Abstract
We analyzed the tuf gene, encoding elongation factor Tu, from 33 strains representing 17 Lactobacillus species and 8 Bifidobacterium species. The tuf sequences were aligned and used to infer phylogenesis among species of lactobacilli and bifidobacteria. We demonstrated that the synonymous substitution affecting this gene renders elongation factor Tu a reliable molecular clock for investigating evolutionary distances of lactobacilli and bifidobacteria. In fact, the phylogeny generated by these tuf sequences is consistent with that derived from 16S rRNA analysis. The investigation of a multiple alignment of tuf sequences revealed regions conserved among strains belonging to the same species but distinct from those of other species. PCR primers complementary to these regions allowed species-specific identification of closely related species, such as Lactobacillus casei group members. These tuf gene-based assays developed in this study provide an alternative to present methods for the identification for lactic acid bacterial species. Since a variable number of tuf genes have been described for bacteria, the presence of multiple genes was examined. Southern analysis revealed one tuf gene in the genomes of lactobacilli and bifidobacteria, but the tuf gene was arranged differently in the genomes of these two taxa. Our results revealed that the tuf gene in bifidobacteria is flanked by the same gene constellation as the str operon, as originally reported for Escherichia coli. In contrast, bioinformatic and transcriptional analyses of the DNA region flanking the tuf gene in four Lactobacillus species indicated the same four-gene unit and suggested a novel tuf operon specific for the genus Lactobacillus.
The members of the genera Lactobacillus and Bifidobacterium are gram-positive organisms considered to belong to the general category of lactic acid bacteria (LAB), even though the genus Bifidobacterium is phylogenetically unrelated and has a unique mode of sugar fermentation (44). These organisms are inhabitants of a wide range of environments, including the gastrointestinal and urogenital tracts of humans and animals. Many LAB strains have a worldwide industrial use as starters in the manufacture of fermented foods. Moreover, some Lactobacillus and Bifidobacterium strains have been shown to have beneficial effects on human and animal health (45).
The evolutionary relationships among LAB have been determined by comparing rRNA gene sequences (mainly 16S rRNA) because of their ubiquity and their resistance to evolutionary changes. Several new genetic approaches for the identification of Lactobacillus and Bifidobacterium species have been used in recent years, including the sequencing of rRNA genes (2, 46, 49, 50, 51, 53), restriction endonuclease fingerprinting (51, 52), analysis with oligonucleotide probes (13, 33, 35), analysis of plasmid content (41), analysis of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis patterns of whole-cell proteins (13, 33), and comparisons of tuf sequences (4, 26, 27). Now, with the advent of the genomics era, this rRNA-based view of bacterial phylogeny is being critically examined. Indeed, many microbial genome sequencing projects are providing phylogenetic markers that supply alternatives for the widely accepted small-subunit rRNA marker.
Many studies emphasize that the present LAB phylogeny, deriving almost entirely from the analysis of only a single gene, may be unsatisfactory; a critical reevaluation of phylogenetic relationships is needed (11, 25). A highly conserved protein, such as RecA, was proposed as an alternative phylogenetic marker for comparative phylogenetic analysis of the genus Bifidobacterium (22) and the Lactobacillus plantarum group (47). Alternative molecules, such as 23S rRNA (26), ATPase subunits (26), RNA polymerase (25), and other proteins (16, 36), recently were used to examine whether phylogenies derived from comparative analysis of 16S rRNA reflect the evolution of microorganisms in general or only their own history. In addition, the significance of 16S rRNA genes as molecular markers sometimes has been questioned, as in the genus Helicobacter, where a large insertion of DNA could change the overall evolutionary scenario. The low rate of 16S rRNA evolution is responsible for the failure of this molecule to provide multiple diagnostic sites for closely related but ecologically distinct taxa. Rates of evolutionary substitution in protein-coding genes are 1 order of magnitude greater than those for 16S rRNA genes. Thus, some pairs of ecologically distinct taxa may have had time to accumulate neutral sequence divergence at rapidly evolving loci but not yet at the 16S rRNA level (11, 30). The highly conserved function and ubiquitous distribution of the gene encoding elongation factor Tu (EF-Tu) may render this gene a valuable phylogenetic marker for eubacteria; this gene already has given satisfying results for enterococcal species (18, 19) and some eubacterial species (27).
EF-Tu is a GTP binding protein playing a central role in protein synthesis. It loads the amino-acyl tRNA molecule onto the ribosome during the translation process. The EF-Tu protein is encoded by the tuf gene in eubacteria and is present in various copy numbers per bacterial genome. The tuf gene belongs to a large transcriptional unit, the str operon, which encodes many ribosomal proteins and related regulatory proteins (5, 21). The str operon of Escherichia coli is composed of four genes: rpsL (coding for ribosomal protein S12), rpsG (ribosomal protein S7), fus (elongation factor G), and tufA (EF-Tu). The order of these genes in this transcriptional unit is similar to that described for many species, including Enterococcus spp., Bacillus subtilis, and Neisseria meningitidis (24). In myxobacteria, EF-Tu is genetically organized in the tRNA-tufB operon, where the tuf gene is preceded by four tRNA genes which are cotranscribed with the tuf gene (3).
In this study, short tuf gene sequences of different LAB strains were obtained and used to analyze the phylogeny of many Lactobacillus and Bifidobacterium species. We also describe the genomic locations of the tuf genes in some Lactobacillus and Bifidobacterium species and their transcription patterns. Moreover, species-specific primers for the identification of members of the L. casei group were designed based on available genome sequences and used successfully in a multiplex PCR assay.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and their origins are summarized in Table 1. All Bifidobacterium strains were grown anaerobically in MRS medium (Difco, Detroit, Mich.) supplemented with 0.05% l-cysteine-HCl and incubated at 37°C for 16 h. Lactobacillus strains were grown aerobically in MRS medium and incubated at 37°C for 16 h.
TABLE 1.
Bacterial strains used
| Species | Straina | PCR results obtained with L. casei group-specific primersb | Origin |
|---|---|---|---|
| L. acidophilus | ATCC 4356T | Human | |
| L. amylovorus | DSM 20531T | Cattle waste (corn silage) | |
| L. crispatus | DSM 20584T | Unknown | |
| NCDO A4 | Unknown | ||
| L. gallinarum | ATCC 33199T | Chicken crop | |
| L. helveticus | ATCC 15009T | Cheese | |
| CNRZ 303 | Cheese | ||
| L. delbrueckii subsp. bulgaricus | ATCC 11842T | Yogurt | |
| L. delbrueckii subsp. delbrueckii | ATCC 9649T | Sour grain mash | |
| L. delbrueckii subsp. lactis | ATCC 12315T | Cheese | |
| L. gasseri | DSM 20243T | ||
| ATCC 19992 | Feces | ||
| L. johnsonii | ATCC 33200T | Human blood | |
| NCC 533 | Human feces | ||
| L. reuteri | DSM 20016T | Human feces | |
| L. fermentum | ATCC 14931T | Unknown | |
| L. rhamnosus | ATCC 11443 | L. rhamnosus | Unknown |
| ATCC 11981 | L. rhamnosus | Unknown | |
| NCC 2504T | L. rhamnosus | Unknown | |
| L. casei | NCDO 173 | L. casei | Unknown |
| NCC 2508T | L. casei | Cheese | |
| L. paracasei subsp. paracasei | ATCC 27216 | L. paracasei subsp. paracasei | Saliva of child |
| NCC 989T | L. paracasei subsp. paracasei | Unknown | |
| NCC 2461 | L. paracasei subsp. paracasei | Infant feces | |
| B. longum | ATCC 15707T | Intestine of adult | |
| NCC 2705 | Infant feces | ||
| B. infantis | ATCC 15697T | Intestine of infant | |
| B. bifidum | ATCC 29521T | Infant feces | |
| B. lactis | DSM 10140T | Yogurt | |
| B. catenulatum | DSM 20103T | Intestine of adult | |
| B. adolescentis | ATCC 15703T | Intestine of adult | |
| B. breve | ATCC 15700T | Intestine of infant | |
| B. animalis | ATCC 25527T | Rat feces | |
| L. casei group | NCC 400 | L. paracasei subsp. paracasei | Unknown |
| NCC 438 | L. paracasei subsp. paracasei | Unknown | |
| NCC 476 | L. paracasei subsp. paracasei | Yogurt | |
| NCC 1002 | L. paracasei subsp. paracasei | Milking machine | |
| NCC 2548 | L. paracasei subsp. paracasei | Adult feces | |
| NCC 2556 | L. paracasei subsp. paracasei | Adult feces | |
| NCC 171 | L. paracasei subsp. paracasei | Pizza | |
| NCC 617 | L. casei | Unknown | |
| NCC 596 | L. rhamnosus | Unknown | |
| NCC 587 | L. rhamnosus | Unknown | |
| NCC 2488 | L. rhamnosus | Infant feces | |
| NCC 534 | L. rhamnosus | Unknown | |
| NCC 2455 | L. rhamnosus | Infant feces | |
| NCC 511 | L. paracasei subsp. paracasei | Wine | |
| NCC 500 | L. paracasei subsp. paracasei | Wine | |
| NCC 443 | L. paracasei subsp. paracasei | Wine | |
| NCC 588 | L. paracasei subsp. paracasei | Unknown | |
| NCC 1813 | L. paracasei subsp.paracasei | Unknown | |
| NCC 2501 | L. paracasei subsp. paracasei | Unknown | |
| NCC 558 | L. paracasei subsp. paracasei | Fermented drink | |
| NCC 2472 | L. paracasei subsp. paracasei | Adult feces | |
| NCC 159 | L. paracasei subsp. paracasei | Pizza | |
| NCC 179 | L. paracasei subsp. paracasei | Pizza | |
| NCC 174 | L. paracasei subsp. paracasei | Pizza | |
| NCC 177 | L. paracasei subsp. paracasei | Pizza | |
| NCC 2511 | L. paracasei subsp. paracasei | Unknown | |
| NCC 2537 | L. paracasei subsp. paracasei | Panenone | |
| NCC 108 | L. casei | Unknown | |
| NCC 546 | L. casei | Italian cheese |
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikroorganismen; NCDO, National Collection of Dairy Organisms; CNRZ, Centre National de Recherches Zootechniques; NCC, Nestlé Culture Collection. L. casei group strains were used for species-specific detection.
Identification by the multiplex PCR assay described in this study.
DNA amplification and cloning of the tuf gene and its locus.
PCR was used to amplify the tuf gene in all investigated Lactobacillus strains. A DNA fragment corresponding to the tuf gene was amplified by using oligonucleotides TUF-1 (5′-GATGCTGCTCCAGAAGA-3′) and TUF-2 (5′-ACCTTCTGGCAATTCAATC-3′). The tuf fragment sequence of Bifidobacterium strains was amplified by using oligonucleotides BIF-1 (5′-GAGTACGACTTCAACCAG-3′) and BIF-2 (5′-CAGGCGAGGATCTTGGT-3′). In order to amplify DNA sequences located upstream of the tuf gene in L. delbrueckii subsp. bulgaricus ATCC BAA-365, we used primers rp (5′-ATAAGACCTTTAGAAGCAGC-3′) and Tu-inv (5′-CACGAGTTTGTGGCATAG-3′), targeting the rpsT gene and the 5′ end of the tuf gene, respectively.
Each PCR mixture (50 μl) contained a reaction cocktail of 20 mM Tris-HCl, 50 mM KCl, 200 μM each deoxynucleoside triphosphate (dNTP), 50 pmol of each primer, 1.5 mM MgCl2, and 1 U of Taq DNA polymerase (Gibco BRL, Paisley, United Kingdom). Each PCR cycling profile consisted of an initial denaturation step (3 min at 95°C), followed by amplification for 30 cycles as follows: denaturation for 30 s at 95°C, annealing for 30 s at 52°C, and extension for 2 min at 72°C. PCR was completed with an elongation phase (10 min at 72°C). The resulting amplicons were separated on 1% agarose gels, followed by ethidium bromide staining. PCR fragments were purified by using a PCR purification kit (Qiagen, West Sussex, United Kingdom) and then cloned in the pGEM-T Easy plasmid vector (Promega, Southampton, United Kingdom) by following the supplier's instructions.
DNA sequencing and phylogeny study.
Nucleotide sequencing of both strands from cloned DNA was performed by using a fluorescence-labeled primer cycle sequencing kit (Amersham Buchler, Braunschweig, Germany) by following the supplier's instructions. The primers used were TUF-1, TUF-2, BIF-1, and BIF-2 labeled with IRD800 (MWG Biotech, Ebersberg, Germany). The sequences determined for the tuf genes of all Lactobacillus and Bifidobacterium strains used in this study and those available in the GenBank database were compared. Sequence alignments were done by using the MultiAlign program and the Clustal-W package. Phylogenetic trees were constructed by using the neighbor-joining program from the PHYLIP software package, version 3.5c (10). Calculation of distance matrices was carried out by using the DNADIST and PROTDIST programs (10) for nucleotide and putative amino acid sequences, respectively, and by using the default models. Dendrograms from gene sequences were also drawn by using the Clustal-X, DNAML (maximum likelihood), and DNAPARS (parsimony) programs (10). The numbers of synonymous substitutions between all possible pairs of tuf genes were determined by applying the method of Nei and Gojobori (29) and by using the MEGA computer program (23). The correction for multiple substitutions was made by using the Jukes-Cantor formula (17).
Reference sequences used.
tuf gene sequences from the following bacteria (GenBank accession numbers) were used for our phylogenetic analysis: L. helveticus ATCC 15009 (AJ418903), L. acidophilus ATCC 4356 (AJ418902), L. amylovorus DSM 20531 (AJ418904), L. delbrueckii subsp. bulgaricus ATCC 11842 (AJ418910), L. delbrueckii subsp. delbrueckii ATCC 9649 (AJ418911), L. delbrueckii subsp. lactis (ATCC 12315), L. reuteri ATCC 23272 (AJ418925), L. fermentum ATCC 14931 (AJ418939), L. rhamnosus ATCC 11443 (AJ459828), L. rhamnosus ATCC 11981 (AJ459829), L. casei NCDO 173 (AJ459390), L. paracasei subsp. paracasei ATCC 27216 (AJ418937), L. paracasei subsp. paracasei ATCC 335 (AJ459399), L. lactis ATCC 11154 (AF274745), Enterococcus faecalis ATCC 29212 (AF124221), E. gallinarum ATCC 49573 (tufA) (AF124223), E. gallinarum ATCC 49573 (tufB) (AF274725), E. faecium ATCC 19434 (tufA) (AF124222), E. faecium ATCC 19434 (tufB) (AF274724), Streptococcus pyogenes ATCC 19615 (AF274743), and S. mutans ATCC 25175 (AF274741).
We extracted the genes surrounding the tuf gene from the Bifidobacterium longum NCC 2705 genome (GenBank accession number NC004307) and from the L. plantarum WCFS1 genome (GenBank accession number AL935263). Preliminary sequence data for the L. gasseri ATCC 33323 genome (Genbank accession number NZAAAB00000000), the L. casei ATCC 334 genome, and the L. delbueckii subsp. bulgaricus ATCC BAA-365 genome were obtained from the U.S. Department of Energy Joint Genome Institute at http://www.jgi.doe.gov/JGI_microbial/html/index.html.
Southern hybridization.
Ten micrograms of bacterial DNA was digested to completion with restriction endonuclease HindIII as recommended by the supplier (Roche, Sussex, United Kingdom). This restriction enzyme was chosen because no restriction sites were observed within the amplified tuf gene fragments. Southern blots of agarose gels were performed with Hybond N+ membranes (Amersham, Little Chalfont, United Kingdom) as described by Sambrook and Russell (37). The filters were hybridized with a PCR-generated probe obtained with primer pairs TUF-1-TUF-2 and BIF-1-BIF-2 and labeled with α-32P by using a random-primer DNA labeling system (Roche) (37) and DNA templates extracted from B. longum NCC 2705 and L. johnsonii NCC 533. Subsequent prehybridization, hybridization, and autoradiography were carried out as described by Sambrook and Russell (37).
RNA isolation and Northern blot analysis.
Total RNA was isolated by resuspending bacterial cell pellets in TRIzol (Gibco BRL), adding 106-μm glass beads (Sigma), and shearing the slurry with a Mini-Beadbeater cell disruptor (Biospec Products) as described by Walker et al. (55). An initial Northern blot analysis of the tuf activity of lactobacilli was carried out with 15-μg aliquots of RNA isolated from 10 ml of Lactobacillus strains collected after 8 or 18 h under the growth conditions described above. The RNA was separated in 1.5% agarose-formaldehyde denaturing gels, transferred to Zeta-Probe blotting membranes (Bio-Rad, Hemel Hempstead, United Kingdom) as described by Sambrook and Russell (37), and fixed by UV cross-linking with a Stratalinker 1800 (Stratagene). PCR amplicons obtained with primers TUF-1 and TUF-2 were radiolabeled (37). Prehybridization and hybridization were carried out at 65°C with 0.5 M NaHPO4 (pH 7.2)-1.0 mM EDTA-7.0% SDS. Following 18 h of hybridization, the membranes were rinsed twice for 30 min each time at 65°C in 0.1 M NaHPO4 (pH 7.2)-1.0 mM EDTA-1% SDS and twice for 30 min each time at 65°C in 0.1 mM NaHPO4 (pH 7.2)-1.0 mM EDTA-0.1% SDS and then exposed to X-Omat autoradiography film (Eastman-Kodak). The sizes of the transcripts were estimated by direct comparison to a molecular RNA ladder (Life Technologies).
Primer extension analysis.
The 5′ ends of the RNA transcripts were determined in the following manner. Separate primer extension reactions were conducted with 15-μg aliquots of RNA isolated as described above and mixed with 1 pmol of primer (IRD800 labeled) and 2 μl of buffer H (2 M NaCl, 50 mM PIPES [pH 6.4]). The mixture was denaturated at 90°C for 5 min and then hybridized for 60 min at 42°C. After the addition of 5 μl of 1 M Tris-HCl (pH 8.2), 10 μl of 0.1 M dithiothreitol, 5 μl of 0.12 M MgCl2, 20 μl of 2.5 mM dNTP mixture, 0.4 μl (5 U) of reverse transcriptase (Sigma), and 49.6 μl of double-distilled water, the enzymatic reaction mixture was incubated at 42°C for 2 h. The reaction was stopped by the addition of 250 μl of ethanol-acetone (1:1), and the mixture was incubated at −70°C for 15 min and centrifuged at 12,000 × g for 15 min. The pellets were dissolved in 4 μl of distilled water and mixed with 2.4 μl of loading buffer from the sequencing kit (Thermosequenase, fluorescence labeled). The cDNA was separated on 8% polyacrylamide-urea gels. Sequencing reactions were conducted with the same primers as those used for the primer extension reactions and detected by using a LiCor sequencer (MWG Biotech). The synthetic oligonucleotides used (designed in this study) were tuf-a (5′-CAAAACAGTAGTAATAGCTGC-3′) and tgf-1 (5′-CGAGAAACGTGACCTTTAC-3′).
Amplification with species-specific primers.
Amplification reactions were performed with a 50-μl (total volume) solution containing 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 200 μM each dNTP (Gibco BRL), 10 pmol each of primers PAR (5′-GACGGTTAAGATTGGTGAC-3′), CAS (5′-ACTGAAGGCGACAAGGA-3′), and RHA (5′-GCGTCAGGTTGGTGTTG-3′), 50 pmol of primer CPR (5′-CAANTGGATNGAACCTGGCTTT-3′), 25 ng of template DNA, and 2.5 U of Taq DNA polymerase. Amplification reactions were performed by using a thermocycler (Perkin-Elmer Cetus 9700) with the following temperature profiles: 1 cycle at 95°C for 5 min; 30 cycles at 95°C for 30 s, 54°C for 1 min, and 72°C for 1.5 min; and 1 cycle at 72°C for 7 min. Primers CAS, PAR, RHA, and CPR were all designed in this study. For routine identification, cells were lysed by using a rapid DNA extraction protocol and were used as direct PCR templates. PCR amplicons were analyzed by 2% (wt/vol) agarose gel electrophoresis in Tris-acetate-EDTA buffer at a constant voltage of 7 V/cm, visualized with ethidium bromide (0.5 μg/ml), and photographed under UV light at 260 nm.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the partial tuf gene sequences generated in this study are as follows: L. gallinarum ATCC 33199 (AY372032), L. helveticus CNRZ 303 (AY372033), L. crispatus DSM 20584 (AY372034), L. crispatus NCDO 4 (AY373256), L. gasseri ATCC 19992 (AY372035), L. johnsonii ATCC 33200 (AY372036), L. johnsonii NCC 533 (AY372049), L. rhamnosus NCC 2504 (AY372037), L. casei NCC 2508 (AY372038), L. paracasei subsp. paracasei NCC 989 (AY372039), L. paracasei subsp. paracasei NCC 2461 (AY372040), B. bifidum ATCC 29521 (AY372041), B. longum ATCC 15707 (AY372042), B. infantis ATCC 15697 (AY372043), B. catenulatum DSM 20103 (AY372044), B. adolescentis ATCC 15703 (AY372045), B. breve ATCC 15700 (AY372046), B. animalis ATCC 25527 (AY370920), and B. lactis DSM 10140 (AY370919). Since the L. gasseri tuf sequence extracted from the ongoing genome sequencing of L. gasseri ATCC 33323 (NZAAAB00000000) contained various reading errors, we decided to sequence this tuf gene again and deposited it in GenBank under accession number AY372047). The DNA region located upstream of the tuf gene of L. delbueckii subsp. bulgaricus ATCC BAA-365 and reported here was deposited in GenBank under accession number AY372048. The GenBank accession number for the tuf locus sequence of L. johnsonii NCC 533 is AY372049.
RESULTS
Identification and alignment of tuf sequences.
The tuf sequences from selected bacterial species for which genome sequences are publically available were aligned and compared. Four conserved regions were identified, and two pairs of primers (BIF-1-BIF-2 and TUF-1-TUF-2) for amplifying regions of 800 bp were designed. These primers allowed the amplification of tuf sequences from different Bifidobacterium and Lactobacillus species. All PCR products were cloned into the vector system pGEMT-Easy. Subsequently, the nucleotide sequence of the inserted DNA fragment was determined by sequencing of three randomly selected clones on both strands for each bacterial species.
A multiple alignment of the tuf sequences determined in our laboratory with those retrieved from databases revealed regions which were conserved in all strains from the same species but which were variable in other species. A similarity comparison of the tuf sequences for lactobacilli and for bifidobacteria demonstrated that the tuf genes were highly conserved among all Lactobacillus species investigated here, with identities ranging from 78 to 98% for DNA (reaching a value of 100% between strains belonging to the same species) and from 76 to 100% for translated gene products (Table 2). Identities among the tuf genes of the bifidobacteria ranged from 89 to 97% (reaching a value of 100% for strains belonging to the same species) for DNA and from 91 to 99% for amino acid sequences (Table 3). Many of the differences observed in DNA sequences among species were silent in terms of their effects on the encoded amino acid sequences. Thus, there were many identical protein sequences, even though their encoding DNAs exhibited substantial divergence.
TABLE 2.
Comparison of nucleotide and amino acid sequence identities for EF-Tu among different Lactobacillus strainsa
| Strain no. | Strain | % Sequence identity for strain no.:
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | ||
| 1 | ATCC 11443 | 80 | 79 | 100 | 79 | 93 | 82 | 82 | 79 | 80 | 81 | 81 | 79 | 100 | 96 | 81 | 80 | 93 | 96 | 79 | 93 | 79 | 79 | 78 | |
| 2 | ATCC 15009 | 87 | 80 | 80 | 96 | 79 | 89 | 89 | 95 | 99 | 84 | 88 | 95 | 80 | 80 | 88 | 97 | 79 | 80 | 96 | 79 | 88 | 88 | 87 | |
| 3 | ATCC 14931 | 86 | 83 | 79 | 80 | 80 | 82 | 82 | 80 | 80 | 89 | 82 | 81 | 79 | 81 | 82 | 80 | 80 | 81 | 81 | 80 | 80 | 80 | 80 | |
| 4 | ATCC 11981 | 108 | 87 | 86 | 79 | 93 | 82 | 82 | 79 | 80 | 81 | 81 | 79 | 100 | 96 | 81 | 80 | 93 | 96 | 79 | 93 | 79 | 79 | 78 | |
| 5 | ATCC 33199 | 78 | 90 | 76 | 80 | 78 | 88 | 88 | 95 | 98 | 83 | 88 | 96 | 79 | 78 | 87 | 96 | 78 | 78 | 96 | 78 | 87 | 87 | 87 | |
| 6 | ATCC 27216 | 98 | 85 | 86 | 96 | 76 | 82 | 82 | 79 | 79 | 80 | 81 | 78 | 93 | 92 | 81 | 80 | 100 | 92 | 78 | 100 | 79 | 79 | 79 | |
| 7 | ATCC 19992 | 87 | 91 | 84 | 87 | 84 | 86 | 99 | 88 | 88 | 87 | 97 | 88 | 82 | 81 | 97 | 88 | 82 | 81 | 89 | 82 | 85 | 85 | 85 | |
| 8 | DSM 20243 | 88 | 91 | 84 | 88 | 84 | 87 | 100 | 88 | 88 | 87 | 97 | 89 | 82 | 81 | 97 | 89 | 82 | 81 | 89 | 82 | 85 | 85 | 85 | |
| 9 | ATCC 4356 | 85 | 97 | 83 | 85 | 89 | 85 | 91 | 92 | 95 | 84 | 87 | 94 | 79 | 79 | 87 | 96 | 79 | 79 | 94 | 79 | 87 | 87 | 87 | |
| 10 | CNRZ 303 | 87 | 100 | 83 | 87 | 90 | 85 | 91 | 91 | 97 | 84 | 88 | 95 | 80 | 80 | 88 | 96 | 79 | 80 | 96 | 79 | 87 | 87 | 87 | |
| 11 | DSM 20016 | 88 | 85 | 94 | 88 | 78 | 86 | 86 | 85 | 84 | 85 | 87 | 84 | 81 | 81 | 87 | 84 | 80 | 81 | 84 | 80 | 81 | 81 | 81 | |
| 12 | ATCC 33200 | 85 | 87 | 82 | 85 | 82 | 84 | 95 | 94 | 87 | 87 | 88 | 88 | 81 | 81 | 100 | 88 | 81 | 81 | 88 | 81 | 86 | 86 | 85 | |
| 13 | DSM 20584 | 100 | 87 | 86 | 100 | 78 | 98 | 87 | 88 | 85 | 87 | 88 | 89 | 79 | 79 | 88 | 95 | 78 | 79 | 100 | 78 | 87 | 87 | 87 | |
| 14 | NCC 2504 | 85 | 96 | 82 | 85 | 90 | 83 | 90 | 90 | 95 | 96 | 84 | 85 | 85 | 96 | 81 | 80 | 93 | 96 | 79 | 93 | 79 | 79 | 78 | |
| 15 | NCC 2508 | 100 | 88 | 85 | 100 | 78 | 97 | 87 | 87 | 86 | 87 | 88 | 84 | 100 | 97 | 81 | 80 | 92 | 100 | 79 | 92 | 79 | 79 | 79 | |
| 16 | NCC 533 | 88 | 90 | 85 | 88 | 82 | 87 | 98 | 98 | 90 | 90 | 87 | 97 | 88 | 89 | 87 | 88 | 81 | 81 | 88 | 81 | 85 | 85 | 85 | |
| 17 | DSM 20531 | 85 | 97 | 83 | 85 | 90 | 85 | 91 | 92 | 98 | 97 | 85 | 87 | 85 | 96 | 86 | 90 | 80 | 80 | 96 | 79 | 88 | 88 | 88 | |
| 18 | NCC 2461 | 98 | 85 | 86 | 98 | 76 | 100 | 86 | 87 | 85 | 85 | 86 | 84 | 98 | 83 | 97 | 87 | 85 | 92 | 78 | 100 | 79 | 79 | 79 | |
| 19 | NCDO 173 | 100 | 88 | 85 | 100 | 78 | 97 | 87 | 87 | 86 | 87 | 88 | 84 | 100 | 85 | 100 | 87 | 86 | 97 | 79 | 92 | 79 | 79 | 79 | |
| 20 | NCDO A4 | 80 | 91 | 77 | 80 | 89 | 78 | 85 | 85 | 89 | 91 | 79 | 84 | 80 | 92 | 80 | 84 | 91 | 78 | 80 | 78 | 87 | 87 | 87 | |
| 21 | NCC 989 | 98 | 85 | 86 | 98 | 76 | 100 | 86 | 87 | 85 | 85 | 86 | 84 | 98 | 83 | 97 | 87 | 85 | 100 | 97 | 79 | 79 | 79 | 79 | |
| 22 | ATCC 9649 | 85 | 91 | 83 | 85 | 84 | 84 | 89 | 89 | 91 | 91 | 85 | 86 | 85 | 91 | 85 | 89 | 91 | 84 | 85 | 85 | 84 | 99 | 99 | |
| 23 | ATCC 12315 | 85 | 91 | 84 | 85 | 84 | 84 | 88 | 88 | 91 | 91 | 85 | 85 | 85 | 90 | 85 | 89 | 91 | 84 | 85 | 85 | 84 | 100 | 99 | |
| 24 | ATCC 11842 | 85 | 90 | 83 | 85 | 83 | 84 | 88 | 88 | 91 | 90 | 85 | 85 | 85 | 90 | 85 | 88 | 91 | 84 | 85 | 84 | 84 | 99 | 100 | |
Data in the upper right triangle represent DNA sequence identities of the tuf genes in Lactobacillus strains, while data in the lower left triangle represent deduced amino acid sequence identities of the corresponding EF-Tu proteins.
TABLE 3.
Comparison of nucleotide and amino acid sequence identities for EF-Tu among different Bifidobacterium strainsa
| Strain no. | Strain | % Sequence identity for strain no.:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
| 1 | DSM 10140 | 90 | 90 | 97 | 91 | 91 | 95 | 89 | 91 | |
| 2 | ATCC 15697 | 92 | 97 | 90 | 95 | 91 | 91 | 96 | 09 | |
| 3 | ATCC 15707 | 92 | 99 | 90 | 94 | 90 | 91 | 100 | 89 | |
| 4 | ATCC 25527 | 99 | 93 | 93 | 91 | 91 | 95 | 89 | 91 | |
| 5 | ATCC 29521 | 95 | 96 | 96 | 95 | 90 | 91 | 94 | 89 | |
| 6 | ATCC 15703 | 93 | 93 | 93 | 93 | 91 | 91 | 90 | 94 | |
| 7 | ATCC 15700 | 97 | 95 | 94 | 98 | 93 | 94 | 91 | 91 | |
| 8 | NCC 2705 | 93 | 99 | 100 | 93 | 96 | 93 | 94 | 89 | |
| 9 | DSM 20103 | 93 | 91 | 91 | 93 | 92 | 93 | 93 | 91 | |
Data in the upper right triangle represent DNA sequence identities of the tuf genes in Bifidobacterium strains, while data in the lower left triangle represent deduced amino acid sequence identities of the corresponding EF-Tu proteins.
Alignment of the amino acid sequences deduced from the tuf genes of lactobacilli and bifidobacteria with other EF-Tu sequences available in databases demonstrated that their gene products are conserved and carry conserved amino acid residues typically found in prokaryotic EF-Tu (18). The portion of the tuf genes of lactobacilli and bifidobacteria described in this study encodes the portion of the EF-Tu protein from residues 104 to 335, according to the numbering for E. coli (42). A secondary structure prediction for this portion included the last four α helices and two β sheets of domain I, the entire domain II, and the N-terminal portion of domain III, on the basis of the experimentally determined structure of EF-Tu of E. coli (42). These domains have been determined to play a crucial role in the correct folding of the protein (42); consequently, the corresponding sequences have remained highly conserved among eubacterial species.
Phylogenetic analysis.
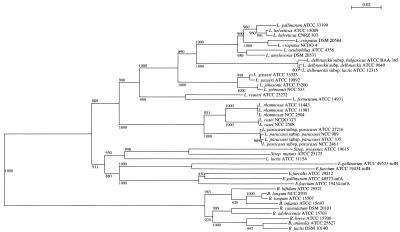
Phylogenetic analysis of the tuf DNA sequences within the genera Lactobacillus and Bifidobacterium by neighbor-joining and maximum-parsimony methods showed clear distinct positions of the two genera (Fig. 1). These data were supported by the reported bootstrap values. For completeness, we included in our analysis the tuf DNA sequences of other strains belonging to different genera representing the LAB group (e.g., Lactococcus, Streptococcus, and Enterococcus). The tree shows two major clusters representing the low-G+C-content gram-positive bacteria (genera Lactobacillus, Lactococcus, Streptococcus, and Enterococcus) and the high-G+C-content gram-positive bacteria (genus Bifidobacterium). Moreover, a further subdivision into three groups corresponding to Lactobacillus, Lactococcus-Streptococcus, and Enterococcus was identified.
FIG. 1.
Phylogenetic tree of Lactobacillus EF-Tu and Bifidobacterium EF-Tu based on nucleotide sequence homology. The phylogenetic tree was constructed by the neighbor-joining method. The scale bar represents sequence divergence. In this tree topology, the phylogenetic distance between organisms is the sum of the horizontal segments. L. acidophilus group A includes the species L. acidophilus, L. amylovorus, L. gallinarum, and L. crispatus; L. acidophilus group B includes only L. gasseri and L. johnsonii. Bootstrap values are reported for a total of 1,000 replicates. tufA and tufB indicate the tufA and tufB genes, respectively.
In order to improve the accuracy of our phylogenetic estimation, we traced trees using different methods. The tree topologies obtained showed similar hierarchical arrangements (data not shown).
The different Lactobacillus and Bifidobacterium species under investigation were unambiguously differentiated by a comparative sequence analysis of a fragment of the tuf gene, as indicated by the phylogenetic tree in Fig. 1. A phylogenetic tree was also constructed on the basis of 16S ribosomal DNA (rDNA) sequences available in databases. The tree topologies obtained with the 16S rDNA sequences showed a phylogenetic arrangement very similar to that of the tuf-based tree (data not shown). A striking feature of the tuf phylogeny is that L. delbrueckii tuf sequences clustered closely with L. acidophilus group A tuf sequences, while L. acidophilus group B tuf sequences clustered more distantly. Interestingly, closely related strains with nearly identical 16S rRNA sequences, e.g., the L. casei group (L. casei, L. paracasei subsp. paracasei, and L. rhamnosus), the L. acidophilus B group (L. gasseri and L. johnsonii), B. animalis-B. lactis, and B. longum-B. infantis, clearly branched separately in the tuf-based tree (Fig. 1).
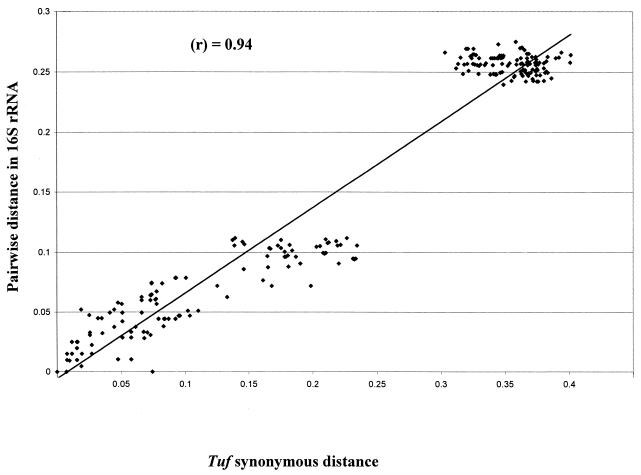
Most of the base substitutions in the tuf genes were synonymous, i.e., did not result in amino acid changes. The synonymous distances calculated from the nucleotide substitution ratios at synonymous positions in the tuf genes were examined for all possible combinations of Lactobacillus and Bifidobacterium tuf genes. The relationships between the pairwise distances for the 16S rRNAs and the synonymous distances for the tuf genes are shown in Fig. 2. There was a significant correlation between the genetic distances for the16S rDNA sequences and those for the tuf sequences. In fact, lactobacilli and bifidobacteria showed a correlation coefficient (r) of 0.94 (Fig. 2). The two groups of dots depicted in Fig. 2 represent the Lactobacillus and Bifidobacterium species in accordance with the different G+C contents of their 16S rRNAs and tuf genes. Therefore, it can be concluded that the base substitutions occurring in the tuf sequences during the evolutionary process render the tuf gene a reliable molecular evolutionary clock.
FIG. 2.
Comparison between genetic distances estimated from 16S rRNA and synonymous distances estimated from tuf genes. The synonymous distances were obtained from the nucleotide sequences of the tuf genes by applying the method of Nei and Gojobori (29). r, coefficient of correlation.
Presence of tuf genes in the Lactobacillus and Bifidobacterium genomes.
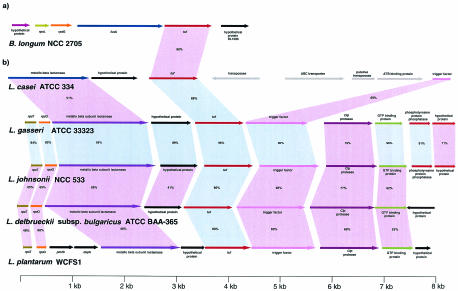
We surveyed all available genomic data for the presence of the tuf gene and its genomic location in various Lactobacillus and Bifidobacterium species and strains (Fig. 3). In B. longum NCC 2705, the tuf gene is directly downstream of the fusA gene (translation elongation factor G), the rpsG gene (30S ribosomal protein S7), and the rpsL gene (30S ribosomal protein S12) and directly upstream of an unidentified open reading frame (Fig. 3a). PCR amplification with a primer targeting a conserved region of the fus gene and the tuf gene yielded the expected amplicons for all nine bifidobacteria tested, indicating the presence of the conserved fus-tuf organization in the Bifidobacterium species tested here (data not shown). The overall organization of the tuf gene of bifidobacteria displayed extensive homology with that of the str operon of E. coli (15) and enterobacteria (19).
FIG. 3.
Comparison of the organization of the tuf loci in bifidobacteria (a) and in lactobacilli (b). (a) B. longum NCC 2705. The DNA sequence fragments of Bifidobacterium represented here are 4.8 kb. (b) L. casei ATCC 334, L. gasseri ATCC 33323, L. johnsonii NCC 533, L. delbrueckii subsp. bulgaricus ATCC BAA-365, and L. plantarum WCFS1. The putative function of a protein is indicated above or below each arrow. Related proteins are linked by blue shading (≥80%) and pink shading (<80%) according to different amino acid similarities; percent amino acid identities are shown. The length of an arrow is proportional to the length of the predicted open reading frame. Corresponding genes are indicated by the same colors of arrows.
Screening of the sequence data from the ongoing genome sequencing projects for L. gasseri ATCC 33323, L. casei ATCC 334, L. johnsonii NCC 533, L. delbrueckii subsp. bulgaricus ATCC BAA-365, and L. plantarum WCFS1 revealed a similar tuf genomic location (Fig. 3). Surprisingly, this tuf arrangement does not resemble any other so far described for tuf loci (3, 5, 6, 14, 15, 19, 20, 21, 38, 54). In L. gasseri ATCC 33323 and L. johnsonii NCC 533, the tuf gene is located downstream of a metallo-beta-lactamase gene, the rpsO gene (30S ribosomal protein S15), and the rpsT gene (30S ribosomal protein S20), while directly downstream of the tuf gene is a transcription regulator trigger factor gene (tig gene), which is followed by genes encoding a Clp protease (clp gene), a GTP binding protein, and a phosphotyrosine protein phosphatase. Both strains showed high sequence identity (from 82 to 96%) within a 9-kb genome fragment. The consensus nucleotide binding domain, Walker motif A (GXXXXGKT), was conserved in the deduced sequences of the tuf and clp genes. An examination of the immediate neighborhood of the Walker sequence indicates that this region is preceded by a β strand and followed by an α helix, an arrangement which complies with the rules for Walker motif A (34). A comparison with various other Lactobacillus species showed a very similar genetic organization of the tuf region. Analysis of the tuf region of L. casei ATCC 334 revealed similar corresponding genes (Fig. 3b). Interestingly, between the tuf gene and the tig gene is an insertion of a 4-kb DNA segment that bears a high resemblance to a mobile element. It contains four genes, the first of which encodes a predicted protein sharing 51% identity with a transposase of Leuconostoc mesenteroides. The next gene encodes a protein which matches an ATP binding cassette transporter. The following genes encode proteins that are identical to a putative transposase of L. rhamnosus and to an ATP binding protein.
Since the ongoing genome sequencing project for L. delbrueckii subsp. bulgaricus ATCC BAA-365 provided incomplete DNA sequences for the tuf gene, these sequences were completed for this study. The DNA sequence located upstream of the tuf gene of L. delbrueckii subsp. bulgaricus ATCC BAA-365 was generated by PCR with two primers targeting conserved regions in the tuf and rpsT genes. The tuf region of L. delbrueckii subsp. bulgaricus ATCC BAA-365 showed the same gene order as that identified for the L. johnsonii, L. gasseri, and L. casei strains examined (Fig. 3b). In L. plantarum WCFS1, except for two gene insertions (pmrB gene, encoding a multidrug resistance efflux pump, and dapA gene, encoding dihydrodipicolinate synthase) upstream of the open reading frame encoding the metallo-beta-lactamase, the gene order surrounding the tuf gene was conserved (Fig. 3b). The degree of sequence conservation in the tuf region of Lactobacillus species reflects the evolutionary distance separating these different species. Bioinformatic analysis suggested a highly conserved DNA module among the Lactobacillus strains investigated here, consisting of the tuf, tig, clp, and GTP binding protein genes.
Estimation of the numbers of tuf genes in the Lactobacillus and Bifidobacterium genomes.
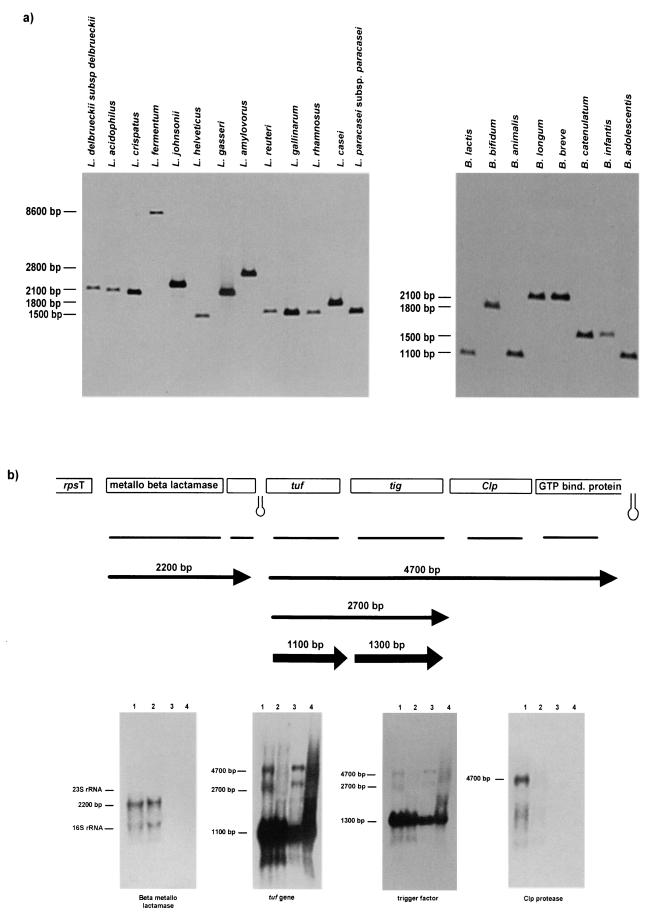
In a Southern hybridization analysis, HindIII-digested genomic DNAs from 13 Lactobacillus species and from 8 Bifidobacterium species were probed with the tuf gene (Fig. 4a). All investigated strains of lactobacilli and bifidobacteria yielded single bands of different sizes (ranging from 1,500 to 8,600 bp in lactobacilli and from 1,100 to 2,100 bp in bifidobacteria), suggesting that only one tuf gene is present in all of the genomes. All bacterial DNAs were also digested with BamHI, and the resulting hybridization patterns again yielded only one band for each bacterial species, confirming the presence of a single tuf gene(data not shown). This result was also confirmed by analysis of the incomplete lactobacillus and bifidobacterium genomes. Sequence analysis of the entire genomes of L. gasseri ATCC 33323, L. johnsonii NCC 533, and B. longum NCC 2705 (39) reveals a unique copy of the tuf gene, whereas other eubacterial taxa (e.g., enterobacteria) have a duplication of EF-Tu.
FIG.4.
Southern and Northern hybridization analyses. (a) Southern hybridization analysis of HindIII-digested genomic DNAs of 13 Lactobacillus and 8 Bifidobacterium species with the tuf gene fragment as a probe. Northern hybridization analysis of Lactobacillus RNA and transcription unit mapping of the tuf locus. In panel a, the sizes of hybridizing fragments are shown. The tested strain was the type strain (Table 1). In the top portion of panel b, all predicted open reading frames are indicated and are annotated with their database matches. The positions of the probes used in Northern blot experiments are indicated under the gene map. The transcripts are depicted as arrows; the arrows point to the 3′ end of the mRNA. The length of the arrow is proportional to the length of the mRNA derived from the Northern blots. The estimated sizes of the mRNAs are indicated. Hairpins indicate possible rho-independent terminators. The transcripts are positioned with respect to the gene map. The width of an arrow indicates the relative abundance of the mRNA species. bind., binding. The bottom portion of panel b shows Northern blot hybridization of RNA isolated from lactobacilli with probes corresponding to the open reading frames in the gene map. The sizes calculated for the hybridization signals are provided.
tuf transcription analysis.
Northern hybridization experiments were performed in order to determine whether the tuf gene is cotranscribed with its flanking genes. Total RNA was extracted from L. johnsonii NCC 533 and L. gasseri ATCC 33323 in the exponential and stationary growth phases. A probe corresponding to the tuf gene hybridized to transcripts of 4.7, 2.7, and 1.1 kb (Fig. 4b). A second probe, overlapping a gene next to the tuf gene that encodes a trigger factor, hybridized to 4.7-, 2.7-, and 1.3-kb transcripts (Fig. 4b). It is highly unlikely that the 1.3-kb transcripts are merely processed products of the 4.7- and 2.7-kb transcripts, since only one band was systematically found with the clp gene probe. In fact, a third probe targeting the gene encoding the Clp protease revealed only one transcript, of 4.7 kb, as did a probe targeting the gene encoding the GTP binding protein (data not shown). These results led us to conclude that the transcripts of 4.7 kb correspond to the tuf gene cotranscribed with the tig and clp genes and with the gene encoding the GTP binding protein and that the transcript of 2.7 kb includes the tuf and tig genes.
The genes located upstream of the tuf gene showed transcription patterns independent of those of the tuf gene. In fact, two probes corresponding to the genes encoding the metallo-beta-lactamase and a hypothetical protein hybridized to a 2.2-kb transcript (Fig. 4b and data not shown). Moreover, these transcripts were present in both exponential and latent growth phases, while the tuf, tig, and clp genes followed different kinetics and appeared to be transcribed only during the exponential growth phase. The amount of transcript corresponding only to the tuf gene is larger than that of the largest transcript species covering the entire tuf gene. These results confirmed what was described earlier for the str operon of B. stearothermophilus and B. subtilis (20). The hybridization signals corresponding to lanes loaded with RNA samples extracted from L. gasseri ATCC 33323 were absent or were weaker than those of L. johnsonii NCC 533 because we used probes for the DNA of L. johnsonii NCC 533 which shared variable degrees of similarity with the DNA of L. gasseri (ranging from 70 to 80%) (Fig. 3b).
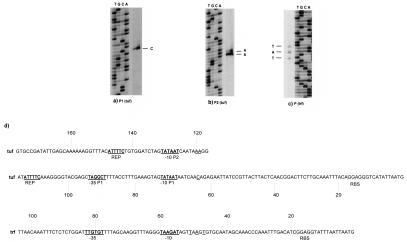
Analysis of the nucleotide sequence of the tuf operon revealed several notable features (Fig. 4b). The tuf operon was delimited at the border by two strong terminator sequences, one located at the 5′ end of a gene upstream of the tuf gene and a second one located at the 5′ end of the GTP binding protein gene. To map precisely the transcription start sites directly upstream of the tuf gene, primer extension experiments were carried out with RNA isolated from exponentially growing L. johnsonii NCC 533 (Fig. 5). Multiple promoter structures have been found preceding the tuf gene. In fact, two transcription start sites were identified at −64 bp (putative promoter P1) and at −119 bp (putative promoter P2) relative to the start site of the coding sequence (Fig. 5a and b). Putative promoter P1 had a −10 region (TATAAT) and a −35 region (TAGGCT), while putative promoter P2 had a −10 box identical to that of putative promoter P1, but no consensus −35 sequences were found (Fig. 5d). Notably, two direct repeats (ATTTTC) were detected in the region upstream of the −10 box for both start sites and could play a role in the recognition of the RNA polymerase. Primer extension experiments confirmed that the gene encoding the trigger factor not only is cotranscribed with the tuf gene but also possesses its own promoter. Primer extension experiments located the 5′ end 47 bp upstream of the start codon of the tig gene (Fig. 5c). An analysis of the putative promoter regions revealed a potential promoter-like sequence having a putative −10 hexamer (TAAGAT) and −35 box (TTGTGT) (Fig. 5d). The promoter sequences comply with all requirements of Lactobacillus promoter sequences necessary for efficient recognition by the σ subunit of the RNA polymerase involved in the transcription of housekeeping genes (7).
FIG. 5.
Primer extension analysis and comparison of the putative promoter sequences of the tuf and trigger factor genes. (a and b) Results of primer extension experiments with oligonucleotides targeting the 5′ end of the tuf gene. P1, putative promoter P1; P2, putative promoter P2. (c) Results of primer extension experiments with an oligonucleotide targeting the 5′ end of the trigger factor gene. The lanes in panels a, b, and c indicate the sequencing products for the tuf and trigger factor genes. (d) Overview of experimentally determined 5′ ends of the tuf gene (tuf) and the trigger factor gene (trf). The endpoints of the primer extension products are underlined. Underlined bold type indicates the −10 and −35 boxes and direct repeats (REP); RBS, Shine-Dalgarno sequences. The start codon is given at the right end of the sequence.
Primer design and PCR assay for Lactobacillus species identification.
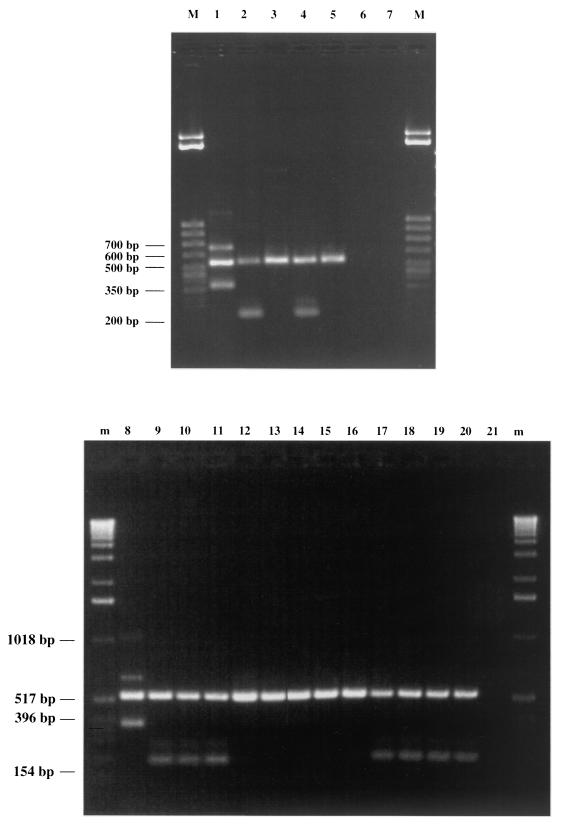
We designed a single reverse primer (CPR) and three forward primers (PAR, CAS, and RHA) for the specific detection of L. paracasei subsp. paracasei, L. casei, and L. rhamnosus. Application of the CPR-PAR-CAS-RHA oligonucleotide mixture (Fig. 6) resulted in PCR amplicons of 700, 540, and 350 bp with DNA extracted from L. casei NCC 2508, amplicons of 540 and 240 bp with DNA derived from L. paracasei subsp. paracasei NCC 989, but only one amplicon of 540 bp with DNA isolated from L. rhamnosus NCC 2504. No PCR product of the above expected sizes was detectable with these primers for any other Lactobacillus or Bifidobacterium strains listed in Table 1. The amplicon sizes were in agreement with those expected from the analysis of the tuf sequences. In fact, the CPR-PAR, CPR-CAS, and CPR-RHA primer pairs must generate amplicons of 240, 350, and 520 bp, respectively. Multiple products are explained by the fact that L. paracasei subsp. paracasei should generate only two amplicons (240 and 520 bp), L. casei should produce two PCR products (350 and 520 bp), and L. rhamnosus should generate only one amplicon (520 bp).
FIG. 6.
Amplification products obtained from the tuf multiplex assay. Lane M, 50-bp DNA molecular marker (Sigma); lane m, 1-kb DNA ladder (Gibco BRL). Lane 1, L. casei NCC 2508; lane 2, L. paracasei subsp. paracasei NCC 989; lane 3, L. rhamnosus NCC 2504; lane 4, L. paracasei subsp. paracasei NCC 2461; lane 5, L. rhamnosus ATCC 11443; lane 6, L. reuteri DSM 2006; lane 7, L. fermentum ATCC 14931; lane 8, L. casei NCC 617; lane 9, L. paracasei subsp. paracasei NCC 438; lane 10, L. paracasei subsp. paracasei NCC 476; lane 11, L. paracasei subsp. paracasei NCC 400; lane 12, L. rhamnosus NCC 596; lane 13, L. rhamnosus NCC 587; lane 14, L. rhamnosus NCC 546; lane 15, L. rhamnosus NCC 2488; lane 16, L. rhamnosus NCC 2455; lane 17, L. paracasei subsp. paracasei NCC 1002; lane 18, L. paracasei subsp. paracasei NCC 2548; lane 19, L. paracasei subsp. paracasei NCC 2556; lane 20, L. paracasei subsp. paracasei NCC 171; lane 21, negative control.
The identities of the PCR fragments were confirmed by sequence analysis (data not shown). The species-specific primer sets based on the tuf gene were also extended to an additional 29 lactobacillus strains (L. casei group strains in Table 1). These strains were originally allocated within the L. casei group on the basis of their fermentative properties and the results of amplified rDNA restriction analysis for Lactobacillus species identification (51). As shown in Table 1 (L. casei group strains) and Fig. 5, 21 strains were clearly allocated within the species L. paracasei subsp. paracasei, 2 strains were identified as belonging to the species L. paracasei subsp. casei, while the remaining 6 strains were found to belong to the species L. rhamnosus. All strains had been previously characterized by ribotyping-hybridization, which produced individual and repeatable profiles for each strain. The heterogeneity among all ribotyping-hybridization patterns clearly demonstrated that all strains investigated with species-specific tuf-based primers were different (data not shown).
DISCUSSION
Significant changes have occurred in bacterial taxonomy since the introduction of molecular techniques. The accurate identification of many bacterial species can be accomplished by reference to rRNA gene sequences (mainly the 16S rRNA gene), which is considered an important molecular marker of modern bacterial taxonomy. The use of other highly conserved macromolecules as evolutionary chronometers might have strong applications in the identification, differentiation, and tracing of bacterial species.
In this study, we have investigated the occurrence of the gene encoding EF-Tu in different species of the genera Bifidobacterium and Lactobacillus. The tuf gene product brings aminoacylated tRNA molecules to the ribosome. This gene represents an ideal target candidate for diagnostic purposes because it is highly conserved and ubiquitous in bacteria (26, 27). It has been already applied to infer phylogeny in the genera Enterococcus (18), Mycoplasma (1), and Staphylococcus (28). In addition, in a very recent study, a comparative analysis of partial tuf sequences was evaluated for the differentiation of some Lactobacillus species (4). It fulfills all prerequisites to server as a suitable phylogenetic marker, such as very high genetic stability and a wide distribution (25). This alternative molecular marker might corroborate and help to complete the evolutionary history of various LAB species. In this report, we demonstrated that there is a high correlation between 16S rDNA sequences and the tuf genes of lactobacilli and bifidobacteria. The use of tuf genes in LAB species as an alternative or complement to the 16S rRNA marker mainly supports the phylogenetic relationships that are revealed by the 16S rRNA-based determination of bacterial phylogeny but also provides more detail that can be used to distinguish closely related species and that can be helpful for inferring phylogeny in closely related species (e.g., B. animalis-B. lactis, B. longum-B. infantis, and L. johnsonii-L. gasseri).
Recently, polyphasic taxonomy (48) was recognized by the International Committee on Systematic Bacteriology as a new tool for the description of species and for the revision of the present nomenclature of some bacterial groups. In view of its demonstrated effectiveness, sequence analysis of protein-coding genes (e.g., tuf genes) as alternative phylogenetic markers could be added to the arsenal of rRNA sequence databases and to the relatively small groEL (16) and recA (9, 22) sequence databases. It has been shown that species having 70% or greater DNA similarity (at the DNA-DNA hybridization or reassociation level) possess in fact more than 97% 16S rDNA sequence identity (43). Consequently, 16S rDNA sequence analysis might not be an appropriate replacement for DNA reassociation to define closely related taxa. Our results and those of previous studies (4, 18, 19, 26, 27) suggested that tuf gene analysis also could be a valid tool for inferring relationships among closely related bacterial species. The use of the tuf gene, as well as the recA gene, as a phylogenetic marker for LAB has the advantage that the amino acid sequences from these genes can be used to infer bacterial phylogenies, avoiding the problems associated with rRNAs and the likely overestimation of the relatedness of taxa with similar nucleotide differences, nonindependence of substitution patterns at different sites, and bias derived from different G+C contents of microorganisms (8). Moreover, at the nucleotide level, EF-Tu can tolerate mutations that do not or only slightly alter it. These mutations can provide information about recent evolutionary history which is too recent to be fixed in slowly diverging sequences such as 16S rRNAs (31).
In this study, we demonstrated that the tuf sequence is a valid molecular marker for inferring interspecies relationships. However, the lack of tuf sequence variations in strains within the same species showed its inadequancy for any intraspecific relationship analysis (e.g., as a typing tool at the strain level). The analysis of variable regions within the tuf genes of the former L. casei group led us to design a set of species-specific PCR primers. We focused our attention on the establishment of a tuf PCR-based assay for the identification of closely related microorganims (e.g., within the L. casei group), which so far cannot be differentiated in a reliable manner by traditional approaches (9).
The tuf genes have been described to be present in the bacterial genome in various copy numbers. Most gram-positive bacteria carry only one tuf gene, and only a few exceptions to this rule have been described (e.g., some Enterococcus [19] and Clostridium [40] species). Since it has been postulated that the second copy of the tuf gene (tufB) in enterococci has been generated from a horizontal gene transfer event (19), caution should be exercised in the interpretation of bacterial evolution when such events occurred. This is not the case for Lactobacillus and Bifidobacterium strains. We have determined that both the low-G+C-content gram-positive bacteria (lactobacilli) and the high-G+C-content gram-positive bacteria (bifidobacteria) investigated here contain only one copy of the tuf gene.
The tuf genes usually are associated with characteristic flanking genes (5, 24). EF-Tu has been described to be part of either the bacterial str operon (20) or the tRNA-tufB operon (3, 5). The arrangement of the tuf gene in the str operon has been described for a variety of bacteria, such as E. coli (15), Bacillus (20, 21), Streptomyces (54), and Enterococcus (19). The arrangement of the tuf gene in the tRNA-tufB operon has been described for Chlamydia trachomatis (5), Thermus thermophilus (38), and Aquifex aeolicus (6), as has that of the tufB gene of E. coli (14). It has been postulated that the widespread EF-Tu gene arrangements might argue in favor of their ancient origins (5). All sequenced gram-positive bacteria with a high G+C content (e.g., bifidobacteria) contain only a single copy of the tuf gene arranged in a manner similar to that of the str operon of E. coli (14). This is the case for B. longum, B. lactis (M. Laloi, personal communication), and all Bifidobacterium species tested in this study.
However, in the available Lactobacillus genomes, the sequences flanking the tuf genes differ from those of any other tuf locus described so far. We have found a common genetic map of the tuf region in all five investigated Lactobacillus species, including rpsT, rpsO, metallo-beta-lactamase, tig, clp, and GTP binding protein genes. However, only the EF-Tu-, trigger factor-, Clp protease-, and GTP binding protein-encoding genes seem to constitute a highly conserved module. Functional analysis of these genes seems to corroborate the hypothesis that these genes constitute the same operon. In fact, the trigger factor is a ribosome-associated protein that interacts with the EF-Tu protein and with a wide variety of nascent polypeptides to catalyze their folding (32). Clp ATP-dependent proteases are stress-induced proteins acting to refold or degrade misfolded or denatured proteins (12). The elongation phase of protein synthesis is promoted by two proteins, EF-Tu and elongation factor G, which binds to the ribosome in its GTP form, hydrolyzes GTP to drive tRNA movement on the ribosome, and is released in its GDP form. We might speculate that the GTP binding protein following the Clp protease could play the role of elongation factor G. The tuf region of Lactobacillus species displays some features that are not found throughout the bacterial world and that could be of great interest from an evolutionary point of view.
We demonstrated that the EF-Tu-, trigger factor-, Clp protease-, and GTP binding protein-encoding genes are cotranscribed and belong to the same transcription unit. Primer extension experiments precisely mapped the start of the transcript species occurring in the tuf operon. Transcripts derived from the tuf promoter and the readthrough tig promoter were present and covered the entire tuf operon. A similar situation in which the Ef-Tu gene is cotranscribed with flanking genes has been described already for Bacillus (20, 21) and E. coli (3). The tuf gene of L. johnsonii has a multiple-promoter structure, which has been described previously for the tuf gene of B. stearothermophilus (20, 21). Transcription directed by a multiple tuf promoter structure in Streptomyces ramocissimus has been demonstrated to be growth phase dependent. Preliminary results regarding the tuf gene of other LAB genera (e.g., Lactococcus and Streptococcus) show a general organization common to their loci but not in common with those of the genus Lactobacillus. The analysis of the flanking regions suggests that in general, the genes surrounding the tuf gene have coevolved with EF-Tu. The still relatively small number of LAB tuf regions available renders attempts to understand the direction of their evolution a challenge. Analysis of a large number of LAB tuf regions may provide important clues to better understanding the biology and the evolutionary history of the tuf region and LAB phylogeny. The tuf locus of Lactobacillus should undergo complementary studies to clarify the role of the 5′-proximal region of the locus in the regulation of expression of the genes and the effects of other possible factors (e.g., growth rate phase) on modulation of the promoter activity of the tuf gene.
In conclusion, in this study we determined the tuf gene sequences of a large number of species of lactobacilli and bifidobacteria, increasing the already existent tuf sequence databases of LAB species. We demonstrated a higher distinctness of the tuf sequences than of the 16S rRNA sequences and offer a valid molecular marker for inferring phylogeny among closely related taxa (e.g., L. casei group). Moreover, we showed for the first time the genetic organization of the tuf operon of lactobacilli, which has no counterpart in any other known bacterial genomes so far.
Acknowledgments
We thank the members of the L. delbrueckii subsp. bulgaricus ATCC BAA-365, L. casei ATCC 364, and L. gasseri ATCC 33323 genome sequencing projects, funded by the U.S. Department of Energy Joint Genome Institute, for providing us the sequence of the tuf loci. We also thank G. Unnlu and J. Broadbent for making raw genome data for L. delbrueckii subsp. bulgaricus ATCC BAA-365 and L. casei ATCC 334 available for tuf locus comparisons (Fig. 3) prior to publication. We are indebted to D. R. Pridmore and M. Laloi (both at NRC) for helpful discussions and comments. Finally, we thank A. Mercenier and F. Arigoni (both at NRC) for constructive and critical reading of the manuscript.
REFERENCES
- 1.Berg, S., E. Luneberg, and M. Frosch. 1996. Development of an amplification and hybridization assay for the specific and sensitive detection of Mycoplasma fermentans DNA. Mol. Cell. Probes 10:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Bourget, N. L., H. Philippe, I. Mangin, and B. Decaris. 1996. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int. J. Syst. Bacteriol. 46:102-111. [DOI] [PubMed] [Google Scholar]
- 3.Bremaud, L., C. Fremaux, S. Laalami, and Y. Cenatiempo. 1995. Genetic and molecular analysis of the tRNA-tufB operon of the myxobacterium Stigmatella aurantica. Nucleic Acids Res. 23:1737-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chavagnat, F., M. Haueter, J. Jimeno, M. G. Casey. 2002. Comparison of partial tuf gene sequences for the identification of lactobacilli. FEMS Microbiol. Lett. 217:177-183. [DOI] [PubMed] [Google Scholar]
- 5.Cousineau, B., C. Cerpa, J. Lefebvre, and R. Cedergren. 1992. The sequence of the gene encoding elongation factor Tu from Chlamydia trachomatis compared with those of other organisms. Gene 120:33-41. [DOI] [PubMed] [Google Scholar]
- 6.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujav, R. Huber, R. A. Felman, J. M. Short, G. J. Olsen, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 7.Djordjevic, G., B. Bojovic, N. Miladinov, and L. Topisirovic. 1997., Cloning and molecular analysis of promoter-like sequences isolated from the chromosomal DNA of Lactobacillus acidophilus ATCC 4356. Can. J. Microbiol. 43:61-69. [DOI] [PubMed] [Google Scholar]
- 8.Eisen, J. A. 1995. The recA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of recAs and 16S rRNAs from the same species. J. Mol. Evol. 41:1105-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felis, G. E., F. Dellaglio, L. Mizzi, and S. Torriani. 2001. Comparative sequence analysis of recA gene fragment brings new evidence for a change in the taxonomy of the Lactobacillus casei group. Int. J. Syst. Evol. Microbiol. 51:2113-2117. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 11.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 12.Gottesman, S., S. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:1338-1347. [DOI] [PubMed] [Google Scholar]
- 13.Hertel, C., W. Ludwig, B. Pot, K. Kersters, and K. H. Schleifer. 1993. Differentiation of lactobacilli occurring in fermented milk products by using oligonucleotide probes and electrophoretic protein profiles. Syst. Appl. Microbiol. 16:463-467. [Google Scholar]
- 14.Hudson, H., J. Rossi, and A. Landy. 1981. Dual function transcripts specifying tRNA and mRNA. Nature 294:422-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaskunas, R. S., L. Lindahl, M. Nomura, and R. R. Burgess. 1975. Identification of two copies of the gene for the elongation factor EF-Tu in E. coli. Nature 257:458-462. [DOI] [PubMed] [Google Scholar]
- 16.Jian, W., L. Zhu, and X. Dong. 2001. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 51:1633-1638. [DOI] [PubMed] [Google Scholar]
- 17.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, N.Y.
- 18.Ke, D., F. J. Picard, F. Martineau, C. Menard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 1999. Development of a PCR assay for rapid detection of enterococci. J. Clin. Microbiol. 37:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ke, D., M. Boissinot, A. Huletsky, F. J. Picard, J. Frenette, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Evidence for horizontal gene transfer in evolution of elongation factor Tu in enterococci. J. Bacteriol. 182:6913-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krasny, L., J. R. Mesters, L. N. Tieleman, B. Kraal, V. Fucik, R. Hilgenfeld, and J. Jonak. 1998. Structure and expression of elongation factor Tu from Bacillus stearothermophilus. J. Mol. Biol. 283:371-381. [DOI] [PubMed] [Google Scholar]
- 21.Krasny, L., T. Vacik, V. Fucik, and J. Jonak. 2000. Cloning and characterization of the str operon and elongation factor Tu expression in Bacillus stearothermophilus. J. Bacteriol. 182:6114-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kullen, M., L. J. Brady, and D. J. O'Sullivan. 1997. Evaluation of using a short region of the recA gene for rapid and sensitive speciation of dominant bifidobacteria in the human large intestine. FEMS Microbiol. Lett. 154:377-383. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, S., K. Tamura, and M. Nei.1993. MEGA: molecular evolutionary genetics analysis, version 1.01. Pennsylvania State University, University Park, Pa.
- 24.Lathe, W. C., III, and P. Bork. 2001. Evolution of tuf genes: ancient duplication, differential loss and gene conversion. FEBS Lett. 502:113-116. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig, W., and K. H. Schleifer. 1999. Phylogeny of bacteria beyond the 16S rRNA standard. ASM News 65:752-757. [Google Scholar]
- 26.Ludwig, W., J. Neumaier, N. Klugbauer, E. Brockmann, C. Roller, S. Jilg, K. Reetz, I. Schachtner, A. Ludvigsen, M. Bachleitner, U. Fischer, and K. H. Schleifer. 1993. Phylogenetic relationships of bacteria based on comparative sequence analysis of elongation factor Tu and ATP-synthase beta subunit genes. Antonie Leeuwenhoek 64:285-305. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig, W., M. Weizenegger, D. Betzl, E. Leidel, T. Lenz, A. Ludvigsen, D. Mollenhoff, P. Wenzig, and K. H. Schleifer. 1990. Complete nucleotide sequences of seven eubacterial genes coding for the elongation factor Tu: functional, structural and phylogenetic evaluations. Arch. Microbiol. 153:241-247. [DOI] [PubMed] [Google Scholar]
- 28.Martineau, F., F. J. Picard, D. Ke, S. Paradis, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2001. Development of a PCR assay for identification of staphylococci at genus and species levels. J. Clin. Microbiol. 39:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and non-synonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 30.Palys, T., E. Berger, I. Mitrica, L. K. Nakamura, and F. M. Cohan. 2000. Protein coding genes as molecular markers for ecologically distinct populations: the case of two Bacillus species. Int. J. Syst. Bacteriol. 50:1021-1028. [DOI] [PubMed] [Google Scholar]
- 31.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 32.Patzelt, H., S. Rudiger, D. Brehmer, G. Kramer, S. Vorderwulbecke, E. Schaffitzel, A. Waitz, T. Hesterkamp, L. Dong, J. Schneider-Mergener, B. Bukau, and E. Deuerling. 1999. Binding specificity of Escherichia coli trigger factor. Proc. Natl. Acad. Sci. USA 98:14244-14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pot, B., C. Hertel, W. Ludwig, P. Descheemaeker, K. Kersters, and K. H. Schleifer. 1993. Identification and classification of Lactobacillus acidophilus, L. gasseri and L. johnsonii strains by SDS-PAGE and rRNA targeted oligonucleotide probe hybridization. J. Gen. Microbiol. 139:513-517. [DOI] [PubMed] [Google Scholar]
- 34.Ramakrishnan, C., V. S. Dani, and T. Ramasarma. 2002. A conformational analysis of Walker motif A [GXXXXGKT(S)] in nucleotide-binding and other proteins. Protein Eng. 15:783-798. [DOI] [PubMed] [Google Scholar]
- 35.Rodtong, S., and G. W. Tannock. 1993. Differentiation of Lactobacillus strains by ribotyping. Appl. Environ. Microbiol. 59:3480-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy, D., S. Sirois. 2001. Molecular differentiation of Bifidobacterium species with amplified ribosomal DNA restriction analysis and alignment of short regions of ldh gene. FEMS Microbiol. Lett. 191:17-24. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Satoh, M., T. Tanaka, A. Kushiro, T. Hakoshima, and K. Tomita. 1991. Molecular cloning, nucleotide sequence and expression of the tufB gene encoding elongation factor Tu from Thermus thermophilus HB8. FEBS Lett. 288:98-100. [DOI] [PubMed] [Google Scholar]
- 39.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sela, S., D. Yogev, S. Razin, and H. Bercovier. 1989. Duplication of the tuf gene: a new insight into the phylogeny of eubacteria. J. Bacteriol. 171:581-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sgorbati, B., V. Scardovi, and D. J. Leblanc. 1982. Plasmids in the genus Bifidobacterium. J. Gen. Microbiol. 128:2121-2131. [DOI] [PubMed] [Google Scholar]
- 42.Song, H., M. R. Parsons, S. Rowsell, G. Leonard, and S. E. V. Phillips. 1999. Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05A resolution. J. Mol. Biol. 285:1245-1256. [DOI] [PubMed] [Google Scholar]
- 43.Stackebrandt, E., and M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 44.Staley, J. T., and N. R. Krieg. 1986. Classification of procaryotic organisms: an overview, p. 1-4. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergery's manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore, Md.
- 45.Tannock, G. W.1994. The acquisition of the normal microflora of the gastrointestinal tract, p. 1-16. In S. A. Gibson (ed.), Human health: the contribution of microorganisms. Springer, London, United Kingdom.
- 46.Tannock, G. W., A. Tilsala-Timisjarvi, S. Rodtong, J. Ng, K. Munro, and T. Alatossava. 1999. Identification of Lactobacillus isolates from the gastrointestinal tract, silage, and yogurt by 16S-23S rRNA gene intergenic spacer region sequence comparison. Appl. Environ. Microbiol. 65:4264-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torriani, S., G. E. Felix, and F. Dellaglio. 2001. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 67:3450-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandamme, P., B. Pot, M. Gillis, P. de Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ventura, M., and R. Zink. 2002. Rapid identification, differentiation, and proposed new taxonomic classification of Bifidobacterium lactis. Appl. Environ. Microbiol. 68:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ventura, M., and R. Zink. 2002. Specific identification and molecular typing analysis of Lactobacillus johnsonii by using PCR-based methods and pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 217:141-154. [DOI] [PubMed] [Google Scholar]
- 51.Ventura, M., I. A. Casas, and L. Morelli. 2000. Rapid amplified ribosomal DNA restriction analysis (ARDRA) identification of Lactobacillus spp. isolated from fecal and vaginal samples. Syst. Appl. Microbiol. 23:504-509. [DOI] [PubMed] [Google Scholar]
- 52.Ventura, M., M. Elli, R. Reniero, and R. Zink. 2001. Molecular microbial analysis of Bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ARDRA). FEMS Microbiol. Ecol. 36:113-121. [DOI] [PubMed] [Google Scholar]
- 53.Ventura, M., R. Reniero, and R. Zink. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex PCR approach. Appl. Environ. Microbiol. 67:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vijgenboom, E., L. P. Woudt, P. W. H. Heinstra, K. Rietveld, J. van Haarlem, G. P. van Wezel, S. Schochat, and L. Bosch. 1994. Three tuf like genes in the kirromycin producer Streptomyces ramocissimus. Microbiology 140:983-998. [DOI] [PubMed] [Google Scholar]
- 55.Walker, D. C., H. S. Girgis, and T. R. Klaenhammer. 1999. The groESL chaperone operon of Lactobacillus johnsonii. Appl. Environ. Microbiol. 65:3033-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]