Abstract
The cyanobacterium Microcystis sp. frequently develops water blooms consisting of organisms with different genotypes that either produce or lack the hepatotoxin microcystin. In order to monitor the development of microcystin (mcy) genotypes during the seasonal cycle of the total population, mcy genotypes were quantified by means of real-time PCR in Lake Wannsee (Berlin, Germany) from June 1999 to October 2000. Standard curves were established by relating cell concentrations to the threshold cycle (the PCR cycle number at which the fluorescence passes a set threshold level) determined by the Taq nuclease assay (TNA) for two gene regions, the intergenic spacer region within the phycocyanin (PC) operon to quantify the total population and the mcyB gene, which is indicative of microcystin synthesis. In laboratory batch cultures, the cell numbers inferred from the standard curve by TNA correlated significantly with the microscopically determined cell numbers on a logarithmic scale. The TNA analysis of 10 strains revealed identical amplification efficiencies for both genes. In the field, the proportion of mcy genotypes made up the smaller part of the PC genotypes, ranging from 1 to 38%. The number of mcyB genotypes was one-to-one related to the number of PC genotypes, and parallel relationships between cell numbers estimated via the inverted microscope technique and TNA were found for both genes. It is concluded that the mean proportion of microcystin genotypes is stable from winter to summer and that Microcystis cell numbers could be used to infer the mean proportion of mcy genotypes in Lake Wannsee.
Water blooms of the cyanobacterium Microcystis sp. have frequently been found to contain the toxic heptapeptide microcystin. In recent decades, environmental problems associated with microcystin in water have been documented (5). It has long been known that microcystin-producing and non-microcystin-producing strains can be isolated from one water sample, and the waxing and waning of microcystin-producing versus non-microcystin-producing strains has been suggested as a most important factor regulating net microcystin production in water (26). However, the quantification of microcystin-producers versus non-microcystin-producers was for a long time impeded because of the morphological similarity between strains differing in microcystin production. The genes involved in microcystin synthesis have been identified and sequenced (8, 28). On this basis, attempts have been made to study the occurrence of microcystin genotypes directly in the field (14, 15). The first approach consisted of the isolation of individual colonies, subsequent morphological characterization, and PCR analysis to detect the microcystin genes (14). This resulted in significant differences in mcyB distribution between morphologically defined species (as described in reference 11); i.e., 73% of the colonies assigned to Microcystis aeruginosa contained mcyB, while only 16% of the colonies assigned to Microcystis ichthyoblabe and none of the Microcystis wesenbergii colonies showed the mcyB gene. In a second attempt, the proportion of microcystin genotypes was quantified in different colony size classes by the ratio of the number of PCR products of the microcystin gene and the number of products of a reference gene obtained for a dilution series of a DNA extract (15). The second approach revealed a more general significant correlation of the frequency of the microcystin gene in regard to colony size, i.e., the largest colonies (>100 μm) had a 10-fold-higher percentage of microcystin genotypes than the smallest colonies (<50 μm). Both approaches were useful to show that there are significant correlations between the frequency of the microcystin genes and morphological characteristics. However, both approaches were time-consuming and were unable to quantify genotypes in absolute terms, i.e., cells of microcystin genotypes in a given volume of lake water.
Recently, the TaqMan PCR, or the Taq nuclease assay (TNA), was introduced to quantify specific genotypes of picocyanobacteria (1) or microcystin-producing cyanobacteria in the field (10). This technique utilizes a sequence specific dually labeled fluorescent probe (TaqMan probe) and primers to quantify the level of DNA template initially present in a sample. The rate of exponential accumulation of the amplicon is monitored by the hydrolysis of the TaqMan probe, in which it generates a fluorescent signal during the amplification process. The threshold cycle (Ct) is the PCR cycle number at which the fluorescence passes a set threshold level and can be used to determine the starting DNA amount in the sample based on a standard curve (based on samples with a known concentration).
The usefulness of the TNA in microbial ecology could be constrained by two factors. Firstly, the DNA content of a specific organism is dependent upon the growth rate which has been shown to be maximal under the highest-growth-rate conditions (16). Consequently, physiological conditions may influence the result on the quantification of genotypes and calibration is usually done based on DNA concentration or genome copy number. However, for population ecology the quantification in terms of cell numbers would be most useful. Secondly, due to the linear-log calibration curves, the technique is very sensitive to variations in the slope induced by minor variations of the Cts. It is not known whether spatial or temporal variations in genotype abundance in nature can be reliably monitored in spite of the noise induced by the semilogarithmic calibration algorithm alone.
It was the aim of the study to develop a quantitative PCR approach useful for quantifying the total population of Microcystis sp. (as defined in reference 20) as well as the subpopulation comprising all microcystin genotypes in terms of cell numbers and for monitoring the seasonal development of microcystin genotypes in Lake Wannsee (Berlin, Germany). We developed two independent TNAs, one to quantify the total population of Microcystis sp. using the intergenic spacer region within the phycocyanin (PC) operon and the other assay to quantify microcystin genotypes using a region of mcyB part of the microcystin synthetase gene cluster (28). To validate the results of the TNA, (i) strains were quantified in batch culture during the growth cycle and compared in amplification efficiency in the laboratory, and (ii) cell numbers estimated by the TNA in the field were compared to cell numbers estimated by counting under an inverted microscope.
MATERIALS AND METHODS
Cultivation of organisms and determination of cell numbers.
Several unicellular strains of Microcystis sp. were chosen to test primer and TaqMan probe sensitivity and specificity, as well as to compare individual strains in amplification efficiency (Table 1). The strain comparison was repeated once. All strains were grown nonaxenically under sterile conditions in batch culture in a medium as described by Zehnder and Gorham (32) and harvested during the stationary phase. Cells were harvested by filtering on glass fiber filters (GF/C; Whatman, Kent, Great Britain) under vacuum pressure and stored frozen (−20°C) until DNA extraction. Aliquots were analyzed for cell numbers by two independent methods, electronic particle counting (Casy 1; Schärfe System, Reutlingen, Germany) and counting of cells by the inverted microscope technique (29). Not all strains were found to sediment consistently in sedimentation chambers. Consequently, for comparing cell numbers between individual strains, cells were filtered onto 0.2-μm-pore-size polycarbonate filters (Millipore, Cork, Ireland) and counted by using autofluorescence. At least 400 specimens of Microcystis were counted at a magnification of ×400, and the results were averaged from at least two transects per sedimentation chamber or filter.
TABLE 1.
Microcystis species and strains used to test primer and TaqMan probe sensitivity and specificity (HUB524), to monitor DNA content over the growth cycle (HUB524), and to compare amplification efficiencies of individual strains
| Organism | Isolation date | Geographic origin | mcyB |
|---|---|---|---|
| HUB524a | 1977 | Pehlitzsee, Brandenburg, Germany | + |
| PCC7806 | 1972 | Braakman Reservoir, The Netherlands | + |
| W368a | 1995 | Wannsee | + |
| W334a | 1995 | Wannsee | + |
| M. aeruginosab | 1982 | Neusiedler See, Austria | + |
| HUB53a | 1977 | Pehlitzsee | − |
| W75a | 1995 | Wannsee | − |
| M. flos-aquaeb | 1982 | Neusiedler See | − |
| W61a | 1995 | Wannsee | − |
| P461a | 1997 | Pehlitzsee | − |
Kindly provided by M. Henning, Humboldt University, Berlin, Germany.
Kindly provided by E. Kusel-Fetzmann, University of Vienna, Vienna, Austria.
To monitor the cellular DNA content during the growth cycle, 2,000 cells of strain HUB524 ml−1 were inoculated and grown under high-light conditions (80 to 90 μmol m−2 s−1; Osram Lumilux Plus Eco L36W/11-860) with a 16-h-8-h light-dark cycle at 20°C for 4 months. Cells were quantified and harvested every third day during the first month and monthly during the rest of the growth phase. Depending on the cell number during the growth cycle, the volume filtered ranged between 2,000 and 4 ml and was adjusted to achieve a cell number of 8.3 × 107 on each filter. The cellular DNA content was assessed twice during the growth cycle. The specific growth rate (μ day−1) was calculated from the following: μ = (ln N2 − ln N1)/(t2 − t1), where N1 and N2 are number of cells per milliliter on two consecutive sampling dates (t1 and t2).
DNA extraction and TNA.
Extraction from frozen filters was performed as described previously (15). The TNA was used to quantify two specific gene regions, the intergenic spacer region within the PC operon and the mcyB region, which encodes one step in microcystin biosynthesis (28). A variable gene region of the PC operon was selected based on an alignment (ClustalW 1.8) of PC genes from several genera of cyanobacteria from GenBank, including Microcystis (AJ003168, AF385388; R. Kurmayer, unpublished data), Planktothrix (AJ132279 and AJ131820), Oscillatoria (AJ401186 and AJ401185), Nostoc (X05239), Synechocystis (AJ003180), Chroococcus (AJ003189), Synechococcus (AF223465, AF223464, and AF223463), and Lyngbya (AJ401187). The variability of the intergenic spacer region of the PC gene was sufficiently high to achieve specificity for amplification of the PC operon of Microcystis even in the presence of other cyanobacteria (e.g., Planktothrix; see below). The mcyB gene region was selected from Microcystis strain HUB524 (Z28338) and was located between core motifs A2 and A3 (17). From those gene regions, optimal primers and TaqMan probes were designed with Primer Express 2.0 software (Applied Biosystems, Vienna, Austria). The primers were 188F (5′-GCTACTTCGACCGCGCC-3′) and 254R (5′-TCCTACGGTTTAATTGAGACTAGCC-3′) for the PC operon and 30F 5′-CCTACCGAGCGCTTGGG-3′ and 108R (5′-GAAAATCCCCTAAAGATTCCTGAGT-3′) for mcyB, with amplicon sizes of 66 and 78 bp, respectively. The TaqMan probes were 5′-CCGCTGCTGTCGCCTAGTCCCTG-3′ for the PC operon and 5′-CACCAAAGAAACACCCGAATCTGAGAGG-3′ for mcyB. These probes each had a fluorescent reporter dye (6-carboxyfluorescein) covalently attached to the 5′ end (5′-FAM) and a 3′-TAMRA fluorescent quencher dye (6-carboxytetramethylrhodamine). PCR was initiated with two holds, one for 2 min at 50°C (AmpErase UNG protection against carryover contamination) and one for 10 min at 95°C. Subsequently, a 95°C denaturation step for 15 s was followed by a 60°C annealing and extension step for 1 min, for 45 cycles. PCRs were performed with a GeneAmp 5700 sequence detection system (ABI, Vienna, Austria) using SDS 1.3 software. TNAs were performed with a volume of 25 μl, containing 12.5 μl of 2× TaqMan universal PCR master mix (ABI), a 300 nM (300 fmol μl−1) concentration of each primer, a 100 nM concentration of the TaqMan probe, and 5 μl of template containing various amounts of genomic DNA and filled up to 25 μl with sterile Millipore water. For mcyB, a 900 nM concentration of each primer and a 250 nM concentration of the TaqMan probe were used. Each measurement was performed in triplicate.
Specificity and sensitivity of the TNA.
For both genes (the PC operon and mcyB), a standard curve based on predetermined cell concentrations was established by relating the known DNA concentrations (in cell equivalents) to the Ct of the diluted samples. The threshold value for the fluorescence of all samples was set manually at 0.1 in accordance with the instruction manual of the GeneAmp 5700 sequence detection system. Five milliliters of a suspension of 2.08 × 107 cells of HUB524 ml−1 (determined by electronic particle counting) were filtered on a GF/C filter, and from the DNA extract, six dilutions ranging from 1:102 to 1:2 × 106 of template DNA (equivalent to 104,000 cells to 5.2 cells) were prepared. To test the specificity of both TNAs in the presence of a highly diversified DNA background from a natural phytoplankton community, water from Lake Wannsee was filtered on 4 May 2000 through a sieve (25-μm mesh size), selectively removing Microcystis but permitting other cyanobacteria to pass through the filter. Three hundred milliliters of the filtrate was filtered on a GF/C filter, and the DNA was extracted and added to the dilutions of DNA obtained from Microcystis strain HUB524. The counting of cyanobacteria under an inverted microscope (3.2-ml sedimentation chamber) revealed the dominance of filamentous cyanobacteria (Aphanizomenon spp., Limnothrix spp., Limnothrix redekei, and Planktothrix agardhii) but no Microcystis (see reference 15 for details on counting and biovolume determination). The dilutions 1:100 (DNA equivalent to a cyanobacterial biovolume of 6.6 × 10−4 mm3 in the template) and 1:1,000 (6.6 × 10−5 mm3) were used, and the maximum and minimum ratios of biovolume of cells of HUB524 to the background biovolume were 66 (at 100,000 cells, 1:1,000) and 6.6 × 10−4 (at 10 cells, 1:100), respectively (calculated with an average Microcystis cell biovolume of 42.2 μm3). To compare background effects, the number of cells estimated in the presence of a natural background was divided by the number estimated in the absence of a natural background, and the ratio of cells with background to cells without background was calculated.
Sample collection from Lake Wannsee.
Sampling was performed at Lake Wannsee (Berlin, Germany) from June 1999 to October 2000. The lake is shallow (mean depth, 5.5 m; maximum depth, 8.5 m; area, 2.82 km2), polymictic, hypertrophic, and regularly dominated by Microcystis sp. during the summer. The phytoplankton was sampled weekly (May to October) and fortnightly (November to April). Water samples were obtained at the deepest part of the lake and integrated into a 30-liter bucket by collecting 2 liters every meter from the surface to the sediment. Cells were harvested and extracted as described above. The cell number and the biovolume were determined by counting via the inverted-microscope technique. Details on counting procedures and ambient abiotic factors are provided in reference 15.
Statistical analyses.
The accuracy and the statistical significance of the measurements obtained by TNA were tested by regressing the TNA variables (Ct or cell numbers) against the respective cell numbers determined via the electronic particle counter or the microscope (as an independent variable). For linear regression analysis, the data were log transformed and tested for normal distribution (Kolmogorov-Smirnov test, P < 0.01) and for constant variance of the dependent variable regardless of the value of the independent variable (Spearman rank correlation, P < 0.01). The residuals were tested for their independence of each other (Durbin-Watson statistic; if the residuals are not correlated, the value will be 2 ± 0.5). All data passed those tests; however, the errors in field data were serially correlated (Durbin-Watson statistic = 1.1 [PC operon] and 1.07 [mcyB]). Consequently, the interval of field observations was increased and every third sampling date was omitted from statistical analysis to achieve the assumption that the error term is a random normal variable. The linear curves were fitted by using the least-square approximation and the associated statistical tests of Sigma Plot 2000 (version 6.10). For the field analysis, the two linear regressions between cell numbers as determined by microscopy and by TNA were compared in slope and intercept using a general factorial model of analysis of variance (ANOVA). The data were modeled as follows: yijz = μ + αj + βz + (αβ)jz + ɛijz, where yijz is the cell number measured by TNA, μ is the overall mean level, αj is the effect of the cell number in the microscope, βz is the effect of the grouping factor as a covariate (PC operon and mcyB), (αβ)jz is the interaction between both factors, and ɛijz is the unexplained part of the variance (27). An SPSS statistical package (release 6.0) was used for ANOVA.
RESULTS
Sensitivity and specificity of the TNA.
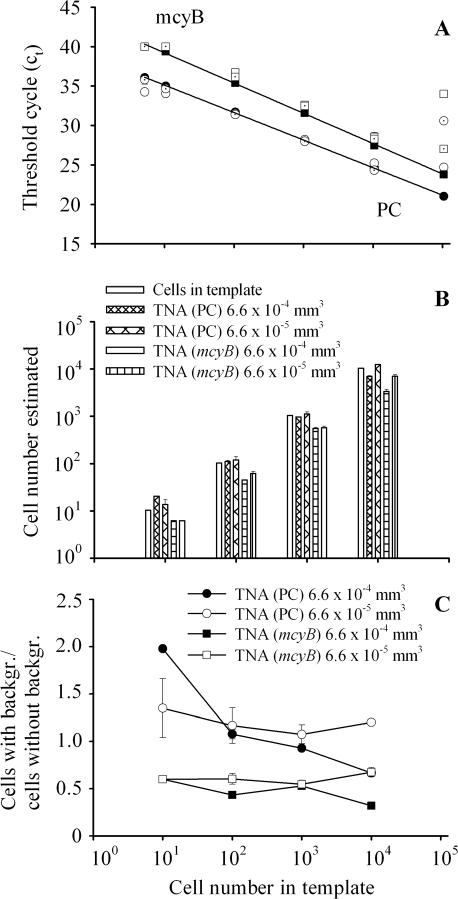
For both the PC operon and mcyB, highly significant linear curves between the amount of starting DNA (in cell numbers) and the Ct were obtained (Fig. 1A). For the PC operon and mcyB, the regression equations were y = 38.61 − 3.49x (R2 = 0.99, n = 6, P < 0.0001) and y = 43.05 − 3.84x (R2 = 0.99, n = 6, P < 0.0001), respectively, where y is the Ct at the set fluorescence threshold level (0.1) and x is the amount of starting DNA (given as log10 cell number equivalents). For both genes the detection limit was less than five cells. Generally, the variation in Cts in the presence of both dilutions of the natural background was small within the central region of the standard curves (100 to 10,000 cells) but more pronounced towards higher (100,000 cells) or lower (<10 cells) cell numbers. Consequently, only cell numbers from 10 to 10,000 corresponding to the amount of starting DNA were considered in further analysis. For both genes and for both dilutions of the natural background, the cell number estimated via TNA correlated perfectly with the cell number initially present in the template on a log10 scale (Fig. 1B). On a linear scale (Fig. 1C), the presence of 6.6 × 10−4 mm3 of the natural background (1:100) resulted in a twofold overestimation of the lowest cell number (10 cells) for the PC operon and gradually shifted to a 0.7-fold underestimation of the highest cell number at 10,000. The influence of the 1:1,000 dilution (6.6 × 10−5 mm3) resulted in a 1.35- to 1.07-fold overestimation of cell numbers. In contrast, both dilutions of the same background consistently shifted the Cts of mcyB upwards and thus underestimated the number of cells in the initial template by a factor of 0.32 to 0.67. This implies that background effects can distort the mcyB results by a factor of 0.55 ± 0.2 (95% confidence limit [CL]). For field analysis, cell quantification of PC genotypes was directed towards the central region of the standard curves (1,000 cells) which were found to be most resistant against background effects.
FIG. 1.
(A) Standard curves (black symbols) for both the PC operon (circles) and mcyB (squares) based on predetermined cell concentrations of Microcystis HUB524 by relating the known DNA concentrations (in cell equivalents) to the Ct of the diluted samples. In addition, both TNAs were tested in the presence of a 1:100 dilution (6.6 × 10−4 mm3, white symbols) and a 1:1,000 dilution (6.6 × 10−5 mm3, dotted symbols) of a natural background (containing other cyanobacteria but no Microcystis) from Lake Wannsee. All data are means of three parallels ± 1 standard error (error bars that are not visible are hidden behind the symbols; however, error bars have been omitted for outliers shown at 105 cells). (B) Comparison between cell numbers estimated for both genes by TNA from the standard curve in the absence (white columns) or in the presence of 6.6 × 10−4 mm3 and 6.6 × 10−5 mm3 of background. (C) Number of cells estimated in the presence of a natural background divided by the cell number estimated in the absence of a natural background for both the PC operon (circles) and mcyB (squares) and both background dilutions (6.6 × 10−4 mm3 [black symbols] and 6.6 × 10−5 mm3 [white symbols]).
Variation in quantification of cell numbers within a single Microcystis strain and among different strains.
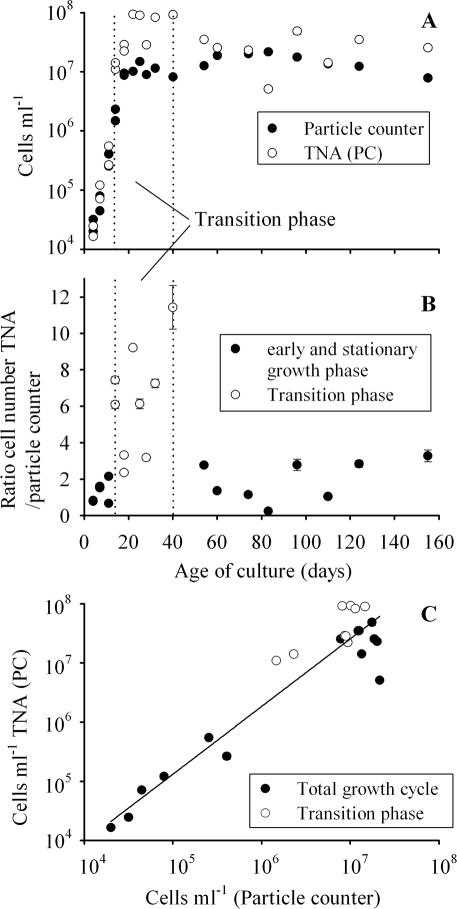
During the growth cycle monitored in batch culture, the cell numbers determined with the electronic particle counter increased from 3,000 to 20,000,000 cells ml−1 at day 80 and started to decline afterwards. The color of the culture gradually changed from green at the beginning to yellow at the end of the experiment. The maximum specific growth rate (μ day−1) was observed from day 11 to day 14 (μ = 0.58 ± 0.003 [standard deviation]). Cell numbers determined with a particle counter correlated with cell numbers determined with a microscope, following the equation y = 0.62 + 0.93x (R2 = 0.99, n = 22, P < 0.0001), where y is log10 cells per milliliter (electronic particle counter) and x is log10 cells per milliliter (inverted microscope). The cell numbers determined by the electronic particle counter correlated significantly with the cell numbers inferred from the standard curve of the PC operon TNA on a logarithmic scale (Fig. 2A and C). For the PC operon and mcyB, the regression equations were y = −0.46 + 1.13x (R2 = 0.89, n = 23, P < 0.0001) and y = −0.02 + 1.04x (R2 = 0.87, n = 23, P < 0.0001), respectively, where y is the log10 cell number determined by the TNA and x is the log10 cell number determined by the particle counter. The most pronounced overestimation of cell numbers by the TNA by a factor of about 10 occurred during the transition from the logarithmic growth phase to the stationary phase (Fig. 2B). For both the PC operon and mcyB, the ratio of cell numbers determined by TNA to cell numbers determined with the particle counter was significantly higher from day 14 to day 40 (25th percentile, median, and 75th percentile were 3.3, 6.1, and 7.9 [PC operon] and 2.5, 4.0, and 7.3 [mcyB]; n = 9) than ratios from earlier and later growth phases (25th percentile, median, and 75th percentile were 0.8, 1.4, and 2.8 [PC operon] and 0.6, 1.0, and 2.0 [mcyB]; n = 14; Mann-Whitney rank sum test, P < 0.001).
FIG. 2.
(A) Cell numbers of Microcystis HUB524 grown under high-light conditions in batch culture and quantified via electronic particle counting and TNA over 4 months (mean ± 1 standard error). Data are from two independent batch culture experiments. For reasons of clarity, only the cell numbers quantified with the PC gene are shown; for details on statistical correlation between cell numbers and both the PC gene and mcyB, see the text. The region within the dotted lines is considered the transition phase from logarithmic growth to the stationary phase. (B) Ratio of cell numbers determined by TNA to cell numbers determined by the particle counter during the same batch culture experiment (mean ± 1 standard error). The transition from the exponential growth phase to the stationary phase differed significantly (P < 0.001, Mann-Whitney rank sum test) from the earlier and later growth phases. (C) Regression of cell numbers (mean ± 1 standard error) determined by the TNA (PC operon) versus cell numbers determined by the electronic particle counter during the same batch culture experiment. Error bars that are not visible are hidden behind the symbols.
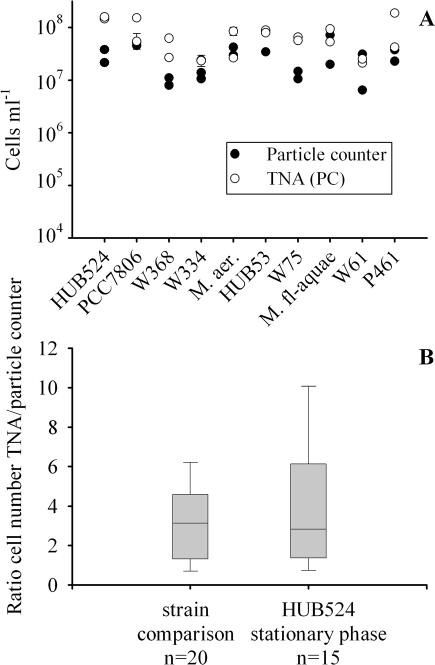
All strains showed cell numbers comparable to those of strain HUB524 when harvested during the stationary phase from day 18 onward (compare Fig. 2A with Fig. 3A). Cell numbers determined by the particle counter correlated with cell numbers determined with the microscope following the equation y = 2.30 + 0.71x (R2 = 0.59, n = 20, P < 0.0001), where y is log10 cells ml−1 (electronic particle counter) and x is log10 cells ml−1 (autofluorescence). All strains were quantified by TNA using the standard curve for the PC operon, and in general the cell numbers were overestimated two- to sixfold (Fig. 3B). Compared to the range of cell number ratios of strain HUB524 during the stationary phase (3.9 ± 1.8 [95% CL], n = 15), the cell number ratios of the strain comparison (3.1 ± 0.9, n = 20) did not differ significantly (paired t test, t = 0.87, df = 35, P = 0.39). It is concluded that the variability in cell quantification found between strains is identical to the variability found with strain HUB524 during the stationary phase and that the variability was induced by physiological differences analogous to those found with strain HUB524. It is noteworthy that all microcystin-containing strains showed a stable ratio of the PCR cycle number set at the threshold level for the PC operon to the PCR cycle number set at the threshold level for mcyB: for strains HUB524 and PCC7806 the ratio was 0.90 ± 0.02 (95% CL), for strain W368 the ratio was 0.90 ± 0.04, for strain W334 the ratio was 0.90 ± 0.01, and for M. aeruginosa the ratio was 0.89 ± 0.01. It is concluded that all microcystin-containing strains were quantified by the TNA with mcyB with equal amplification efficiencies.
FIG. 3.
(A) Cell numbers of 10 Microcystis strains grown under high-light conditions in batch culture and quantified by the electronic particle counter and by the TNA (mean ± 1 standard error). Data are from two independent parallels. (B) Ratios of cell numbers determined by TNA to cell numbers determined by the particle counter observed among strains and during the stationary phase of strain HUB524. The boxes show the median (line) and the 25th and the 75th percentiles, and the whiskers indicate the 5th and the 95th percentiles.
Microcystin genotype proportion in Lake Wannsee.
For mcyB, two TaqMan probes with identical sequences were synthesized. For unknown reasons, the second TaqMan probe (VBC, Vienna, Austria) which was used for field analysis had a lower sensitivity than the previously synthesized TaqMan probe (ABI). For five dilutions ranging from 10 to 100,000 cells of template DNA of strain HUB524, the mean difference in Ct between the two probes was 2.08 ± 0.24 (95% CL, n = 10). The new regression equation was y = 46.14 − 4.07x (R2 = 0.99, n = 5, P < 0.0001), where y is the Ct at the set fluorescence threshold level (0.1) and x is the amount of starting DNA (given as log10 cell number equivalents).
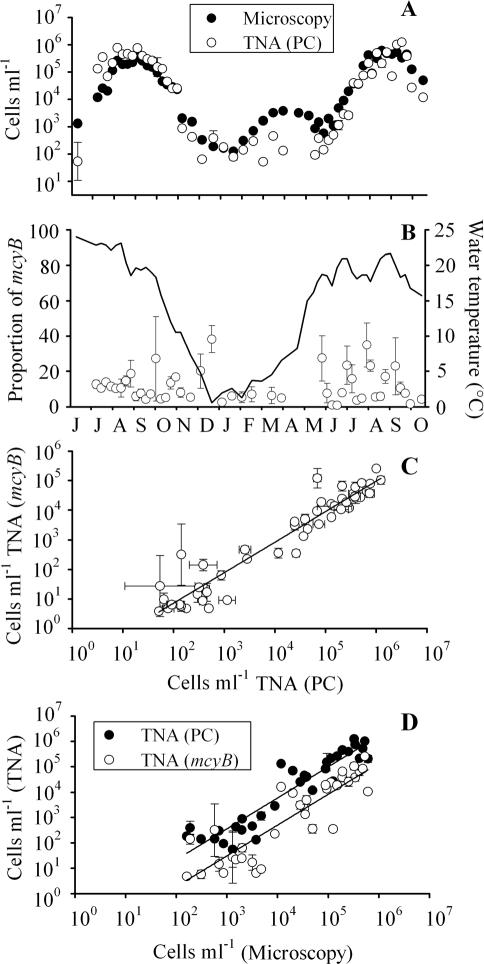
Throughout the study period, Microcystis cells were detected during counting in the microscope. On 29 of 55 sampling dates, Microcystis made up more than two-thirds of the cyanobacterial biovolume. On both sampling dates in April 2000, the Cts were out of the range of the standard curve of the TNA for the PC operon, and on 22 August 2000, no result was obtained. Those values were omitted from further analysis. Over the entire study period a significant relationship between cell numbers estimated via the inverted microscope technique and TNA of the PC gene was found (Fig. 4A and D). For the PC operon the regression equation was y = −1.11 + 1.22x (R2 = 0.88, n = 34, P < 0.0001), where y is the log10 cell number as determined by the TNA and x is the log10 cell number counted in the microscope. In general, mcyB-containing cells were always found when the PC operon genotypes were detected (Fig. 4B). On six sampling dates (22 February, 12 and 25 April, 4 and 9 May, and 22 August 2000), the concentration of mcyB was lower than the detection limit (Ct > 45). The mean mcyB proportion over the study period was 11.4% ± 2.6% (95% CL, n = 46), with a minimum and maximum of 0.9 and 38.3%, respectively (one outlier on the 8 June 1999, 114%). The proportion of mcyB genotypes from June 1999 to September 1999 (11.9% ± 3.6% [95% CL]) did not differ significantly from that in the corresponding period in the summer of 2000 (12.4% ± 5.5%; Mann-Whitney rank sum test, n = 28, P = 0.88). It is concluded that microcystin genotypes constituted only the smaller part of the Microcystis population in Lake Wannsee. The cell number of mcyB genotypes was found to be one-to-one related to the cell number of the PC operon genotypes (Fig. 4C). The regression equation was y = −1.20 + 1.03x (R2 = 0.95, n = 50, P < 0.0001), where y is the log10 cell number as determined by the TNA (mcyB) and x is the log10 cell number determined by TNA (the PC operon). The linear regression between cell numbers determined by microscopy and by TNA for mcyB was y = −2.17 + 1.22x (R2 = 0.80, n = 34, P < 0.0001), where y is the log10 cell number determined by TNA and x is the log10 cell number counted in the microscope. When the regressions for both genes (Fig. 4D) in intercept and slope were compared via the ANOVA model, the influence of the microscopically determined cell numbers (αj) on the TNA estimation of cell numbers and the influence of the grouping variable (βz) were found to be significant (P < 0.0001 [αj] and P < 0.1 [βz], n = 69). In contrast, no significant interaction effect between both factors [(αβ)jz] was found (P = 0.98), indicating that both regressions differed significantly in intercept (βz) but not in slope [(αβ)jz]. It is concluded that the mean proportion of microcystin genotypes was stable during the period of seasonal population growth from the lowest cell numbers in winter to the highest cell numbers in summer and did not depend on seasonal influences during the study period.
FIG. 4.
(A) Numbers of Microcystis cells in Lake Wannsee from June 1999 to October 2000, determined by counting under an inverted microscope or by TNA with the PC gene (mean ± 1 standard error). (B) Proportion of mcyB genotypes during the same period (one outlier on 8 June 1999, 114%, was omitted). The solid line indicates the mean water temperature integrated every meter over the total water column. (C) Dependence of cell numbers determined by TNA of the mcyB gene on cell numbers determined by TNA for PC. Error bars show the mean ± 1 standard error. (D) Comparison between cell number determined in the microscope and determined via TNA for PC (black) and mcyB (white) during the same study period. For details on statistical regression parameters, see the text.
DISCUSSION
Methodology.
This study is the first demonstrating that the results of cell quantification obtained by the TNA do correlate significantly with cell numbers determined by a more widespread cell counting method, both for laboratory cultures and for field samples. This validation is necessary to estimate the reliability of quantitative real-time PCR in field analysis. To date, no method for quantifying microcystin genotypes that is comparable in accuracy and feasibility to the technique developed in this study has been published. Due to the linear-log calibration curves, TNA cell quantification is sensitive to minor variations of the Cts, and it was demonstrated that the amplification efficiency of both TNAs was influenced by the presence of the environmental background on a linear scale. The results on background effects (Fig. 1C) indicate that mcyB genotypes were probably consistently underestimated by 45% ± 2% (95% CL); however, on a logarithmic scale, the background influence was negligible. Multiplying all mcyB cell numbers by a factor of 1.8 increases the mean mcyB proportion to 21% ± 4.8% (minimum, 1.7%; maximum, 71%; n = 46). This study represents the first data on the mcyB genotype proportion for the total Microcystis population in Lake Wannsee. In the study by Kurmayer et al. (15), the mcyB genotype proportions were estimated among size classes of Microcystis by the ratio of the number of PCR products of mcyB to the number for the PC operon obtained for a dilution series of the DNA extract. While this dilution assay was primarily designed to be used in a relative way, the main conclusion that the larger colonies (>100 μm) always have the highest percentage of mcyB genotypes was also tested with both TNAs at monthly intervals from June to September for both years. Using the correction factor 1.8 as above, the mean mcyB genotype percentages were 7.2% ± 2.5% (95% CL, n = 9) for the smallest size class (<50 μm), 12.6% ± 5.2% (n = 9) for the intermediate size class (50 to 100 μm), and 29.4% ± 12% (n = 9) and 37.4% ± 24% (n = 4) for the two larger size classes, >100 μm and >340 μm, respectively. From these data, the mean mcyB genotype percentage for the total population of Lake Wannsee can be calculated by multiplying the percentages for each size class with the corresponding proportions in biovolume (Fig. 3B in reference 15). Averaged over both years, this estimation results in a mean mcyB genotype proportion of 24% ± 9% (n = 9; minimum, 9.4; maximum, 48.2). This estimate corroborates the data on mcyB genotype percentage for the total population obtained directly from the water samples without filtration.
The high correlation coefficients between cell numbers in the microscope and those determined by TNA in the field strongly suggest the robustness of both TNAs under natural conditions. Because the phytoplankton of Lake Wannsee typically consists of several cyanobacterial genera (Microcystis, Planktothrix, Aphanizomenon, Anabaena, Limnothrix, and Pseudanabaena) (9), it is expected that both TNAs will work with sufficient accuracy in any body of water. In addition, the population of Microcystis typically varies in abundance by 3 to 4 orders of magnitude over the year, and cell quantification on a logarithmic scale has sufficient accuracy to monitor the population cycle over time for most research and monitoring objectives. Compared with the time involved with the counting of cells of Microcystis under the inverted microscope, TNA can be considered less time-consuming and labor-intensive. The relatively high acquisition and running costs in addition to the costs for acquisition and maintenance of standard laboratory equipment (usually comprising an inverted microscope) may constitute a drawback in applying TNA on a broader scale.
There was a large significant increase in TNA fluorescence signal during the transition from the logarithmic phase to the stationary phase. The reason might be an uncoupling between DNA replication and subsequent cell division when resources become limiting. This result is in contrast to the data obtained by Mann and Carr (16) for Anacystis nidulans demonstrating that faster-growing cells have a higher DNA content per cell. However, Anacystis has a much shorter mean generation time (3 h) than Microcystis (in this study, the maximum generation time was 41 h), and for Microcystis the time needed for DNA replication is probably never a limiting factor during cell division as observed for Anacystis.
Seasonal stability of microcystin genotype proportion.
In this study the averaged proportion of microcystin genotypes was considered constant from 1999 to 2000, as shown by the parallel regression lines between cell numbers in the microscope and cells containing or lacking the microcystin gene as differentiated by the TNA. In addition, the correlation between PC genotypes (as the independent variable) and microcystin genotypes (the dependent variable) was close to 1, and 95% of the variance of mcyB was attributable to the influence of population growth (the increase in PC concentration). The results are in agreement with the conclusion obtained from the genetic analysis of different colony size classes (15) that shifts of microcystin genotype proportions between and within size classes are stable over the growing season.
The stability in microcystin genotype proportion between years may depend on inoculation and reinvasion events after strong winters when the total population was removed from the water column. Microcystis sp. are known to rely on reinvasion into the water column from the sediments (23). It has been reported elsewhere (15) that the Microcystis population of Lake Wannsee consists mainly of the morphospecies M. aeruginosa/Microcystis flos-aquae and M. ichthyoblabe. Those morphospecies also differ significantly in the percentage of microcystin genotypes; i.e., 73% of colonies assigned to M. aeruginosa contain mcyB, while only 16% of colonies assigned to M. ichthyoblabe contain it. M. aeruginosa has frequently been reported to form large and firm colonies, while the colonies of M. ichthyoblabe are typically small and fragile (31). It remains unknown whether there are differences in survival ability in the sediment between morphospecies (e.g., see references 2 and 3), and it cannot be excluded that the proportion of microcystin genotypes could be changed by reinvasion events from the sediment. In this study, Microcystis cells were always found during counting, even in winter, and the persisting population probably preserved the mcyB genotype composition from one year to the next.
Microcystin gene distribution and microcystin net production.
For the population of Lake Wannsee a close relationship between the occurrence of mcyB and microcystin net production has been observed (14, 15). In addition, 322 individual colonies sampled from numerous bodies of water in Europe were tested for mcyB gene distribution and microcystin net production by sensitive matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in parallel, and only three individual colonies (1%) were found to contain mcyB but failed to show any detectable microcystin (L. Via-Odorika et al., unpublished data). Correspondingly, a significant relationship between the population growth rate and the microcystin net production rate for the same population in the summer of 2000 has been reported (15). Significant correlations between surrogate parameters such as chlorophyll a or algal biovolume and microcystin net production by Microcystis sp. have been reported by other authors as well (6, 12, 13, 18). Taken together, those results support the conclusion that it is possible to infer microcystin concentrations from surrogate parameters, for example, Microcystis cell numbers.
Over the entire study period the proportion of mcy genotypes made up the smaller part of the PC operon genotypes only. It is noteworthy that a number of field studies obtained similar results by the isolation of clonal strains from field samples. The percentage of hepatotoxic strains versus nonhepatotoxic strains isolated by Ohtake et al. (19) from Lake Kasumigaura (Japan) was 10% (n = 20), that obtained by Rohrlack et al. (24) from Lake Wannsee and Lake Pehlitzsee (Germany) was 45% (n = 22), that obtained by Shirai et al. (25) from Lake Kasumigaura was 34% (n = 68), and that obtained by Vezie et al. (30) from Lake Grand-Lieu (France) was 16% (n = 98). From the data obtained by PCR analysis of individually isolated colonies in a previous study (14), a higher mcyB genotype proportion for the total population could be expected. However, only the largest colonies of Microcystis (>100 μm) were sampled individually in the study of Kurmayer et al. (14), and it has also been found that the larger colonies (>340 μm) had the highest mcyB proportion but contributed the smallest fraction to the total cell number, <10% (15).
On the other hand, Microcystis populations have been found to vary in hepatotoxicity within a few weeks, e.g., in 50% lethal dose (expressed in milligrams [dry weight] per kilogram of body weight) (7) or in microcystin concentration (12, 21), and a mosaic structure for toxic cyanobacterial blooms has been suggested (4, 22, 30). In the present study, with one exception, short-term variation ranged from 1 to 38%, and the data on mcyB genotype proportions showed maxima on a weekly or biweekly scale (Fig. 4B). Both TNAs showed variability in cell number estimation compared to microscopy (Fig. 4D). While there was a statistically significant correlation between overestimation or underestimation of cell numbers determined in the microscope between both TNAs, the larger part of the variance remained unexplained (Kurmayer, unpublished). This unexplained part of the variance might be due to short-term shifts in mcyB genotype proportion. However, it cannot be excluded that a difference in precision between both TNAs may bias the mcyB genotype proportion in the short term, and the data on short-term variability need to be interpreted with caution. Further studies are needed to determine whether the data on short-term variability in mcyB genotype proportion are a real phenomenon or must be attributed to the noise induced by the real-time PCR approach.
Acknowledgments
We thank Gamila Ali Ahmed, Astrid Baldus, Karina Laskus, and Sara Monka for assistance in phytoplankton counting. Eva Schober assisted in performing the TNA. We thank the technical staff of the Institute for Limnology (Hannes Höllerer, Kurt Mayrhofer, Harald Ployer, and Peter Stadler) for numerous valuable assistance. We are grateful to Ingrid Chorus for her helpful discussion, the UBA team for sampling and field work, and the crew of the Wasserschiffahrtsamt (Harald Becker) for assistance in sampling. The comments of two anonymous reviewers improved the manuscript.
This work was financed by the EU project TOPIC (Toxin Production in Cyanobacteria, CT 38-0246), the FWF project CYTOGENE (Linking Cyanotoxin Production to Genetic Diversity, P15709), and the EU project PEPCY (Toxic and Other Bioactive Peptides in Cyanobacteria, QLK4-CT-2002-02634).
REFERENCES
- 1.Becker, S., P. Böger, R. Oehlmann, and A. Ernst. 2000. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Appl. Environ. Microbiol. 66:4945-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunberg, A.-K., and P. Blomquist. 2002. Benthic overwintering of Microcystis colonies under different environmental conditions. J. Plankton Res. 24:1247-1252. [Google Scholar]
- 3.Brunberg, A.-K., and P. Blomqvist. 2003. Recruitment of Microcystis (Cyanophyceae) from lake sediments: the importance of littoral inocula. J. Phycol. 39:58-63. [Google Scholar]
- 4.Carmichael, W. W., and P. R. Gorham. 1981. The mosaic nature of toxic blooms of cyanobacteria, p. 161-172. In W. W. Carmichael (ed.), The water environment: algal toxins and health. Plenum Press, New York, N.Y.
- 5.Chorus, I., and J. Bartram. 1999. Toxic cyanobacteria in water. E & FN Spon, London, United Kingdom.
- 6.Chorus, I., V. Niesel, J. Fastner, C. Wiedner, B. Nixdorf, and K.-E. Lindenschmidt. 2001. Environmental factors and microcystin levels in water bodies, p. 159-177. In I. Chorus (ed.), Cyanotoxins: occurrences, causes, consequences. Springer Verlag, Berlin, Germany.
- 7.Codd, G. A., and S. G. Bell. 1985. Eutrophication and toxic cyanobacteria in freshwaters. Water Pollut. Control 84:225-232. [Google Scholar]
- 8.Dittmann, E., B. A. Neilan, M. Erhard, H. von Döhren, and T. Börner. 1997. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol. Microbiol. 26:779-787. [DOI] [PubMed] [Google Scholar]
- 9.Fastner, J. 1999. Ph.D. thesis. Microcystins (cyanobacterial hepatotoxins) in German fresh waters: extraction, occurrence, and influence of environmental factors on microcystin production. Freie Universität Berlin, Berlin, Germany.
- 10.Foulds, I. V., A. Granacki, C. Xiao, U. J. Krull, A. Castle, and P. A. Horgen. 2002. Quantification of microcystin-producing cyanobacteria and E. coli in water by 5′-nuclease PCR. J. Appl. Microbiol. 93:825-834. [DOI] [PubMed] [Google Scholar]
- 11.Komárek, J., and K. Anagnostidis. 1999. Cyanoprokaryota, 1. Teil Chroococcales, Gustav Fischer Verlag, Jena, Germany.
- 12.Kotak, B. G., A. K.-Y. Lam, E. E. Prepas, S. L. Kenefick, and S. E. Hrudey. 1995. Variability of the hepatotoxin microcystin-LR in hypereutrophic drinking water lakes. J. Phycol. 31:248-263. [Google Scholar]
- 13.Kotak, B. G., A. K. Y. Lam, E. E. Prepas, and S. E. Hrudey. 2000. Role of chemical and physical variables in regulating microcystin-LR concentration in phytoplankton of eutrophic lakes. Can. J. Fish. Aquat. Sci. 57:1584-1593. [Google Scholar]
- 14.Kurmayer, R., E. Dittmann, J. Fastner, and I. Chorus. 2002. Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. in Lake Wannsee (Berlin, Germany). Microb. Ecol. 43:107-118. [DOI] [PubMed] [Google Scholar]
- 15.Kurmayer, R., G. Christiansen, and I. Chorus. 2003. The abundance of microcystin-producing genotypes correlates positively with colony size in Microcystis and determines its microcystin net production in Lake Wannsee. Appl. Environ. Microbiol. 69:787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mann, N., and N. G. Carr. 1974. Control of macromolecular composition and cell division in the blue-green alga Anacystis nidulans. J. Gen. Microbiol. 83:399-495. [DOI] [PubMed] [Google Scholar]
- 17.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 18.Oh, H.-M., S. J. Lee, J.-H. Kim, H.-S. Kim, and B.-D. Yoon. 2001. Seasonal variation and indirect monitoring of microcystin concentrations in Daechung Reservoir, Korea. Appl. Environ. Microbiol. 67:1484-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohtake, A., M. Shirai, T. Aida, N. Mori, K. I. Harada, K. Matsuura, M. Suzuki, and M. Nakano. 1989. Toxicity of Microcystis species isolated from natural blooms and purification of the toxin. Appl. Environ. Microbiol. 55:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otsuka, S., S. Suda, S. Shibata, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 2001. A proposal for the unification of five species of the cyanobacterial genus Microcystis Kützing ex Lemmermann 1907 under the rules of the bacteriological code. Int. J. Syst. Evol. Microbiol. 51:873-879. [DOI] [PubMed] [Google Scholar]
- 21.Park, H.-D., M. F. Watanabe, K.-I. Harada, H. Nagai, M. Suzuki, M. Watanabe, and H. Hayashi. 1993. Hepatotoxin (microcystin) and neurotoxin (anatoxin-a) contained in natural blooms and strains of cyanobacteria from Japanese freshwaters. Nat. Toxins 1:353-360. [DOI] [PubMed] [Google Scholar]
- 22.Park, H.-D., M. F. Watanabe, K.-I. Harada, M. Suzuki, H. Hayashi, and T. Okino. 1993. Seasonal variations of Microcystis species and toxic heptapeptide microcystins in Lake Suwa. Environ. Toxicol. Water Qual. 8:425-435. [Google Scholar]
- 23.Reynolds, C. S. 1973. Growth and buoyancy of Microcystis aeruginosa Kütz. emend. Elenkin in a shallow eutrophic lake. Proc. R. Soc. Lond. B 184:29-50. [Google Scholar]
- 24.Rohrlack, T., M. Henning, and J. G. Kohl. 2001. Isolation and characterization of colony-forming Microcystis aeruginosa strains, p. 152-158. In I. Chorus (ed.), Cyanotoxins: occurrences, causes, consequences. Springer Verlag, Berlin, Germany.
- 25.Shirai, M., A. Ohtake, T. Sano, S. Matsumoto, T. Sakamoto, A. Sato, T. Aida, K. I. Harada, T. Shimada, M. Suzuki, and M. Nakano. 1991. Toxicity and toxins of natural blooms and isolated strains of Microcystis spp. (cyanobacteria) and improved procedure for purification of cultures. Appl. Environ. Microbiol. 57:1241-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivonen, K., and G. Jones. 1999. Cyanobacterial toxins, p. 41-111. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water. E & FN Spon, London, United Kingdom.
- 27.Sokal, R. R., and F. J. Rohlf. 1995. Biometry. The principles and practice of statistics in biological research. W. H. Freeman and Company, New York, N.Y.
- 28.Tillett, D., E. Dittmann, M. Erhard, H. vonDöhren, T. Börner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 29.Utermöhl, H. 1958. Zur Vervollkommnung der quantitativen Phytoplanktonmethodik. Mitt. Int. Ver. Theor. Angew. Limnol. 2:1-38. [Google Scholar]
- 30.Vezie, C., L. Brient, K. Sivonen, G. Bertru, J.-C. Lefeuvre, and M. Salkinoja-Salonen. 1998. Variation of microcystin content of cyanobacterial blooms and isolated strains in Lake Grand-Lieu (France). Microb. Ecol. 35:126-135. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe, M. 1996. Isolation, cultivation and classification of bloom-forming Microcystis in Japan, p. 13-34. In M. F. Watanabe, K. I. Harada, W. W. Carmichael, and H. Fujiki (ed.), Toxic Microcystis. CRC Press, Boca Raton, Fla.
- 32.Zehnder, A., and P. R. Gorham. 1960. Factors influencing the growth of Microcystis aeruginosa Kütz. emend. Elenkin. Can. J. Microbiol. 6:645-660. [DOI] [PubMed] [Google Scholar]