Abstract
Culture-independent DNA fingerprints are commonly used to assess the diversity of a microbial community. However, relating species composition to community profiles produced by community fingerprint methods is not straightforward. Terminal restriction fragment length polymorphism (T-RFLP) is a community fingerprint method in which phylogenetic assignments may be inferred from the terminal restriction fragment (T-RF) sizes through the use of web-based resources that predict T-RF sizes for known bacteria. The process quickly becomes computationally intensive due to the need to analyze profiles produced by multiple restriction digests and the complexity of profiles generated by natural microbial communities. A web-based tool is described here that rapidly generates phylogenetic assignments from submitted community T-RFLP profiles based on a database of fragments produced by known 16S rRNA gene sequences. Users have the option of submitting a customized database generated from unpublished sequences or from a gene other than the 16S rRNA gene. This phylogenetic assignment tool allows users to employ T-RFLP to simultaneously analyze microbial community diversity and species composition. An analysis of the variability of bacterial species composition throughout the water column in a humic lake was carried out to demonstrate the functionality of the phylogenetic assignment tool. This method was validated by comparing the results generated by this program with results from a 16S rRNA gene clone library.
A variety of culture-independent methods have been developed (recently reviewed in reference 21) to carry out comparative analyses of microbial communities and to relate community composition to environmental parameters. Use of a culture-independent method requires a trade-off between phylogenetic resolution and sample throughput. Community fingerprint methods offer rapid analysis of samples and readily lend themselves to comparison of phylotype richness among many samples (11, 16, 32, 34); however, relating these community profiles to species composition is not straightforward. Clone libraries offer the highest degree of phylogenetic resolution available for culture-independent methodologies but can be cumbersome for analysis of the large numbers of samples that may be produced by studies of temporal or spatial variability of microbial communities. In addition, comparison of community composition between samples by using clone libraries can be problematic if the libraries offer incomplete coverage of a community, although progress has been made in this area (38).
Community analysis by terminal restriction fragment length polymorphism (T-RFLP) offers a compromise between sample throughput and phylogenetic resolution (24, 29). T-RFLP can be used to compare and contrast microbial community structure (4, 6-8, 12-14, 25, 30). Restriction fragment length is determined by the sequence of the fragment to be digested. Terminal restriction fragment (T-RF) lengths can be predicted from known sequences; thus, the T-RFLP method can potentially identify specific organisms in a community based on their T-RF length. There are many instances where the same T-RF length is predicted for multiple species of bacteria, but increased specificity can result from analysis of digests with multiple enzymes (9, 29).
Web-based resources available through the Ribosomal Database Project (http://rdp.cme.msu.edu) (31) or through the Microbial Community Analysis (MiCA) website at the University of Idaho (http://hermes.campus.uidaho.edu) allow prediction of T-RFs from 16S rRNA gene sequences presently in the database based on user input of PCR primers and restriction enzymes. Users are able to compare fragments obtained from T-RFLP analysis to the fragment sizes predicted from known 16S rRNA gene sequences. This comparison is accomplished by manually scanning the predicted fragment sizes to find a subset of species that produce a fragment size similar to one obtained experimentally. A species list can then be refined by comparison with additional digests. This is a reasonable procedure to carry out for uncomplicated profiles (e.g., individual unknown species or mixtures of very few species). However, such assignments are considerably more difficult in complex communities when each individual peak from each digest has the potential to represent multiple species (22, 31). Phylogenetic assignment for complex community profiles involves finding the intersection of the species sets represented by each peak. This is a daunting task when an individual T-RF may correspond to 15 or more species (31).
Discrepancies between observed and predicted fragment sizes may occur, an issue that further increases the list of species associated with each fragment (17, 22). This necessitates specification of size tolerances for matching. The large number of samples generated by studies of spatial or temporal variability magnifies this complexity. As a result, few studies are making use of the full potential of T-RFLP. Those studies which do utilize the T-RFLP method for identification of species from mixed communities often do so in conjunction with sequence analysis of a clone library (3, 6-8, 12-14, 19, 25, 27, 28, 30, 33) or by determination of T-RFs from cultured isolates (35). Others have coupled T-RFLP with Southern hybridization to assign T-RFs to specific phylogenetic groups (17). These variations are all designed to allow phylogenetic assignment from a single T-RFLP profile. A recent study assessing the value of T-RFLP profiles for phylogenetic inference similarly sought to maximize the information provided by a single digest by examining the phylogenetic specificity that can be achieved with different restriction enzymes (9).
In this work, the functionality of the T-RFLP method is expanded by automating the task of phylogenetic assignment from T-RF profiles produced by multiple digests. The use of multiple digests increases the specificity of phylogenetic inferences derived from T-RFLP profiles, and automation of this task makes this type of analysis accessible for analysis of complex communities. The effectiveness of the phylogenetic assignment tool is demonstrated with an analysis of aquatic microbial communities collected from a humic lake compared with the results of a 16S rRNA gene library.
MATERIALS AND METHODS
T-RFLP phylogenetic assignment tool.
The T-RFLP phylogenetic assignment tool (PAT) enables investigators to quickly find possible phylogenetic assignments based on data from a series of restriction enzyme digests. It is designed to accept any number of digests for use in generating phylogenetic matches and associated statistics.
Database.
The default database for PAT includes T-RFs predicted from 16S rRNA gene sequences by using the forward primer 8F (5′-AGAGTTTGATCMTGGCTCAG-3′) (23) and a selection of tetrameric restriction enzymes. The database was generated by using the MiCA query function found at the MiCA website. PAT users may supply a custom database for analysis of their T-RFLP profiles. The MiCA website can be used to generate a database of predicted T-RFs for each species by using different primers or restriction enzymes, or such a database could be generated from sequence data obtained from a clone library. The database file is an array of T-RF lengths for each species, with restriction enzyme names as column headings and bacterial species designations as row labels. The column headings are used to generate the restriction enzyme list used by the program. This file must be formatted as a tab-delimited text file for use with the PAT program. There is no sequence analysis function included with the PAT algorithm.
Program implementation.
The program prompts the researcher for the necessary data and configuration files prior to computation. Input data files are the tab-delimited six-column output tables generated by automated sequencers such as the ABI Genetic Analyzer instruments. Each file includes the data obtained from a single restriction enzyme digest for a series of samples (e.g., data obtained from all HhaI digests for a batch of samples would be contained in one file, MspI digests would be contained in another file, and RsaI digest data would be contained in a third file). One data file is loaded into the program for each enzyme digest, and corresponding samples from each digest are required to have the same lane identification (ID) in order that the algorithm may know that the fragment data are derived from the same sample. This is easily accomplished by editing the lane ID labels prior to analysis in the event that corresponding samples do not have the same lane ID in a given data set. Each record in these data files contains a specific terminal-length size, lane ID, peak height, and a peak area found in the sample. The program also requires a database of known organisms with known T-RF lengths for each specific enzyme used. User input specifies the names of the enzymes associated with each uploaded digest file and the size tolerances to be used during the matching process.
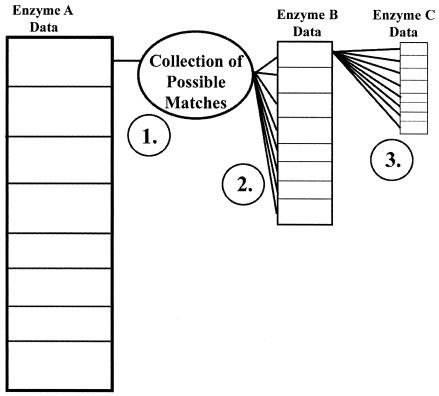
The program uses a filtering approach for phylogenetic assignment. It performs a series of passes through the database of possible organisms to discover all possible phylogenetic assignments consistent with a given set of microbial community data. This series of passes is illustrated in Fig. 1.
FIG. 1.
Cycle of the matching algorithm. For each digest, each individual fragment is assigned a collection of species from the database that are predicted to have a T-RF length that matches the observed fragment length (within the user-specified size tolerance) (step 1). Records that do not match fragments found in additional digests (steps 2 and 3) are discarded. Steps 1 through 3 are repeated for each digest file.
The program starts by computing the possible phylogenetic matches by using the data from the first digest as a base. For each T-RF length present in the first enzyme digest, the program creates an object called a collection to hold possible matches found within the size tolerance. In step 1, the program places known organisms from the database that match the first fragment length into the collection as a possible match.
For each subsequent restriction enzyme digest, possible matches present in the collection are compared to the T-RF lengths for the digest. Records that do not match a T-RF length found in subsequent enzyme digests are discarded from the collection. Steps 2 and 3 in the diagram represent this filtering technique. The collection then contains a list of species from the known organism database that have matched T-RF lengths for all enzyme digests.
The collection creation and filtering cycle is carried out for each fragment length present in the first digest, and steps 1 through 3 are repeated for each digest file. The final result is a series of collections of phylogenetic assignments representing the matches from a given sample. Each possible match contains the organism's name, the observed T-RF lengths that generated each assignment, and the lane ID and peak area of matched fragments.
Output.
After the calculations have been performed on a series of input digestions, the program produces several tab-delimited output files. The first file contains the phylogenetic matches as determined by the program. The record for each match contains the lane ID, peak area, and length of the fragment from each digest. A second file contains an analysis of the completeness of the matching algorithm and the abundance of unmatched fragments from each restriction digest. The final file contains a list of unmatched fragments. It includes lane IDs and peak areas for T-RFs that were not matched to a known organism by the program. All of these files are compatible with Excel or other common spreadsheet applications.
Portability.
This program was written in Java to allow portability across multiple computer platforms. The Java implementation also provided the opportunity to quickly create a web-based interface through the use of Java servlets and jsp pages.
Web interface.
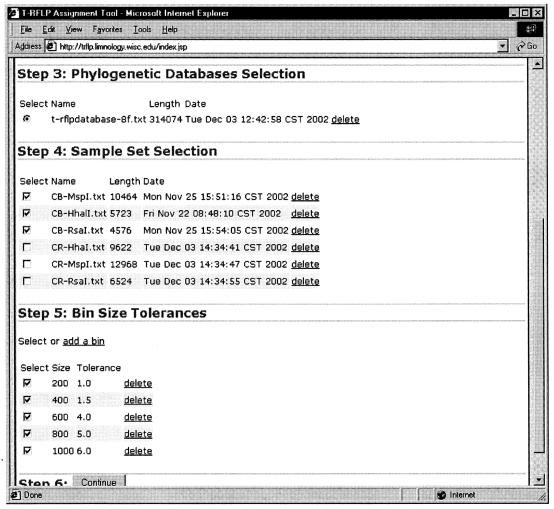
The web interface for the PAT tool allows investigators to submit, process, and manage every aspect of the phylogenetic assignment. It can be accessed at http://trflp.limnology.wisc.edu. The web-based PAT allows for each user of the system to register and maintain an account for data and configuration information. A user can upload digest and database files for use with the account. Users can also elect to use the default database of T-RFs produced by the in silico digestion of known 16S rRNA gene sequences by 27 separate restriction enzymes. The web interface provides management of a user's enzyme digests, known organism database files, and bin size configuration (Fig. 2).
FIG. 2.
The PAT web interface allows users to manage uploaded digest files, known organism database files, and T-RF bin size configuration.
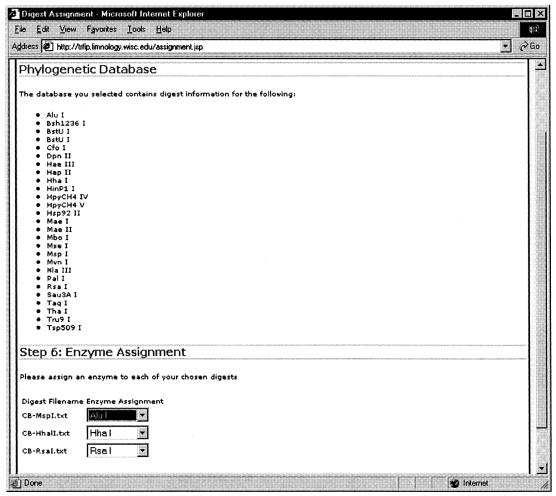
To generate phylogenetic assignments from the supplied data, the user selects a database file, associated digest files, and the bin sizes to use in the computation. The PAT program then prompts the user to assign a specific restriction enzyme present in the selected database to each of the digest files. This enzyme assignment process can be seen in Fig. 3.
FIG. 3.
The PAT web interface prompts users to select the restriction enzymes used to generate their T-RF digest files from a list of enzymes found in the selected database.
After the enzyme assignment, the program runs the PAT filtering algorithm to determine phylogenetic matches. The phylogenetic matches, matching statistics, and unmatched fragment information can then be downloaded from the results page.
Community analysis.
T-RFLP data produced from aquatic samples were analyzed to demonstrate the functionality of the PAT. Samples were obtained from Devil's Lake (45°31′N, 88°52′W), a humic lake located in northern Wisconsin with a surface area of 12.5 ha and a maximum depth of 7 m.
Sample collection.
Whole water samples were collected on 19 August 2002 from discrete depths throughout the water column. Samples for culture-independent analyses of community composition were immediately concentrated in aliquots of 250 ml onto 0.2-μm-pore-size filters (Supor-200; Gelman). Filters were placed in cryovials, frozen immediately, and stored at −80°C.
DNA extraction.
DNA was extracted from aquatic samples by using the method described by Fisher and Triplett (11).
T-RFLP.
PCRs to amplify 16S rRNA genes for T-RFLP analysis (24) contained PCR buffer consisting of 50 mM Tris (pH 8.0), 250 μg of bovine serum albumin per ml, 3.0 mM MgCl2 (catalog no. 1770; Idaho Technology), 250 μM (each) deoxynucleoside triphosphate, 10 pmol of each primer, 1.25 U of Taq polymerase (Promega), and 1 μl of extracted DNA in a final volume of 25 μl. The primers used were 8F (labeled with 6-FAM) and 1492R (23). Reactions were cycled in an Eppendorf MasterCycler Gradient (Eppendorf) with an initial denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 35 s, 55°C for 45 s, and 72°C for 2 min, with a final extension carried out at 72°C for 2 min. PCR products were digested with HhaI, MspI, and RsaI. Multiple single digests were carried out to increase the specificity of the phylogenetic assignments. Denaturing capillary electrophoresis was carried out for each digest by using an ABI 310 genetic analyzer (PE Applied Biosystems). Electrophoresis conditions were 60°C and 15 kV, with a run time of 50 min with POP-4 polymer. A custom 200- to 2,000-bp rhodamine X-labeled size standard (Bioventures) was used as the internal size standard for each sample. The data were analyzed using GeneScan 3.1 software (Perkin-Elmer).
Analysis.
Data tables containing fragment size and abundance data for each digest of the aquatic samples were exported from GeneScan, and the resulting text file was uploaded to the PAT website for phylogenetic assignment.
Clone library analysis.
Amplification of the 16S rRNA genes by PCR for clone library analysis of microbial community samples collected at 4.9 and 5.5 m was carried out as described for T-RFLP except that the 8F primer was unlabeled. Three μl of PCR product from each community was ligated into the pGEM-T vector (Promega). Clone libraries were produced with the pGEM-T Easy Vector System II (no. A1380; Promega) according to the manufacturer's instructions. Sequencing of 48 clones from each sample was carried out as described by Vergin et al. (39). Briefly, insert DNA was sequenced by using the ABI PRISM Big Dye Terminator cycle sequencing kit and 30 pmol of the 8F primer according to standard cycle sequencing parameters. The sequences were edited by using ABIView and submitted to BLAST (1) for initial phylogenetic assignment. Additional information regarding taxonomic assignment of cloned sequences was obtained by using the Hierarchy Browser function of the Ribosomal Database Project II at http://rdp.cme.msu.edu (5).
RESULTS
T-RFLP analysis.
An example of the output from the phylogenetic assignment tool is presented in Table 1. Note that the program considers matches of a single HhaI fragment with all possible MspI and RsaI fragments generated from the same sample.
TABLE 1.
PAT output for samples collected from Devil's Lake (northern Wisconsin) in August 2002a
| Species match | Length of fragment (bp) for:
|
||
|---|---|---|---|
| HhaI | MspI | RsaI | |
| 4.9-m sample | |||
| Chlorobium phaeovibrioides strain DSM 270 | 83.47 | 488.97 | 445.63 |
| Desulfuromonas acetoxidans strain 11070 DSM 684 | 94.67 | 162.93 | 189.42 |
| Campylobacter lanienae NCTC 13004 | 96.44 | 472.35 | 452.83 |
| Campylobacter sp. | 96.44 | 472.35 | 448.95 |
| Campylobacter mucosalis CCUG 6822 | 96.44 | 472.35 | 452.83 |
| Quinella ovalis | 98.12 | 150.00 | 452.83 |
| Campylobacter coli | 98.12 | 472.35 | 452.83 |
| Campylobacter jejuni | 98.12 | 472.35 | 452.83 |
| Microbacterium arborescens IFO 3750 | 141.51 | 161.42 | 452.83 |
| Streptomyces sp. NRRL 3890 | 172.94 | 159.37 | 448.95 |
| Streptomyces neyagawaensis ATCC 27449 | 172.94 | 159.37 | 448.95 |
| Fusobacterium gonidiaformans ATCC 25563 | 200.45 | 151.29 | 445.63 |
| Fusobacterium mortiferum ATCC 25557 | 200.45 | 151.29 | 445.63 |
| Marinospirillum minutulum ATCC 19193 | 203.59 | 302.29 | 646.34 |
| Achromatium strain JD13 | 203.59 | 488.97 | 646.34 |
| Achromatium strain HK13 | 203.59 | 488.97 | 646.34 |
| Methylomicrobium buryaticum strain 5G | 203.59 | 488.97 | 740.23 |
| Methylomicrobium buryaticum strain 7G | 203.59 | 488.97 | 740.23 |
| Pseudomonas mendocina ATCC 25411 | 205.72 | 497.64 | 166.47 |
| Pseudomonas flavescens strain B62 NCPPB 3063 | 205.72 | 488.97 | 646.34 |
| Pseudomonas plecoglossicida strain FPC951 | 205.72 | 488.97 | 646.34 |
| Marinospirillum minutulum ATCC 19193 | 205.72 | 302.29 | 646.34 |
| Oceanospirillum multiglobuliferum ATCC 33336 | 205.72 | 302.29 | 646.34 |
| 5.5-m sample | |||
| Desulfuromonas acetoxidans: | |||
| Strain 11070 DSM 684 | 94.55 | 162.84 | 189.32 |
| Strain BD3-7 | 94.55 | 164.89 | 448.84 |
| Strain JTB36 | 94.55 | 164.89 | 455.96 |
| Campylobacter lanienae NCTC 13004 | 96.37 | 472.36 | 448.84 |
| Campylobacter mucosalis CCUG 6822 | 96.37 | 472.36 | 455.96 |
| Campylobacter sp. | 96.37 | 472.36 | 448.84 |
| Uncultured Verrucomicrobiales strain ESR 25 | 96.37 | 604.5 | 273.53 |
| Fusobacterium gonidiaformans ATCC 25563 | 200.45 | 151.28 | 448.84 |
| Fusobacterium mortiferum ATCC 25557 | 200.45 | 151.28 | 448.84 |
| Achromatium strain HK13 | 203.57 | 488.77 | 646.56 |
| Achromatium strain JD13 | 203.57 | 488.77 | 646.56 |
| Achromatium strain JD2 | 203.57 | 488.77 | 646.56 |
| Clone OM241 | 203.57 | 490.21 | 81.28 |
| Methylomicrobium buryaticum strain 5G | 203.57 | 490.21 | 743.54 |
| Methylomicrobium buryaticum strain 7G | 203.57 | 490.21 | 743.54 |
| Oceanospirillum multiglobuliferum ATCC 33336 | 205.80 | 302.73 | 646.56 |
| Pseudomonas flavescens strain B62 NCPPB 3063 | 205.80 | 488.77 | 646.56 |
| Pseudomonas plecoglossicida strain FPC951 | 205.80 | 488.77 | 646.56 |
| Achromatium oxaliferum | 207.36 | 488.77 | 646.56 |
| Acinetobacter Hop10 strain Hop10 | 207.36 | 491.12 | 754.41 |
| Pelodictyon clathratiforme | 207.36 | 440.74 | 448.84 |
| Pseudomonas flavescens strain B62 NCPPB 3063 | 207.36 | 488.77 | 646.56 |
| Pseudomonas plecoglossicida strain FPC951 | 207.36 | 488.77 | 646.56 |
| Thauera T1 strain T1 | 207.36 | 82.58 | 153.76 |
| Achromatium oxaliferum | 209.15 | 491.12 | 646.56 |
| Clone env. OPS 1 | 209.15 | 98.14 | 448.84 |
| Clone SJA-121 | 211.83 | 164.89 | 455.96 |
| Clone SJA-176 | 211.83 | 302.73 | 455.96 |
| Clone Sva0115 | 211.83 | 497.52 | 797.46 |
| Uncultured Holophaga sp. clone GuBH2-AD-9 | 211.83 | 186.71 | 448.84 |
| Haemophilus influenzae-murium NCTC 11146 | 356.64 | 491.12 | 646.56 |
| Oxobacter pfennigii DSM 3222 | 356.64 | 517.46 | 448.84 |
| Strain AS2987 | 356.64 | 141.07 | 452.3 |
| Strain AS2988 | 356.64 | 138.97 | 448.84 |
| Haemophilus influenzae-murium NCTC 11146 | 358.24 | 488.77 | 646.56 |
| Clostridium malenominatum ATCC 25776 | 359.96 | 517.46 | 448.84 |
| Clostridium tetanomorphum DSM 528 | 359.96 | 517.46 | 448.84 |
| Clostridium tetanomorphum strain H1 NCIMB 11547 | 359.96 | 517.46 | 448.84 |
| Achromatium HK9 | 363.92 | 455.23 | 646.56/PICK> |
| Actinobacillus salpingitidis CCUG 23139 | 363.92 | 497.52 | 646.56 |
| Actinomycetales | 363.92 | 159.63 | 455.96 |
| Eubacterium lentum JCM 9979 | 363.92 | 133.14 | 452.3 |
| Eubacterium lentum JCM 9979 | 363.92 | 133.14 | 455.96 |
| Haemophilus haemoglobinophilus NCTC 1659 | 363.92 | 491.12 | 646.56 |
| Haemophilus haemolyticus NCTC 10659 | 363.92 | 497.52 | 646.56 |
| Haemophilus influenzae ATCC 33391 | 363.92 | 497.52 | 646.56 |
| Haemophilus parainfluenzae ATCC 33392 | 363.92 | 497.52 | 743.54 |
| Mycobacterium chubuense ATCC 27278 | 363.92 | 72.47 | 452.3 |
| Oceanospirillum kriegii ATCC 27133 | 363.92 | 488.77 | 646.56 |
| Pasteurella sp. strain CCUG 19794 | 363.92 | 497.52 | 646.56 |
| Pasteurella sp. strain Bisgaard Taxon 15 CCUG 16500 | 363.92 | 497.52 | 646.56 |
| Streptomyces bikiniensis DSM 40581 | 363.92 | 159.63 | 452.3 |
| Marinomonas communis ATCC 27118 | 372.36 | 497.52 | 646.56 |
| Oceanospirillum linum ATCC 11336 | 372.36 | 128.58 | 646.56 |
| Uncultured bacterium; ASL8 | 372.36 | 72.47 | 96.23 |
| Vibrio diazotrophicus strain NS ATCC 33466 | 372.36 | 497.52 | 646.56 |
| Vibrio vulnificus ATCC 27562 | 372.36 | 497.52 | 646.56 |
The phylogenetic assignments are sorted by HhaI fragment size. The output indicates which three T-RFs present in the sample were used to generate each assignment.
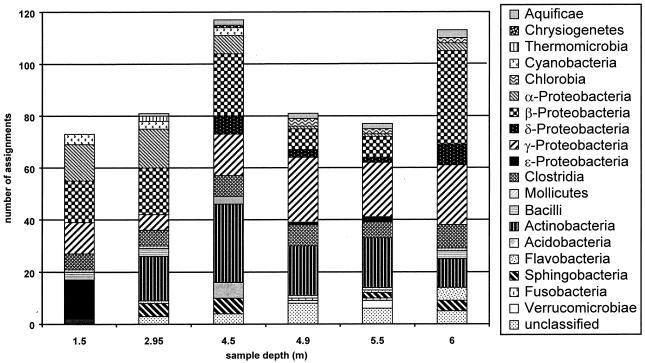
An analysis of the variability of bacterial community composition over the entire water column in Devil's Lake on 19 August 2002 was performed using this tool (Fig. 4). The diversity of bacterial classes identified by T-RFLP varied throughout the water column, although the communities at all depths appear to be dominated by species classified as γ- and β-Proteobacteria and Actinobacteria.
FIG. 4.
Bacterial classes detected by T-RFLP in Devil's Lake (northern Wisconsin) at various depths in August 2002. The plots indicate the number of fragments in each sample that could be assigned to each indicated phylogenetic class.
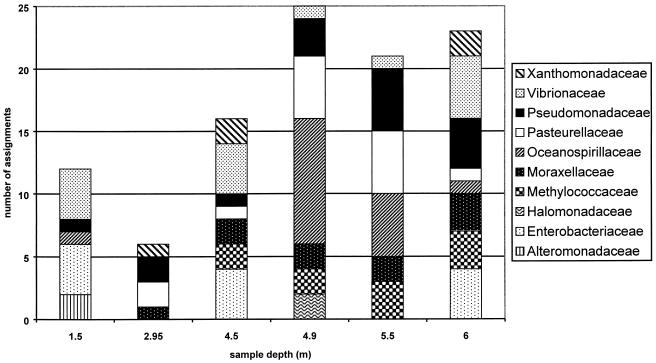
Finer-scale phylogenetic comparisons can also be carried out by using the PAT output. While γ-Proteobacteria were detected at every sample depth (Fig. 4), the PAT assignments can be used to describe the variability of T-RFs within this class. T-RFs classified as Oceanospirillaceae, for example, are detected at only a few depths, while Vibrionaceae are detected throughout the water column (Fig. 5).
FIG. 5.
Bacterial diversity of families within the γ-Proteobacteria in Devil's Lake (northern Wisconsin) throughout the water column in August 2002. The y axis plots the number of fragments in each sample that could be assigned to a particular phylogenetic group within the γ-Proteobacteria.
Comparison of PAT results with clone library results.
The bacterial community composition determined by T-RFLP analysis was compared with the results of sequencing clone libraries generated from two of the samples (Table 2). For the 4.9-m depth sample at the class level of taxonomic discrimination, the T-RFLP and clone library analyses detected Chlorobia, four classes of proteobacteria, clostridia, bacilli, actinobacteria, acidobacteria, and Verrucomicrobiae. Two classes, Aquificae and Fusobacteria, were detected by the T-RFLP approach but were not detected in the clone library. Five classes (Cyanobacteria, Bacilli, Planctomycetacia, Bacterioidetes, and Sphingobacteria) were detected in the clone library that were not observed in the T-RFLP analysis. For the 5.5-m depth sample, two classes (Bacilli and Bacterioidetes) were unique to the clone library, while five classes (Aquificae, ɛ-Proteobacteria, Acidobacteria, Flavobacteria, and Fusobacteria) were found only in the T-RFLP analysis. Eight classes were found in both analyses.
TABLE 2.
Comparison of taxonomic groups detected by T-RFLP using PAT with those detected by clone library analysisa
| Phylum and class | 4.9-m sample
|
5.5-m sample
|
||
|---|---|---|---|---|
| Clone library | T-RFLP | Clone library | T-RFLP | |
| Aquificae | ||||
| Aquificae | x | x | ||
| Cyanobacteria | ||||
| Cyanobacteria | x | |||
| Chlorobi | ||||
| Chlorobia | x | x | x | x |
| Proteobacteria | ||||
| α-Proteobacteria | x | x | x | x |
| β-Proteobacteria | x | x | x | x |
| γ-Proteobacteria | x | x | x | x |
| δ-Proteobacteria | x | x | x | x |
| ɛ-Proteobacteria | x | x | x | |
| Firmicutes | ||||
| Clostridia | x | x | x | x |
| Bacilli | x | |||
| Actinobacteria | ||||
| Actinobacteria | x | x | x | x |
| Planktomycetes | ||||
| Planctomycetacia | x | |||
| Acidobacteria | ||||
| Acidobacteria | x | x | x | |
| Bacteroidetes | ||||
| Bacteroidetes | x | x | ||
| Flavobacteria | x | |||
| Sphingobacteria | x | x | x | |
| Fusobacteria | ||||
| Fusobacteria | x | x | ||
| Verrucomicrobia | ||||
| Verrucomicrobiae | x | x | x | x |
| Total | 14 | 12 | 11 | 14 |
Samples were collected at 4.9 m and 5.5 m in Devil's Lake (northern Wisconsin) in August 2002.
DISCUSSION
The use of T-RFLP for community analysis offers some of the advantages of both community fingerprint methods and clone libraries. Comparisons of community diversity and composition can be coupled with phylogenetic information available from resources that predict terminal restriction fragment sizes produced by 16S rRNA gene sequences. While phylogenetic assignment of a fragment using the predicted T-RF length appears to be relatively uncomplicated when using the profiles produced by a single digest, the capacity of this approach for phylogenetic inference at the species level is limited, due to the overlap in T-RF size among related organisms (9).
Attempts to conduct phylogenetic assignment for T-RFs by using a single digest are effective only when the amplified PCR products are produced from a single bacterial division (or smaller taxonomic group) (9). The predictive power of a single digest is also diminished by inaccuracies in fragment size measurement that require assignment of each T-RF to a bin of contiguous fragment sizes (which will almost certainly correspond to more organisms than are represented by a single observed T-RF). The phylogenetic information derived from a single digest is further reduced when T-RFs are generated from multiple bacterial subdivisions and also as a result of the ever-increasing number of sequences available for reference (9, 22). Thus, the use of multiple digests is recommended to accomplish any degree of phylogenetic resolution (9, 20, 22, 29, 31). However, one of the reasons that researchers have sought to determine the phylogenetic specificity of T-RFs from a single digest is the difficulty in correlating peaks from different digests produced from complex mixtures of bacteria.
This and other issues associated with database matching of T-RFLP profiles were reviewed by Kitts (22). Phylogenetic assignment uncertainties that arise due to discrepancies between predicted and observed T-RF sizes are multiplied when several digests are considered and contribute to the difficulty in manually interpreting community composition from multiple T-RFLP profiles.
Automation of T-RFLP analysis resolves many of the issues involved with analysis of complex profiles. Our PAT incorporates all of the recommendations found in the literature intended to maximize identification of T-RF peaks. First, it utilizes T-RFLP profiles from multiple digests (9, 20, 22, 29, 31). Second, it incorporates a user-defined window of size tolerances to accommodate discrepancies between predicted and observed T-RF lengths (22). In addition to these recommendations, we have incorporated the ability to define an increasing bin size for this matching window in recognition of the observation that uncertainty in size calling increases with increasing fragment length. Automation of T-RFLP analysis greatly reduces the time involved in phylogenetic assignment of T-RF peaks; in addition, it ensures that the size tolerance windows are uniformly applied and that all possible species matches are considered, thus reducing user-introduced bias. Also included is the option to compare the T-RFLP profiles to a user-defined database, allowing the phylogenetic assignments to be restricted to specific taxonomic groups or to a species list generated from a clone library. A database of T-RFs generated for a gene other than the 16S rRNA gene [e.g., nifH (36, 40), mer (2), elongation factor Tu (29), heat shock proteins (10, 15), glutamine synthetase (37), ATPases (26), and topoisomerases (18)] could also be used for analysis by the PAT algorithm.
The phylogenetic assignment tool generates a list of peaks that are not matched to species in the database. One explanation for the unmatched T-RFs is that these peaks are derived from previously uncharacterized bacteria. However, it is also possible that these peaks merely represent instances where there was insufficient information to make a match. This could occur when a 16S rRNA gene sequence in the database is not full length and therefore fails to match the input primer used for prediction of T-RFs. Unmatched T-RFs may also represent instances where one or more of the peaks produced by an organism falls below the peak detection threshold of the electrophoresis instrument and thus is not included in the PAT analysis.
Analysis of aquatic microbial community samples throughout the water column in a humic lake was used to demonstrate the functionality of PAT. The species lists generated by the PAT program were used to examine the variability in bacterial community composition at different levels of phylogenetic resolution. Many of the T-RFs were classified as γ- and β-Proteobacteria and Actinobacteria, suggesting that these taxa may dominate the microbial communities in this lake.
The phylogenetic assignments generated by using PAT were compared to the results obtained through clone library analysis of the same sample. For the two lake samples compared, 64.3 and 61.5% of the bacterial classes identified were found by both approaches. However, the remainder were found by only one approach. This suggests that the two approaches give similar results and can also complement each other in the identification of taxa not found by using only one method. For those taxa found only by the T-RFLP approach, taxon-specific primers can be designed to screen clones in a 16S rRNA library for these groups. The taxa identified only through the clone library analysis may indicate that some phylogenetic groups are not well represented in the T-RF database.
T-RFLP has demonstrated its utility as a community fingerprint method for comparisons of bacterial community composition between environments or treatments. The phylogenetic assignment tool described here extends this utility by offering a rapid, automated approach for phylogenetic analysis of T-RFs.
Acknowledgments
This work was supported by a Microbial Observatories grant from the National Science Foundation (grant DEB 9977903) to the Center for Limnology at the University of Wisconsin—Madison. The North Temperate Lakes Long-Term Ecological Research project (grants DEB 9632853 and DEB 0217533) at the University of Wisconsin also supported this work.
REFERENCES
- 1.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruce, K. D., and M. R. Hughes. 2000. Terminal restriction fragment length polymorphism monitoring of genes amplified directly from bacterial communities in soils and sediments. Mol. Biotechnol. 16:261-269. [DOI] [PubMed] [Google Scholar]
- 3.Chin, K. J., T. Lukow, and R. Conrad. 1999. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin, K. J., T. Lukow, S. Stubner, and R. Conrad. 1999. Structure and function of the methanogenic archaeal community in stable cellulose-degrading enrichment cultures at two different temperatures (15 and 30 degrees C). FEMS Microbiol. Ecol. 30:313-326. [DOI] [PubMed] [Google Scholar]
- 5.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covert, J. S., and M. A. Moran. 2001. Molecular characterization of estuarine bacterial communities that use high- and low-molecular weight fractions of dissolved organic carbon. Aquat. Microb. Ecol. 25:127-139. [Google Scholar]
- 7.Derakshani, M., T. Lukow, and W. Liesack. 2001. Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 67:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emmerhoff, O. J., H. P. Klenk, and N. K. Birkeland. 1998. Characterization and sequence comparison of temperature-regulated chaperonins from the hyperthermophilic archaeon Archaeoglobus fulgidus. Gene 215:431-438. [DOI] [PubMed] [Google Scholar]
- 11.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich, M. W., D. Schmitt-Wagner, T. Lueders, and A. Brune. 2001. Axial differences in community structure of Crenarchaeota and Euryarchaeota in the highly compartmentalized gut of the soil-feeding termite Cubitermes orthognathus. Appl. Environ. Microbiol. 67:4880-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong, J. H., R. J. Forster, H. Yu, J. R. Chambers, P. M. Sabour, R. Wheatcroft, and S. Chen. 2002. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 208:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Gong, J. H., R. J. Forster, H. Yu, J. R. Chambers, R. Wheatcroft, P. M. Sabour, and S. Chen. 2002. Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microbiol. Ecol. 41:171-179. [DOI] [PubMed] [Google Scholar]
- 15.Gupta, R. S. 1998. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62:1435-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heuer, H., and K. Smalla. 1997. Application of denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis for studying soil microbial communities. In J. D. van Elsas, E. M. H. Wellington, and J. T. Trevors (ed.), Modern soil microbiology. Marcel Dekker Inc., New York, N.Y.
- 17.Hiraishi, A., M. Iwasaki, and H. Shinjo. 2000. Terminal restriction pattern analysis of 16S rRNA genes for the characterization of bacterial communities of activated sludge. J. Biosci. Bioeng. 90:148-156. [DOI] [PubMed] [Google Scholar]
- 18.Huang, W. M. 1996. Bacterial diversity based on type II DNA topoisomerase genes. Annu. Rev. Genet. 30:79-107. [DOI] [PubMed] [Google Scholar]
- 19.Inagaki, F., Y. Sakihama, A. Inoue, C. Kato, and K. Horikoshi. 2002. Molecular phylogenetic analyses of reverse-transcribed bacterial rRNA obtained from deep-sea cold seep sediments. Environ. Microbiol. 4:277-286. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan, C. W., J. C. Astaire, M. E. Sanders, B. S. Reddy, and C. L. Kitts. 2001. 16S ribosomal DNA terminal restriction fragment pattern analysis of bacterial communities in feces of rats fed Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 67:1935-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent, A. D., and E. W. Triplett. 2002. Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu. Rev. Microbiol. 56:211-236. [DOI] [PubMed] [Google Scholar]
- 22.Kitts, C. L. 2001. Terminal restriction fragment patterns: a tool for comparing microbial communities and assessing community dynamics. Curr. Issues Intest. Microbiol. 2:17-25. [PubMed] [Google Scholar]
- 23.Lane, D. J. 1991. 16S/23S rRNA sequencing. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley & Sons, Chichester, United Kingdom.
- 24.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludemann, H., I. Arth, and W. Liesack. 2000. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl. Environ. Microbiol. 66:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 27.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 28.Lueders, T., and M. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 30.Marsh, T. L., W. T. Liu, L. J. Forney, and H. Cheng. 1998. Beginning a molecular analysis of the eukaryal community in activated sludge. Water Sci. Technol. 37:455-460. [Google Scholar]
- 31.Marsh, T. L., P. Saxman, J. Cole, and J. Tiedje. 2000. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 66:3616-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massol-Deya, A. A., D. A. Odelson, R. F. Hickey, and J. M. Tiedje. 1995. Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA), p. 1-8. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 2. Kluwer Academic Publishers, Boston, Mass.
- 33.Moeseneder, M. M., C. Winter, J. M. Arrieta, and G. J. Herndl. 2001. Terminal-restriction fragment length polymorphism (T-RFLP) screening of a marine archaeal clone library to determine the different phylotypes. J. Microbiol. Methods 44:159-172. [DOI] [PubMed] [Google Scholar]
- 34.Muyzer, G. A., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsson, W. B., and M. S. Strom. 2002. Detection and identification of bacterial pathogens of fish in kidney tissue using terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes. Dis. Aquat. Org. 48:175-185. [DOI] [PubMed] [Google Scholar]
- 36.Poly, F., L. Ranjard, S. Nazaret, F. Gourbičre, and L. J. Monrozier. 2001. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 67:2255-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saccone, C., C. Gissi, C. Lanave, and G. J. Pesole. 1995. Molecular classification of living organisms. Mol. Evol. 40:273-279. [DOI] [PubMed] [Google Scholar]
- 38.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vergin, K. L., M. S. Rappe, and S. J. Giovannoni. 2001. Streamlined method to analyze 16S rRNA gene clone libraries. BioTechniques 30:938-940, 943-944. [DOI] [PubMed] [Google Scholar]
- 40.Widmer, F., B. T. Shaffer, L. A. Portous, and R. J. Seidler. 1999. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade mountain range. Appl. Environ. Microbiol. 65:374-380. [DOI] [PMC free article] [PubMed] [Google Scholar]