Abstract
The medicinal value associated with complex polyketide and nonribosomal peptide natural products has prompted biosynthetic schemes dependent upon heterologous microbial hosts. Here we report the successful biosynthesis of yersiniabactin (Ybt), a model polyketide-nonribosomal peptide hybrid natural product, using Escherichia coli as a heterologous host. After introducing the biochemical pathway for Ybt into E. coli, biosynthesis was initially monitored qualitatively by mass spectrometry. Next, production of Ybt was quantified in a high-cell-density fermentation environment with titers reaching 67 ± 21 (mean ± standard deviation) mg/liter and a volumetric productivity of 1.1 ± 0.3 mg/liter-h. This success has implications for basic and applied studies on Ybt biosynthesis and also, more generally, for future production of polyketide, nonribosomal peptide, and mixed polyketide-nonribosomal peptide natural products using E. coli.
Polyketides and nonribosomal peptides represent two natural product classes with tremendous therapeutic value (1). Erythromycin (a polyketide antibiotic), vancomycin (a nonribosomal peptide antibiotic), and epothilone (a mixed polyketide-nonribosomal peptide antitumor agent) represent just a few of these important natural compounds. Harnessing this medicinal relevance, however, is often limited by the rudimentary knowledge or poor growth and culturability profiles associated with the original microbial hosts. In addition, the complex structure of these compounds limits efficient synthetic production and leaves microbial fermentation as the only economical route to these products. Complex nonribosomal peptides and polyketides derive from complex or modular enzymatic systems that show the potential for controlled manipulation, given the appropriate cellular host (7). To this end, an initiative to transfer the genetic and biochemical pathways responsible for these natural products from their original microbial hosts to heterologous (and often more amenable) hosts has been implemented (12). This transfer would then offer the best of both worlds: an ideal (or significantly better) and potentially universal heterologous host for the controlled production of therapeutic natural products. However, this simple concept faces significant challenges posed by the nature of the complex natural products being produced. To generalize, the heterologous host must generate active enzymatic complexes (either a polyketide synthase [PKS] or nonribosomal peptide synthetase [NRPS]) and the substrates that these complexes use for polyketide or nonribosomal peptide biosynthesis. The individual gene size and overall gene number for most modular PKS or NRPS systems pose problems for successful and coordinated gene expression (a problem compounded by the need for posttranslational modification of PKS and NRPS via phosphopantetheinylation). In addition, the individual PKS or NRPS will need a pool of intracellular substrates to biosynthesize the respective natural product; PKS systems require small acyl coenzyme A substrates, while NRPS complexes use both proteinogenic and nonproteinogenic amino acids. These substrates may or may not be native to the heterologous host. Recently, the use of Escherichia coli for the heterologous production of complex polyketide natural products has been described (11). E. coli offers many advantages as a heterologous host, and the successful production of a complex polyketide opens future biosynthetic potential for other polyketide products with this “user-friendly” organism. There also exists the potential to use this heterologous host to generate other complex natural products such as nonribosomal peptides or mixed polyketide-nonribosomal peptides.
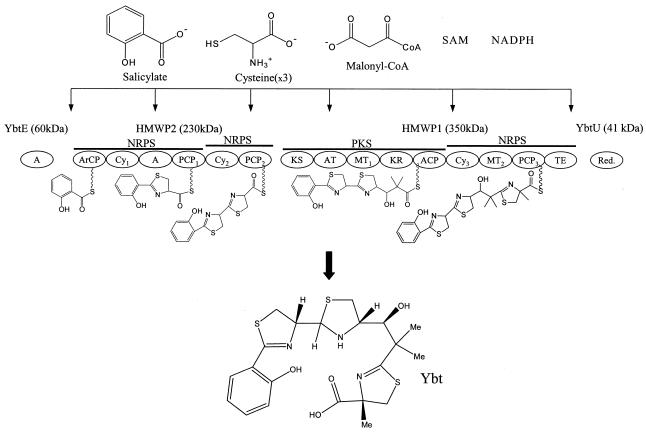
A candidate system to test the heterologous potential of E. coli further is that for yersiniabactin (Ybt), a siderophore naturally produced by Yersinia pestis (the causative agent for bubonic plague) during times of iron starvation (9). Ybt is biosynthesized intracellularly by the modular Ybt synthetase composed of four primary enzymes that were recently used to reconstitute Ybt production in vitro (8) (Fig. 1). Initially, YbtE adenylates a salicylate unit for addition to the second biosynthesis enzyme, HMWP2. HMWP2 consists of two multidomain NRPS modules; as the first module accepts the activated salicylate unit through an aryl carrier protein, an adenylation domain attaches two cysteine units through thioester bonds to the first and second peptidyl carrier proteins. HMWP2 then catalyzes the successive addition and cyclization of each cysteine unit as the growing Ybt chain is then passed to HMWP1. HMWP1 contains both a polyketide and nonribosomal peptide module. The ketosynthase domain initially accepts the partial Ybt chain from HMWP2. The PKS portion of HMWP1 then catalyzes the addition of a malonate unit (preloaded onto the acyl carrier protein through the acyltransferase domain), and the resultant enzyme-tethered product is reduced (through an NADPH-dependent ketoreductase domain) and methylated (through the first methyltransferase domain using S-adenosylmethionine as a substrate). The HMWP1 NRPS module then catalyzes the addition, cyclization, and methylation of a final cysteine unit (presumably loaded through the A domain of HMWP2) before the final thioesterase domain catalyzes the release of Ybt from HMWP1. YbtU acts as a reductase, reducing the second thiazoline ring of the growing enzyme-tethered product during biosynthesis. YbtT (not depicted in Fig. 1) represents an auxiliary enzyme thought responsible for in vivo biosynthetic editing (3). HMWP1, as indicated above, has both an NRPS function (through the addition of a cysteine unit) and a PKS activity (through the incorporation of a malonyl coenzyme A unit). Hence, Ybt is a mixed polyketide-nonribosomal peptide natural product.
FIG. 1.
The Ybt synthetase and its substrates needed to reconstitute Ybt production in E. coli. ArCP, aryl carrier protein; A, adenylation; PCP, peptidyl carrier proteins; Cy, cyclization; KS, ketosynthase; ACP, acyl carrier protein; AT, acyltransferase; KR, NADPH-dependent ketoreductase; MT, methyltransferase; SAM, S-adenosylmethionine; TE, thioesterase. See the text for details on biosynthesis.
In this work, we report the heterologous production of Ybt using E. coli and the implications for this success in both basic and applied studies. Successful production of Ybt further expands the potential to use E. coli as a route to therapeutic polyketide, nonribosomal peptide, and mixed polyketide-nonribosomal peptide products. For Ybt production, the high cell density titers achieved compare favorably to the industrial production of other heterologously produced complex natural products. A robust route to Ybt opens the possibility of using this high-affinity molecular ligand to specifically target or inhibit pathogenic microbes dependent upon Ybt for survival.
MATERIALS AND METHODS
Strains and plasmids.
E. coli strain K207-3 [BL21(DE3) ΔprpRBCD::T7 promoter-sfp-T7promoter-prpE panD::panD25A ygfG::T7 promoter-accA1-T7 promoter-pccB-T7terminator] was used for all experiments (K207-3 is a derivative of BAP1 [11]). Plasmids containing the genes for Ybt biosynthesis as well as ybtT were provided on individual expression plasmids as described previously (8). Standard molecular biology protocols were then used to couple these individual genes on multicystronic expression plasmids. pBP198 (carbenicillin resistant and derived from pET21c [Novagen, Milwaukee, Wis.]) contained (in order) HMWP2 and ybtU, with each gene under the control of its own T7 promoter. Likewise, pBP205 (kanamycin resistant and derived from pET28a) contained ybtE followed by HMWP1, with each gene under individual T7 promoters. An additional plasmid, pBP200, contained ybtT under a T7 promoter on a chloramphenicol-resistant plasmid derived from pGZ119EH (5, 10).
In vivo gene expression and biosynthesis. Strains K207-3/pBP198/pBP205 and K207-3/pBP198/pBP205/pBP200 were used for Ybt biosynthesis with all plasmids or plasmid combinations introduced to K207-3 via electroporation. All cultures used Luria-Bertani (LB) broth media and contained 100 μg of carbenicillin/ml, 50 μg of kanamycin/ml, and 34 μg of chloramphenicol/ml where needed. Cultures (typically 5 to 10 ml) were inoculated 3% (vol/vol) with a previous starter culture, and growth was carried out at 37°C on a rotary shaker (250 rpm) to an optical density at 600 nm (OD600) between 0.6 and 0.8.
Cultures were then cooled at 20°C for 10 min. At this point, 75 μM isopropyl-β-d-thiogalactopyranoside (IPTG) was added together with 1 mM salicylate. The culture was then incubated between 12 and 30 h at either 13 or 22°C on a rotary shaker (200 rpm).
For analysis, samples were centrifuged and 5 mM FeCl3 was then added to the resultant supernatant. The supernatant was then either extracted with ethyl acetate or submitted to the Stanford mass spectrometry facility for liquid chromatography-mass spectrometry (LC-MS) analysis directly. Samples extracted were done so twice with an equal volume of ethyl acetate each time. These samples were dried and also submitted for LC-MS analysis. The gradient used for the LC-MS was a linear method from 2 to 98% acetonitrile (balance water) with Ybt-Fe3+ observed by MS at ∼60% acetonitrile. The fragmentation pattern for Ybt-Fe3+ was also determined and compared to previous reports (2, 4). Finally, samples taken experimentally were compared by retention times and fragmentation patterns to an authentic sample of Ybt-Fe3+. Negative controls included K207-3/pBP198, K207-3/pBP205, and K207-3/pBP198/pBP205 without added salicylate. Cell pellets were sonicated, clarified, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to examine the extent of gene expression for both positive and negative controls.
High cell density fed-batch fermentation for yersiniabactin purification and quantification.
The fermentation procedure derives from a similar effort used for complex polyketide production (10). Briefly, the fermentation medium (termed F1 medium) contained KH2PO4 at 1.5 g/liter, K2HPO4 at 4.34 g/liter, (NH4)2SO4 at 0.4 g/liter, MgSO4 at 150.5 mg/liter, glucose at 5 g/liter, trace metal solution at 1.25 ml/liter, and vitamin solution at 1.25 ml/liter. The feed medium contained (NH4)2SO4 at 110 g/liter, MgSO4 at 3.9 g/liter, glucose at 430 g/liter; trace metal solution at 10 ml/liter; vitamin solution at 10 ml/liter. The trace metals solution consisted of FeCl3 · 6H2O at 27 g/liter, ZnCl2 · 4H2O at 2 g/liter, CaCl2 · 6H2O at 2 g/liter, Na2MoO4 · 2H2O at 2 g/liter, CuSO4 · 5H2O at 1.9 g/liter, H3BO3 at 0.5 g/liter, and concentrated HCl at 100 ml/liter. The vitamin solution consisted of riboflavin at 0.42 g/liter, pantothenic acid at 5.4 g/liter, niacin at 6 g/liter, pyridoxine at 1.4 g/liter, biotin at 0.06 g/liter, and folic acid at 0.04 g/liter.
Fed-batch aerated fermentations were conducted by use of an Applikon 3L Biobundle system (Applikon Inc., Foster City, Calif.). A starter culture of K207-3/pBP198/pBP205 was grown in 1.5 ml of LB medium (with 100 mg of carbenicillin/liter and 50 mg of kanamycin/liter). After reaching late exponential phase at 37°C and 250 rpm, the culture was centrifuged and resuspended in 50 ml of LB (at 100-mg of carbenicillin/liter and 50 mg of kanamycin/liter). The culture grew overnight at 30°C and 200 rpm to stationary phase, was centrifuged, and was resuspended in 20 ml of phosphate-buffered saline for inoculation into the 3-liter vessel containing 2 liters of F1 medium. Growth was conducted at 37°C with pH maintained throughout the experiment at 7.1 with 1 M H2SO4 and concentrated NH4OH. Aeration was maintained at 2.8 liters/min with agitation controlled at 600 to 900 rpm to maintain dissolved oxygen over 50% of air saturation. The fermentation apparatus including a salt solution [KH2PO4, K2HPO4, and (NH4)2SO4] was autoclaved, whereas the additional components (MgSO4, glucose, trace metals, and vitamins) were filter sterilized and added aseptically prior to inoculation along with carbenicillin at 150 mg/liter and kanamycin at 75 mg/liter. The feed medium was also filter sterilized. Once the glucose was exhausted from the starting medium (as indicated by a sudden decrease in the oxygen requirement of the culture), the temperature was reduced to 22°C, and IPTG (75 μM) and salicylate (0.160 g/liter) were added. At that point a peristaltic pump started to deliver 0.1 ml of the feed medium/min, and samples were typically taken twice daily thereafter.
Initially, a final fermentation broth was extracted with ethyl acetate (2 liters, twice). The extract was dried, resuspended in water and 10% acetonitrile, and loaded onto a preparatory high-performance liquid chromatography (HPLC) instrument. A 10 to 100% acetonitrile (balance water) gradient at an 8-ml/min flow rate was used to obtain pure Ybt (isolated as an Fe3+ chelate). Due to Ybt acid sensitivity, no trifluoroacetic acid was used in the HPLC purification process.
The Ybt fractions eluted at 75% acetonitrile, and these fractions were pooled, dried, and confirmed to contain Ybt-Fe3+ by MS. The remaining purified product was resuspended in water and quantified at 385 nm by using the known extinction coefficient (ɛ = 2884) for Ybt-Fe3+ (2). This initial purified batch of Ybt-Fe3+ was then used to generate a calibration curve to quantify production from subsequent fermentation time point samples that were first clarified before directly loading the fermentation broth onto a preparatory HPLC instrument for analysis.
RESULTS
Gene expression and biosynthesis.
Based upon earlier polyketide biosynthetic schemes, the potential to produce Ybt in E. coli was evaluated. The Ybt synthetase genes were introduced into E. coli strain K207-3 by using the multicystronic plasmids previously developed (11). Initially, gene expression was analyzed at 13 or 22°C based on previous work using similar postinduction temperatures (8, 11). Qualitatively, the large gene products were more abundant at 22°C (data not shown), and this postinduction temperature was subsequently used.
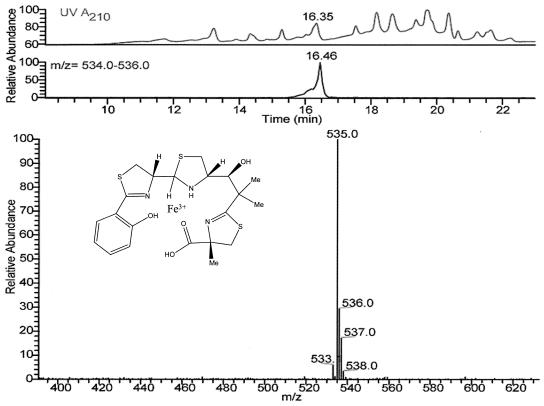
After successful gene expression was confirmed, in vivo experiments commenced for Ybt biosynthesis. The main difference between these and previous experiments was the addition of salicylate upon induction with IPTG. The rest of the Ybt synthetase substrates were presumed to be native to E. coli. Isolation efforts indicated the production of Ybt found to be chelated to Fe3+. This compound was analyzed by LC-MS (Fig. 2) with LC retention times nearly identical to those indicated in previous reports (2, 4) and an authentic Ybt-Fe3+ standard. Furthermore, MS and MS fragmentation patterns for the proposed Ybt-Fe3+ compound also matched those of previous reports and the authentic standard. Relative comparisons showed increased Ybt production at 22°C compared to that observed at 13°C. Production could be eliminated by excluding either of the two expression plasmids needed to reconstitute the Ybt pathway or by omitting the addition of salicylate to cultures that had been induced for gene expression.
FIG. 2.
LC-MS analysis of Ybt from K207-3/pBP198/pBP205. The top LC trace shows the Ybt-Fe3+ compound eluting at 16.35 min as analyzed by UV at A210. The bottom trace presents the elution profile for compounds within the mass range of 535 to 536 (molecular weight of Ybt-Fe3+, 535). Finally, the MS pattern for the Ybt-Fe3+ peak is presented.
Finally, the addition of YbtT, an enzyme thought to edit the biosynthetic production of yersiniabactin, resulted in increased levels of Ybt. Again, based on relative comparisons, the inclusion of YbtT increased yersiniabactin production ∼2.5 times over that observed for strains without this enzyme.
High-cell-density yersiniabactin production and quantification.
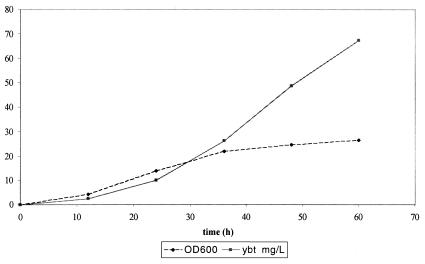
A high-cell-density fermentation was performed to generate Ybt in sufficient quantities needed to readily quantify cellular productivity and overall titers. The fed-batch fermentation was initially used as a way to obtain pure Ybt. Upon completion of the fermentation run, Ybt was purified from the fermentation broth by using a combination of organic phase extraction and preparatory HPLC. The purified product was then quantified at 385 nm by using the known extinction coefficient (2). This purified Ybt then served as a standard to quantify the final titers for subsequent fermentations. Figure 3 shows the results for a typical fermentation run. Final titers of 67 ± 21 mg/liter were reached; the maximum specific productivity of the recombinant E. coli strain was 1.2 ± 0.3 mg/liter-h. Both parameters compare well to the biosynthesis of 6-deoxyerythronolide B in the same host (final 6-deoxyerythronolide B titer, 95.2 ± 7.7 mg/liter; maximum 6-deoxyerythronolide B specific productivity, 1.1 ± 0.007 mg/liter-h) (10).
FIG. 3.
Representative fed-batch fermentation for Ybt production. The time courses of OD600 and Ybt titers for strain K207-3/pBP198/pBP205 are shown. pBP198 (carbenicillin resistant) contained HMWP2 and ybtU, whereas pBP205 (kanamycin resistant) held ybtE and HMWP1. Together, pBP198 and pBP205 expressed the needed genes for complete Ybt reconstitution within the cellular environment of K207-3, a strain genetically engineered to support biosynthesis.
DISCUSSION
The development of E. coli-derived Ybt marks an important step forward for heterologously produced complex nonribosomal peptide products. Although E. coli has the ability to generate simple nonribosomal peptide products (for example, the native E. coli siderophore enterobactin), Ybt biosynthesis represents an advance due to the complex, modular nature of the Ybt biosynthetic pathway. This biochemical complexity is a hallmark for many important polyketide and nonribosomal peptides (including those examples cited at the beginning of this report), and the present example of heterologous production through E. coli helps support the notion that this host has the capacity to support production of both natural product classes. In addition, hybrid polyketide-nonribosomal peptide natural products like Ybt should also then be viable options for heterologous production using E. coli.
Complex natural product heterologous biosynthesis requires both an active PKS or NRPS and intracellular substrates required by these enzymes to catalyze their respective products. For Ybt, the yersiniabactin synthetase enzyme complex (containing two large, modular proteins in HMWP1 and HMWP2) was introduced with multicistronic expression plasmids that facilitated the coordinate expression of all the needed biosynthetic genes upon induction with IPTG. Induction by IPTG also called for the expression of a chromosomally located sfp gene needed to activate the biosynthetic enzymes through posttranslational modification. (The sfp gene encodes an Sfp phosphopantetheinyltransferase that was transferred to K207-3 from Bacillus subtilus to facilitate complex natural product biosynthesis [11].) Unlike most complex polyketide systems, many of the substrates needed for Ybt biosynthesis are native to E. coli. The exception was a salicylate unit used to initiate biosynthesis. To overcome this, salicylate was simply fed to the media after gene induction. Thus, for biosynthetic substrates that are not native to E. coli or that cannot be metabolically engineered for intracellular production, exogenous substrate feeding is yet another option.
This study also indicated the enhanced effect that YbtT has on in vivo Ybt production. YbtT is thought to act as an auxiliary thioesterase enzyme that edits the biosynthetic process, perhaps by removing “misprimed” units that have been incorrectly attached to the Ybt synthetase complex. In these experiments, YbtT increased Ybt production ∼2.5 times. This result supports previous observations that YbtT increases Ybt production in vivo (3). Interestingly, analogous polyketide systems employing a YbtT analog also show an approximately twofold increase in in vivo polyketide production (10).
Finally, the biosynthetic capabilities of this Ybt system were expanded by using the high-cell-density capabilities of E. coli in the context of a fed-batch fermentation. Using this route produced Ybt at levels comparable to those of heterologous host systems that produce the macrolide 6-deoxyerythronolide B (6, 10) and highlights the potential for future heterologous (and therapeutic) polyketide, nonribosomal peptide, or mixed polyketide-nonribosomal peptide product biosynthesis in E. coli. Moreover, given the role of Ybt in establishing virulence for pathogenic microbes, its heterologous production (from a nonpathogenic organism) provides a source of this compound for inhibitor design or targeted drug delivery applications. For example, Ybt analogs, generated through manipulation of the modular Ybt synthetase, or Ybt-drug conjugates might present unique and high-affinity ways to inhibit and target pathogens (such as Yersinia species or other pathogens [13]) that have a distinct dependence on Ybt for survival.
Acknowledgments
We thank Sumati Murli at Kosan Biosciences for providing E. coli K207-3.
Research in the authors' laboratories was supported by grants from the NIH (GM067937 to C.K. and AI 42708 to C.T.W.). B.A.P. and C.C.C.W. were recipients of an ARCS predoctoral fellowship and an NIH postdoctoral fellowship, respectively.
REFERENCES
- 1.Cane, D. E., and C. T. Walsh. 1999. The parallel and convergent universe of polyketide synthases and nonribosomal peptide synthetases. Chem. Biol. 6:R319-R325. [DOI] [PubMed] [Google Scholar]
- 2.Drechsel, H., H. Stephan, R. Lotz, H. Haag, H. Zahner, K. Hantke, and G. Jung. 1995. Structural elucidation of yersiniabactin, a siderophore from highly virulent Yersinia strains. Liebigs Ann. 10:1727-1733. [Google Scholar]
- 3.Geoffroy, V. A., J. D. Fetherston, and R. D. Perry. 2000. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect. Immun. 68:4452-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haag, H., K. Hantke, H. Drechsel, I. Stojiljkovic, G. Jung, and H. Zahner. 1993. Purification of yersiniabactin: a siderophore and possible virulence factor of Yersinia enterocolitica. J. Gen. Microbiol. 139:2159-2165. [DOI] [PubMed] [Google Scholar]
- 5.Lessl, M., D. Balzer, R. Lurz, V. L. Waters, D. G. Guiney, and E. Lanka. 1992. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 174:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombo, F., B. Pfeifer, T. Leaf, S. Ou, Y. S. Kim, D. E. Cane, P. Licari, and C. Khosla. 2001. Enhancing the atom economy of polyketide biosynthetic processes through metabolic engineering. Biotechnol. Prog. 17:612-617. [DOI] [PubMed] [Google Scholar]
- 7.McDaniel, R., A. Thamchaipenet, C. Gustafsson, H. Fu, M. Betlach, and G. Ashley. 1999. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel “unnatural” natural products. Proc. Natl. Acad. Sci. USA 96:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller, D. A., L. Luo, N. Hillson, T. A. Keating, and C. T. Walsh. 2002. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem. Biol. 9:333-344. [DOI] [PubMed] [Google Scholar]
- 9.Perry, R. D., P. B. Balbo, H. A. Jones, J. D. Fetherston, and E. DeMoll. 1999. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 145:1181-1190. [DOI] [PubMed] [Google Scholar]
- 10.Pfeifer, B., Z. Hu, P. Licari, and C. Khosla. 2002. Process and metabolic strategies for improved production of Escherichia coli-derived 6-deoxyerythronolide B. Appl. Environ. Microbiol. 68:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeifer, B. A., S. J. Admiraal, H. Gramajo, D. E. Cane, and C. Khosla. 2001. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291:1790-1792. [DOI] [PubMed] [Google Scholar]
- 12.Pfeifer, B. A., and C. Khosla. 2001. Biosynthesis of polyketides in heterologous hosts. Microbiol. Mol. Biol. Rev. 65:106-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]