzyme activities involved in the l-proline biosynthetic pathway due to gene dosage caused the accelerated conversion of l-glutamate to l-proline (Fig. 1). We then constructed four high-copy-number plasmids for the S. cerevisiae PRO1 encoding γ-GK (the wild type and the Asp154Asn mutant), PRO2 encoding γ-GPR, and PRO3 encoding P5CR as described in Materials and Methods. In these plasmids, pAD-WTPRO1, pAD-D154NPRO1, pTV-PRO2, and pUV-PRO3, each gene was expressed under the control of the ADH1 promoter in S. cerevisiae.
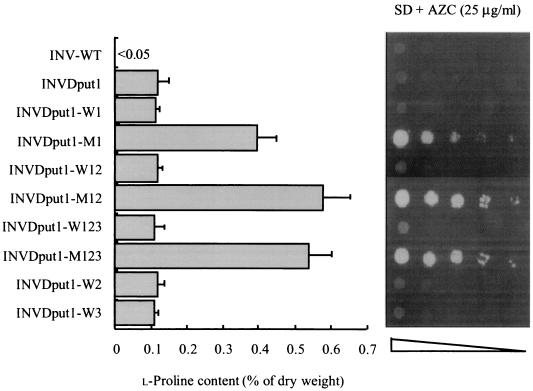
Strain INVDput1, which lacked the proline oxidase required for l-proline utilization, was transformed with these plasmids or with the vector only as controls. The Leu+, Trp+, and Ura+ transformants were cultivated in SD medium, and the cellular l-proline levels were examined (Fig. 3). In agreement with the results of our previous report (32), the put1-disrupted strain INVDput1 accumulated higher l-proline levels (0.10% of the dry weight) than did the control strain INV-WT (<0.05%). l-Proline content was virtually unchanged in the strains which overexpress the wild-type enzymes (INVDput1-WT, INVDput1-W12, INVDput1-W123, INVDput1-W2, and INVDput1-W3). In contrast, strain INVDput1-M1 carrying the mutated PRO1 gene showed a prominent fourfold increase in l-proline content (0.40%), probably due to an increase in enzyme activity. It is noteworthy that when the wild-type γ-GPR was co-overexpressed with the mutant γ-GK, the l-proline level in strain INVDput1-M12 was approximately 1.5-fold that of strain INVDput1-M1. However, the gene dosage effect of PRO3 on l-proline accumulation was not significantly observed in strain INVDput1-M123.
FIG. 3.
Intracellular l-proline contents and the growth phenotypes on AZC-containing medium of S. cerevisiae strains overexpressing l-proline-biosynthetic enzymes. (Left) Intracellular l-proline content was measured after cultivation in liquid SD medium at 30°C for 2 days. Each bar represents the mean result and standard deviation from five independent experiments. (Right) Approximately 106 cells of each strain and serial dilutions of 10−1 to 10−4 (from left to right) were spotted onto SD plates containing 25 μg of AZC/ml. The plates were incubated at 30°C for 3 days.
Overproduction of l-proline is believed to dilute the toxic l-proline analogue AZC, which is incorporated into proteins competitively with l-proline (28). We examined the growth of yeast strains on SD agar plates containing toxic AZC (Fig. 3). Strains INVDput1-M1, INVDput1-M12, and INVDput1-M123 clearly showed AZC resistance, whereas the rest of the strains were sensitive to AZC. Also, the increased l-proline level reflects greater resistance to AZC. These results indicate that overexpression of mutant γ-GK and wild-type γ-GPR and disruption of the proline oxidase gene are effective for l-proline accumulation in yeast cells.
Yeast strains with l-proline accumulation showed higher tolerance to freezing stress.
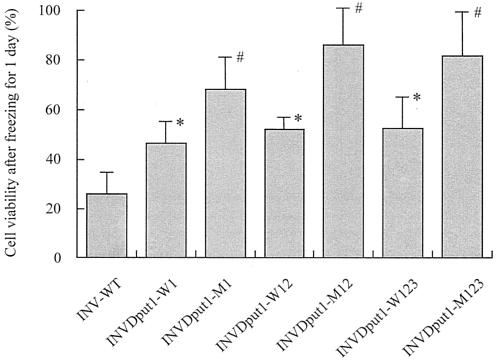
To test the freeze tolerance, yeast strains were cultured in liquid SD medium. As shown in Fig. 4, in proportion to the cellular l-proline level, strains INVDput1-M1, INVDput1-M12, and INVDput1-M123 exhibited increased cell viability compared with the rest of the strains when the cell suspensions were exposed to freezing at −20°C for 1 day. Prolonged storage of the cells at −20°C caused a gradual loss of freeze tolerance in all of the strains, although a significant cryoprotective effect was observed (data not shown). These results are consistent with the finding that there is a positive correlation between intracellular l-proline levels and resistance to freezing stress in S. cerevisiae (17, 18, 31, 32).
FIG. 4.
Freezing stress tolerance of S. cerevisiae strains overexpressing l-proline-biosynthetic enzymes. Cell viability was expressed as a percentage of the number of colonies after freezing at −20°C for 1 day relative to the number of colonies before freezing. The total number of cells corresponding to 100% was approximately 107. Each bar represents the mean result and standard deviation from three independent experiments. Asterisks indicate significant difference from strain INV-WT by Student's t test (P < 0.05). Number signs indicate significant difference from strains INV-WT, INVDput1-WT, INVDput1-W12, and INVDput1-W123 by Student's t test (P < 0.05).
Intracellular l-proline protects yeast cells from damage by oxidative stress.
The processes of freezing and thawing are known to result in oxidative stress to cells (22). In particular, free radicals and reactive oxygen species are generated and cause oxidative damage to cellular components (23). Also, elevated l-proline in plants has been shown to reduce the levels of free radicals in response to osmotic stress (11). We therefore compared yeast cell viabilities after the addition of 3 mM H2O2 to liquid SD medium. Wild-type strain INV-WT contained a trace amount of l-proline (<0.05% of the dry weight), and its cell viabilities at 2 and 4 h after the addition of H2O2 decreased dramatically (24.7 and 3.11%, respectively). In contrast, the typical l-proline-accumulating strain INVDput1-M12 (0.55% of the dry weight) was shown to be much more tolerant of H2O2 (cell viabilities of 35.0 and 8.69%, respectively) than was the wild-type strain INV-WT. This finding suggests that a high concentration of l-proline plays a crucial role in protecting yeast cells under oxidative stress.
DISCUSSION
To accelerate the conversion of l-glutamate to l-proline, we overexpressed three enzymes (γ-GK, γ-GPR, and P5CR) required for S. cerevisiae l-proline biosynthesis by using high-copy-number plasmids. Interestingly, so long as the wild-type γ-GK was overexpressed, l-proline did not accumulate in the cells overexpressing the wild-type γ-GPR and/or P5CR enzymes. In contrast, it should be noted that a combination of the Asp154Asn mutant γ-GK and the wild-type γ-GPR resulted in an approximately 50% increase in the intracellular content of l-proline. Position 154 in γ-GK may be important for the formation of the γ-GK-γ-GPR complex, and the replacement of Asp154 by Asn may facilitate an intermolecular interaction that stabilizes the complex. This mutation at position 154 in the S. cerevisiae γ-GK is novel in that different mutations in other amino acid residues required for feedback inhibition of γ-GK by l-proline were identified in bacterial and plant genes that have been studied (5, 6, 13, 21, 35). Based on the prediction of the secondary structure by the method of Chou and Fasman (3), Asp154 in the yeast γ-GK protein is believed to be located in a turn-like region between an α-helix (Val147-Phe152) and a β-sheet (Thr157-Thr182), which is probably on the molecular surface. It is possible that mutant γ-GK has a stronger interaction with γ-GPR than does wild-type γ-GK. We now analyze the complex formation or protein-protein interaction of γ-GK and γ-GPR by using a yeast two-hybrid assay. Further, the gene dosage effect of PRO3 on l-proline accumulation was not found (Fig. 3). The PRO3 gene encoding P5CR, which converts P5C into l-proline, is constitutively expressed (2). Also, high expression of the P5CR cDNA in tobacco has not been shown to alter the l-proline level in transgenic plants (30). These results strongly suggest that γ-GK and γ-GPR, not P5CR, are rate-limiting enzymes in l-proline biosynthesis in S. cerevisiae.
Injuries to cells due to freezing can be categorized into two types (8, 16). Low cooling rates cause the osmotic shrinkage of cells. Hydration then occurs, and biological macromolecules and/or membrane components denature. More-rapid freezing does not permit the transport of intracellular water through the membrane and impairs the membrane structure or function as ice crystals form in the cells. Therefore, freezing, desiccation, and osmotic stresses, so-called water stresses, are believed to cause common deleterious damage to the cell membrane and functional proteins (25). It is considered preferable for the natural compounds called cryoprotectants to have the capacity to form strong hydrogen bonds with free water (25).
Using yeast mutants defective in antioxidant functions, Park et al. have reported that superoxide anions formed during the aerobic freezing-thawing process induce oxidative stress and injure yeast cells (23). Osmotic stress also induces free-radical production in plant cells, suggesting that oxidative stress is at least partly responsible for the damage caused to plant cells by osmotic stress (11). In general, oxygen-free radicals and other reactive oxygen species could attack vulnerable proteins containing iron sulfur centers (14). It has been proposed that l-proline may act as a free-radical scavenger to protect plants from damage by oxidative stress caused during osmotic stress (11). We found that overexpression of the mutant form of PRO1 could protect yeast cells from oxidative stress during exposure to H2O2. Similar results were obtained when yeast cells were exposed to heat shock treatment at 50°C, which also causes oxidative stress to yeast cells (data not shown). Our findings suggest that, in addition to its cryoprotective activity, l-proline has an important role in reducing the oxidative stress induced during freezing and thawing. l-Proline was found to enhance the stability of proteins and membranes in environments of low water activity or high temperature (4) and to inhibit aggregation during protein refolding. These observations suggest the possibility that l-proline acts as a protein-folding chaperone (27). Hence, l-proline has promising biotechnological potential as a protective agent for industrial microorganisms and enzymes.
Acknowledgments
We thank S. Harashima (Osaka University, Osaka, Japan) and J. Nikawa (Kyushu Institute of Technology, Fukuoka, Japan) for providing the plasmids. We greatly appreciate the technical assistance provided by Y. Morita and M. Nomura of our laboratory.
This work was supported in part by a grant-in-aid for scientific research to H.T. from the Ministry of Education, Culture, Sports, Science and Technology of Japan (14037262).
REFERENCES
- 1.Baich, A. 1969. Proline synthesis in Escherichia coli. A proline inhibitable glutamic acid kinase. Biochim. Biophys. Acta 192:462-467. [DOI] [PubMed] [Google Scholar]
- 2.Brandriss, M. C., and D. A. Falvey. 1992. Proline biosynthesis in Saccharomyces cerevisiae: analysis of the PRO3 gene, which encodes Δ1-pyrroline-5-carboxylate reductase. J. Bacteriol. 174:3782-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou, P. Y., and G. D. Fasman. 1978. Prediction of the secondary structures of proteins from their amino acid sequence. Adv. Enzymol. 47:45-148. [DOI] [PubMed] [Google Scholar]
- 4.Csonka, L. N. 1989. Physical and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53:121-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Csonka, L. N., S. B. Gelvin, B. W. Goodner, C. S. Orser, D. Siemieniak, and J. L. Slightom. 1988. Nucleotide sequence of a mutation in the proB gene of Escherichia coli that confers proline overproduction and enhanced tolerance to osmotic stress. Gene 64:199-205. [DOI] [PubMed] [Google Scholar]
- 6.Dandekar, A. M., and S. L. Uratsu. 1988. A single base pair change in proline biosynthesis genes causes osmotic stress tolerance. J. Bacteriol. 170:5943-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delauney, A. J., and D. P. S. Verma. 1993. Proline biosynthesis and osmoregulation in plants. Plant J. 4:215-223. [Google Scholar]
- 8.Griffiths, J. B. 1978. Effect of hypertonic stress on mammalian cell lines and its relevance to freeze-thaw injury. Cryobiology 15:517-529. [DOI] [PubMed] [Google Scholar]
- 9.Hahn, Y. S., and H. Kawai. 1990. Isolation and characterization of freeze-tolerant yeasts from nature available for the frozen-dough method. Agric. Biol. Chem. 54:829-831. [Google Scholar]
- 10.Hino, A., H. Takano, and Y. Tanaka. 1987. New freeze-tolerant yeast for frozen dough preparations. Cereal Chem. 64:269-275. [Google Scholar]
- 11.Hong, Z., K. Lakkineni, Z. Zhang, and D. P. S. Verma. 2000. Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol. 122:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, C. A., A. J. Delauney, and D. P. S. Verma. 1992. A bifunctional enzyme (Δ1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc. Natl. Acad. Sci. USA 89:9354-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kosuge, T., and T. Hoshino. 1998. Construction of a proline-producing mutant of the extremely thermophilic eubacterium Thermus thermophilus HB27. Appl. Environ. Microbiol. 64:4328-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longo, V. D., L. L. Lion, J. S. Valentine, and E. B. Gralla. 1999. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch. Biochem. Biophys. 365:131-142. [DOI] [PubMed] [Google Scholar]
- 15.Matsutani, K., Y. Fukuda, K. Murata, A. Kumura, I. Nakamura, and N. Yajima. 1990. Physical and biochemical properties of freeze-tolerant mutants of a yeast Saccharomyces cerevisiae. J. Ferment. Bioeng. 70:275-276. [Google Scholar]
- 16.Mazur, P. 1977. The role of intracellular freezing in the death of cells cooled at supraoptimal rates. Cryobiology 14:251-272. [DOI] [PubMed] [Google Scholar]
- 17.Morita, Y., S. Nakamori, and H. Takagi. 2002. Effect of proline and arginine metabolism on freezing stress of Saccharomyces cerevisiae. J. Biosci. Bioeng. 94:390-394. [DOI] [PubMed] [Google Scholar]
- 18.Morita, Y., S. Nakamori, and H. Takagi. 2003. l-Proline accumulation and freeze tolerance in Saccharomyces cerevisiae are caused by a mutation in the PRO1 gene encoding γ-glutamyl kinase. Appl. Environ. Microbiol. 69:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagawa, S., and H. Ouchi. 1994. Improvement of freeze tolerance of commercial baker's yeasts in dough by heat treatment before freezing. Biosci. Biotechnol. Biochem. 58:2077-2079. [Google Scholar]
- 20.Oda, Y., K. Uno, and S. Ohta. 1986. Selection of yeasts for breadmaking by the frozen-dough method. Appl. Environ. Microbiol. 52:941-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omori, K., S. Suzuki, Y. Imai, and S. Komatsubara. 1992. Analysis of the mutant proBA operon from a proline-producing strain of Serratia marcescens. J. Gen. Microbiol. 138:693-699. [DOI] [PubMed] [Google Scholar]
- 22.Park, J.-I., C. M. Grant, P. A. Attfield, and I. W. Dawes. 1997. The freeze-thaw stress response of the yeast Saccharomyces cerevisiae is growth phase specific and is controlled by nutritional state via the RAS-cyclic AMP signal transduction pathway. Appl. Environ. Microbiol. 63:3818-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, J.-I., C. M. Grant, M. J. Davies, and I. W. Dawes. 1998. The cytoplasmic Cu, Zn superoxide dismutase of Saccharomyces cerevisiae is required for resistance to freeze-thaw stress. J. Biol. Chem. 273:22921-22928. [DOI] [PubMed] [Google Scholar]
- 24.Rose, M., and J. R. Broach. 1991. Cloning genes by complementation in yeast. Methods Enzymol. 194:195-230. [DOI] [PubMed] [Google Scholar]
- 25.Rudolph, A. S., and J. H. Crowe. 1985. Membrane stabilization during freezing: the role of two natural cryoprotectants, trehalose and proline. Cryobiology 22:367-377. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Samuel, D., T. K. S. Kumar, G. Ganesh, G. Jayaraman, P.-W. Yang, M.-M. Chang, V. D. Trivedi, S.-L. Wang, K.-C. Hwang, D.-K. Chang, and C. Yu. 2000. Proline inhibits aggregation during protein refolding. Protein Sci. 9:344-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shichiri, M., C. Hoshikawa, S. Nakamori, and H. Takagi. 2001. A novel acetyltransferase found in Saccharomyces cerevisiae Σ1278b that detoxifies a proline analogue, azetidine-2-carboxylic acid. J. Biol. Chem. 276:41998-42002. [DOI] [PubMed] [Google Scholar]
- 29.Smith, C. J., A. H. Deutch, and K. E. Rushlow. 1984. Purification and characteristics of a γ-glutamyl kinase involved in Escherichia coli proline biosynthesis. J. Bacteriol. 157:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szoke, A., G.-H. Miao, Z. Hong, and D. P. S. Verma. 1992. Subcellular localization of Δ1-pyrroline-5-carboxylate reductase in root/nodule and leaf of soybean. Plant Physiol. 99:1642-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takagi, H., F. Iwamoto, and S. Nakamori. 1997. Isolation of freeze-tolerant laboratory strain of Saccharomyces cerevisiae from proline-analogue-resistant mutants. Appl. Microbiol. Biotechnol. 47:405-411. [DOI] [PubMed] [Google Scholar]
- 32.Takagi, H., K. Sakai, K. Morida, and S. Nakamori. 2000. Proline accumulation by mutation or disruption of the proline oxidase gene improves resistance to freezing and desiccation stresses in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 184:103-108. [DOI] [PubMed] [Google Scholar]
- 33.Tomenchok, D. M., and M. C. Brandriss. 1987. Gene-enzyme relationship in the proline biosynthetic pathway of Saccharomyces cerevisiae. J. Bacteriol. 169:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, S.-S., and M. C. Brandriss. 1986. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT1 gene. Mol. Cell. Biol. 6:2638-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, C.-S., Q. Liu, and D. P. S. Verma. 1995. Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase, a bifunctional enzyme catalyzing the first two steps of proline biosynthesis in plants. J. Biol. Chem. 270:20491-20496. [DOI] [PubMed] [Google Scholar]