Abstract
Synapse formation involves reciprocal interactions between cells resulting in formation of a structure optimized for efficient information transfer. Recent work has implicated constituents of the cadherin–catenin cell-adhesion complex in both synapse formation and plasticity. In this review, we describe recent interesting discoveries on mechanisms of cadherin complex function, in addition to regulating adhesion, that are relevant for understanding the role of this complex in synaptogenesis and plasticity. We describe how this complex acts via (i) recruitment/stabilization of intracellular partners; (ii) regulation of intracellular signaling pathways; (iii) regulation of cadherin surface levels, stability and turnover; (iv) stabilization of receptors; and (v) regulation of gene expression. These exciting discoveries provide insights into novel functional roles of the complex beyond regulating cell adhesion.
Introduction
Synapses of the central nervous system are specialized asymmetric cell-adhesion junctions that mediate directional information transfer. Pre- and postsynaptic compartments are morphologically distinct with specialized functional roles. Presynaptic terminals contain synaptic vesicles and machinery for neurotransmitter release, whereas postsynaptic sites include neurotransmitter receptors and a variety of scaffolding and signaling proteins. Together, these molecules ensure rapid and directional information transfer. Moreover, the synaptic compartments are dynamic, allowing for synaptic modulation in response to neural activity, a property believed to underlie the ability of the nervous system to learn and retain memory [1,2]. The processes that underlie synapse assembly, maintenance and plasticity need to be precisely regulated during development and in response to synaptic activity to optimize functioning of synapses and neural circuits.
In this review, we focus on the role of a cell-adhesion complex, the cadherin–catenin complex, in synapse morphogenesis and plasticity within the mammalian central nervous system. This complex has been extensively studied in epithelial cells. It is becoming increasingly clear that it also has very interesting functional roles in neurons. Several studies indicate that components of the complex regulate multiple aspects of synapse morphogenesis and plasticity. Many of the insights obtained in epithelia concerning mechanisms through which adhesion and signaling by the cadherin complex are regulated appear to be useful in understanding the functions of these complexes in synapse formation, function and plasticity [3].
Contact-mediated adhesion in synaptogenesis
The formation of a synapse requires target recognition followed by recruitment of pre- and postsynaptic elements in precise apposition (Figure 1). Synapse morphogenesis is initiated by formation of contacts between filopodia that arise from dendrites and axon segments, followed by contact stabilization and acquisition of appropriate pre- and postsynaptic elements, together with spine maturation at many excitatory synapses. Synaptic maturity is characterized by morphological and molecular specializations in both compartments that are optimized for fast and efficient information transfer.
Figure 1.
Schematic representation of hippocampal excitatory synapse formation and localization of the cadherin–catenin complex at different stages. (a) Synaptogenesis is initiated by the formation of nascent contacts between dendritic filopodia and axons. At these stages, N-cadherin in distributed evenly along the synaptic structure. This is followed by (b) contact stabilization and clustering of cadherin and (c) maturity. A mature synapse is characterized by a stable presynaptic terminal that contains vesicles primed for neurotransmitter release and a postsynaptic density that contains the receptors and scaffolding proteins. In the adult, cadherin is localized to distinct regions bordering the mature active zone, termed as the puncta adherentia. Both β-catenin and α-catenin are known to colocalize with cadherin at these junctions. Recent studies in nonneuronal cells indicate that the binding of α-catenin to β-catenin and actin are mutually exclusive, with the monomer form having a higher affinity for β-catenin and the dimer having a higher affinity for actin. Although p120ctn and δ-catenin are known to be at synapses, their localization at the electron microscope level and during development is unclear.
In central neurons, the majority of excitatory synapses are formed on dendritic spines. Dendritic spine density, morphology and size are important regulators of their roles in information transfer, learning and memory [4]. The ability of spines to regulate shapes and content in response to synaptic activity is vital for their function. Regulation of cadherin-complex-associated functions contributes to spine morphogenesis, plasticity and function [5,6].
The cadherin–catenin cell-adhesion complex
The cadherin family is composed of more than 80 members in a single species that has been classified into several subfamilies, including classical cadherins, protocadherins, Fat cadherins, CNRs and seven-pass transmembrane cadherins [7]. This review is restricted to classical cadherins. The roles for cadherins and catenins, cytosolic partners of cadherins, in regulating cell adhesion in epithelial cells have been extensively explored. It is now clear that these proteins regulate synaptogenesis and plasticity in central neurons. More interestingly, the cadherin–catenin complex regulates synapses through both cell-adhesion-dependent and -independent mechanisms.
Structurally, classical cadherins, the type I and II cadherins, include a series of five extracellular repeat structures which bind calcium, a single transmembrane domain and an intracellular tail. The cadherin repeats mediate cis and trans interactions leading to homophilic cis and trans cadherin dimerization [8]. Cytoplasmic domains of cadherins include binding sites for a variety of proteins, including catenins. The catenins are cytosolic proteins that can be subdivided into three separate groups – two β-catenin-like proteins (β-catenin and plakoglobin), three α-catenins and four p120catenin-related proteins. The distal region of cadherin cytoplasmic domains includes a binding site for β-catenin, whereas the membrane proximal region contains a binding site for members of the p120ctn family that includes δ-catenin, ARVCF and p0071. α-Catenins bind the complex via interactions with β-catenin (Figure 2).
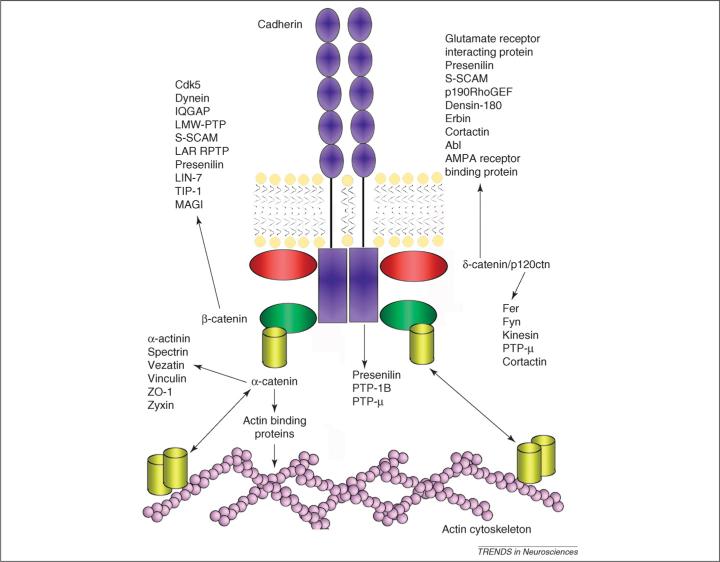
Figure 2.
A partial list of proteins that interact with components of the cadherin–catenin complex. Components of the complex bind to a variety of scaffolding proteins, receptors, kinases and phosphatases.
Structurally, β-catenin includes N- and C-terminal regions flanking a series of 12 armadillo (arm) repeats [9]. Cadherins bind to β-catenin and plakoglobin via an extended groove formed by the arm repeats. The C-terminal domain of β-catenin includes a transcriptional activation motif. The PDZ motif at the C terminus of β-catenin interacts with a variety of synapse-associated PDZ-domain-containing proteins.
Three α-catenins, encoded by different genes, have been characterized. These include αE-catenin, predominantly expressed in epithelia; αN-catenin, restricted to neural tissue; and αT-catenin, expressed in the heart. α-Catenins contain several regions that are homologous to domains in vinculin and include N- and C-terminal regions flanking an M (adhesion modulation) domain [10]. They are not structurally related to other catenins. The N-terminal regions include β-catenin-binding and dimerization domains. The C-terminal regions include an actin-binding site. In addition, α-catenin binds several actin-binding proteins, including vinculin, eplin, α-actinin and formins, proteins that promote F-actin polymerization [10]. In addition, α-catenin inhibits Arp2/3-mediated actin polymerization. Hence, α-catenin can regulate the actin cytoskeleton by multiple mechanisms.
The structures of p120ctn family members share some similarity with that of β-catenin with a central domain that includes nine armadillo repeats plus N- and C- terminal flanking sequences [9,11]. The armadillo repeat region includes the cadherin-binding site and a putative nuclear localization signal. In addition, δ-catenin, ARVCF and p0071 include a C-terminal PDZ-binding motif. p120ctn and δ-catenin function as RhoGDIs (guanine dissociation inhibitor); p120ctn also binds p190RhoGAP. p120ctn binds the cytoplasmic tyrosine kinase Fer which, through phosphorylation, stabilizes interactions between cadherins and β-catenin [12]. p120ctn and δ-catenin bind kaiso, suppressing its ability to repress transcription [13].
Until recently, the cadherin–catenin complex was believed to link cadherins to the actin cytoskeleton via binding of β-catenin to α-catenin, which in turn links to the actin cytoskeleton. By contrast, recent studies have demonstrated that α-catenin binds to β-catenin or F-actin in a mutually exclusive manner [14,15], with the monomeric form preferentially binding β-catenin and the homodimeric form preferentially binding F-actin. As α-catenin binds numerous actin-binding proteins, these observations do not challenge the function of α-catenin as a link between cadherins and the actin cytoskeleton.
The ability of cadherins to bind to catenins serves to link the cell-adhesion complex to a multitude of intracellular signaling pathways, affecting the cytoskeleton, cytoplasmic signaling pathways and gene expression.
The cadherin–catenin complex at central synapses
Multiple cadherins are expressed in the central nervous system [16] with distinct spatial and temporal expression patterns. Our understanding of the contribution of specific cadherins to synapse development and maintenance remains incomplete, in part because many neurons express more than one cadherin.
The most widely distributed neuronal cadherin is N-cadherin, whose distribution changes during development [7]. At young hippocampal synapses in culture (DIV [days in vitro] 5−6), N-cadherin is distributed evenly along the nascent synapse [17]. With maturation (DIV 14), however, N-cadherin is clustered within the active zone. A similar distribution is seen in developing hippocampal synapses in vivo and in calyx-type synapses in the chick ciliary ganglion [18]. At mature hippocampal mossy fiber–CA3 synapses in vivo, cadherin is localized to distinct regions bordering the mature active zone, termed the puncta adherentia [19,20]. In cultured hippocampal neurons, N-cadherin is progressively lost from inhibitory synapses during development and preferentially retained at excitatory synapses [21]. The distributions of αN-catenin and β-catenin at synapses at different developmental stages closely mirrors that of N-cadherin (Figure 1) [19].
In neurons, p120ctn is localized to both membrane and cytosolic fractions. In cultured hippocampal neurons, p120ctn is associated with N-cadherin clusters at DIV 7; this distribution is maintained in older neurons [22]. During development (DIV 7−14), the proportion of synapses that contain p120ctn decreases slightly. Interestingly, the proportion of p120ctn associated with N-cadherin decreased with synaptic maturity in calyx-type synapses in the chick ciliary ganglion [18]. It is not clear how this alteration in distribution with synaptic maturity controls the cadherin complex at the synapse.
δ-Catenin expression appears to be more specific to the central nervous system [23]. In cultured hippocampal neurons, δ-catenin is observed in dendritic compartments and synapses [24]. ARVCF [25] and p0071 [26] are widely expressed, but their expression patterns in the central nervous system remain poorly characterized.
The cadherin–catenin complex in excitatory synapse morphogenesis and plasticity
In neurons, the cadherin–catenin cell-adhesion complex regulates multiple aspects of synaptogenesis and plasticity. Interestingly, several recent studies have examined the mechanistic bases of these functional roles. Cadherins are well suited to promote adhesive functions and, by virtue of association with catenins, link cell adhesive interactions to a variety of intracellular pathways. The effects of the loss or overexpression of the cadherins and catenins are summarized in Table 1.
Table 1.
Effects of overexpression or loss of cadherins and catenins on synapse morphogenesis and plasticity
| N-cadherin | β-catenin | αN-catenin | p120ctn | |
|---|---|---|---|---|
| Spine morphology | DN: Filopodia-like elongation, bifurcation of spine head [53] LOF: (siRNA knockdown) DIV 6−12: Decrease in spine density; DIV 14−18: no alterations in spine density [56] LOF (CRE-mediated recombination): No alterations in spines [57] | LOF: Thin, elongated spines, reduced mushroom spines, no change in density [35] | LOF: Enhanced spine motility [36] OE: Promotes spine stability and increase in density [36] | LOF (DIV 10−14): Reduction in spine density, headwidth and length [37] LOF (DIV 14−17): No change in overall number of protrusions, decreased number of mature spines, increased protrusion length, decreased headwidth, increase in number of longer protrusions [37] OE: Filopodia-like elongation, no mature mushroom-shaped spine heads [37] |
| Postsynaptic organization | DN: Reduction in psd95 puncta, diffuse distribution of psd95 in dendrites LOF: (siRNA knockdown) Decrease in frequency of mEPSCs [56] | LOF: No change in psd95 [31] | LOF: No change in density of spines and synapsin-positive puncta, but slight reduction in synapsin-positive spines [36] | LOF: Decrease in psd95 density [37] |
| Presynaptic organization | DN: Reduction in synapsin puncta, diffused distribution of synapsin in axons [53] | DN: Diffused synaptic vesicle localization [31] | ?? | LOF: Decrease in synaptophysin density [37] |
| Synapse function | DN: Reduced synaptic vesicle recycling [53] | LOF: Reduced amplitude of spontaneous events, no change in frequency, block in quantum scaling associated with inhibition of synaptic activity [35] | ?? | LOF: ?? |
Abbreviations: DN, dominant negative; LOF, loss of function; OE, overexpression.
Cadherins and catenins are localized on both sides of the synapse and use binding partners present in each compartment to accomplish their compartment-specific roles. A partial list of these binding partners is depicted in Figure 2. Components of the complex bind to a variety of PDZ-domain-containing proteins (β-catenin and δ-catenin), protein phosphatases (cadherin, β-catenin and p120ctn), protein kinases (β-catenin, p120ctn and δ-catenin) and F-actin- and microtubule-associated proteins (α-catenin, p120ctn and δ-catenin).
Several lines of evidence indicate roles for cadherins in synaptogenesis. Cadherins are clustered at nascent synapses early during development and colocalize with synaptic markers [21]. N-cadherin is transported in association with active zone components to sites of synapse formation [27]. Recent studies suggest that functional roles of the cadherin–catenin complex in neurons are accomplished by adhesion-dependent and -independent mechanisms, as follows.
Regulation of adhesion
As a cell-adhesion molecule, N-cadherin is well suited to provide a link between structural changes and synaptic plasticity. Consistently, activity promotes dimerization of N-cadherin, acquisition of protease resistance and strong adhesion across the synapse [28]. These responses can be differentially modulated by activity, because KCl depolarization, but not NMDA stimulation, leads to dispersion of cadherins, but in both cases cadherin dimerization is observed. Hence, N-cadherin is dynamic and cadherin-mediated adhesion can respond to local changes in synaptic activity.
N-cadherin is required for induction of late-phase long-term potentiation (LTP) [29]. Inhibition of N-cadherin-mediated adhesion by use of blocking antibodies blocks late LTP, suggesting that the ability of N-cadherin to mediate adhesion is required for this functional role.
Morphological changes in spines have been linked to synaptic plasticity. Activation of AMPA receptors causes a lateral expansion of the spine head and redistribution of cadherins along the expanding spine head. Cadherin adhesion and actin polymerization are essential for this process, because overexpression of a cadherin mutant that lacks the adhesive activity prevents spine head expansion [30].
Recruitment and stabilization of intracellular partners
β-Catenin is a multifunctional protein with different preand postsynaptic functions. At presynaptic sites, β-catenin regulates localization of synaptic vesicles [31]. In the absence of β-catenin, the number of reserve pool vesicles is reduced, with concomitant reduction in exocytosis in response to repetitive stimulation. β-Catenin promotes synaptic vesicle localization through binding to cadherin and recruitment of PDZ-containing proteins via its C-terminal PDZ-binding motif. Although several PDZ-containing proteins bind to β-catenin, including LIN-7/Mint [32], TIP-1 [33] and MAGI [34], the importance of each in promoting synaptic assembly is not known.
Postsynaptically, loss of β-catenin after synaptogenesis results in profound alteration in spine morphology [35], leading to spines that are thin and elongated with no alterations in spine density. This is accompanied by reduction in amplitude, but not frequency, of mEPSCs. The ability of β-catenin to modulate synapse structure and function requires both its ability to bind cadherin and PDZ proteins. In addition, loss of β-catenin inhibits chronic activity blockade-regulated quantum scaling.
Regulation of intracellular signaling pathways
αN-catenin is crucial for spine stability in hippocampal neurons [36]. In the absence of αN-catenin, spines are more motile. Overexpression of α-catenin promotes spine stability and increases spine and synapse density. The ability of αN-catenin to bind both β-catenin and cytoskeletal proteins is necessary for αN-catenin to regulate spine stability. Overexpression of αN-catenin also prevents conversion of spines to filopodia when neuronal activity is inhibited, suggesting that αN-catenin can function to coordinate activity to structural alterations in spine structure.
Loss of p120ctn leads to a profound decrease in spine and synapse density in hippocampal neurons. This is accompanied by a global increase in active RhoA and decrease in active Rac1. The ability of p120ctn to regulate spine density does not require cadherin binding, but does require a domain in p120ctn required for RhoA inhibition [37]. The reduction in spine density associated with loss of p120ctn can be restored by constitutively active Rac or Rho inhibition. The ability of p120ctn to regulate the activity of the Rho family of small GTPases is hence crucial to its ability to regulate spine and synapse density. Loss of p120ctn also leads to a reduction in length and headwidth of spines. The regulation of spine length requires the ability of p120ctn to regulate Rho, whereas control of headwidth appears to require interactions with cadherin.
In epithelial cells it is known that cadherin-mediated adhesion promotes the localization of the products of PI3 kinase at cell–cell contacts. Interestingly, PI3 kinase has been shown to induce synapses in Drosophila motor neurons and adult projection neurons. It would be interesting to examine whether one of the pathways through which the cadherin–catenin complex regulates synaptogenesis is through regulation of PI3 kinase.
At the neuromuscular junction, β-catenin promotes agrin-mediated acetylcholine receptor clustering. The ability of β-catenin to link to the actin cytoskeleton via α-catenin is critical for β-catenin to promote receptor clustering [38]. Absence in muscle of β-catenin at the neuromuscular junction increases acetylcholine clusters, and these clusters are localized over a larger region of the myotube surface. This is accompanied by a reduction in spontaneous and evoked neurotransmitter release. Mice that lack β-catenin in the motor neuron do not have these deficits, suggesting that β-catenin is involved in retrograde signaling from the muscle to the nerve [39].
At the Xenopus neuromuscular junction, p120ctn expression promotes the formation of filopodia-like processes, termed myopodia [40]. Agrin induces phosphorylation of p120ctn and results in its dissociation from cadherin. Agrin- and p120ctn-dependent promotion of myopodia formation is not seen with a p120ctn mutant unable to inhibit Rho, indicating that free p120ctn promotes myopodial formation through inhibition of Rho activity.
Regulation of cadherin surface levels, stability and turnover
NMDA receptor activation reduces endocytosis of N-cadherin, resulting in retention of N-cadherin at the cell surface [6]. This is mediated by a decrease in phosphorylation of tyrosine 654 of β-catenin, which then enhances association of N-cadherin with β-catenin, stabilizing surface N-cadherin. Stabilization of N-cadherin–β-catenin association through mutation of tyrosine 654 to phenylalanine prevents NMDAR-dependent LTD in hippocampal neuron culture, indicating that N-cadherin endocytosis controls synaptic efficacy [6].
Arcadlin, a protocadherin induced by activity [41], binds to and promotes endocytosis of N-cadherin. Loss of arcadlin leads to increase in spine density that can be rescued by concomitant knockdown of N-cadherin [42]. Thus, activity-promoted regulation of arcadlin expression provides a mechanism through which synaptic density and stability can be regulated through control of cadherin surface stability. Although these two results appear contradictory, with activity either promoting or inhibiting endocytosis, these results describe effects of two different phases of synaptic stimulation. The surface stabilization of cadherin is a more rapid (∼100 min) and protein-synthesis-independent response to activity, whereas activity-induced cadherin internalization is initiated 4 h after stimulation and requires protein synthesis.
Members of the p120ctn family of proteins have also been shown to stabilize the surface levels of cadherins through inhibition of endocytosis [43]. Consistent with this, the expression of N-cadherin is reduced in the hippo-campi of mice lacking p120ctn [37]. Loss of p120ctn in neurons results in a reduction of spine headwidth that can be partially overcome by overexpression of N-cadherin. Thus, regulation of p120ctn family functions provides another pathway for control of cadherin levels at synapses, which in turn might regulate synaptic structure and function.
Stabilization of receptors
δ-Catenin, unlike p120ctn, includes a PDZ-binding motif at its C terminus which can interact with a plethora of PDZ-domain-containing proteins concentrated at the synapse. For example, recent studies have demonstrated that δ-catenin interacts with glutamate receptor interacting protein (GRIP) and AMPA receptor binding protein (ABP) [24], proteins that interact with GluR3 and GluR2, respectively. In association with ABP, δ-catenin stabilizes GluR2-containing AMPA receptors at the surface. In addition, several PDZ-interacting partners of δ-catenin have been identified, including erbin [44], densin-180 [45], S-SCAM [46] and PAPIN [47], although the functional significance of these interactions remains unclear. Both ARVCF [44] and p0071 [48] include a PDZ-binding motif.
Regulation of gene expression
N-cadherin is cleaved in response to activity to generate a C-terminal fragment that represses CBP/CREB-mediated transcription [49]. CREB-mediated transcription has been implicated in multiple aspects of development, including plasticity [50]. Activity also induces calpain-dependent cleavage of the N terminus of β-catenin. The resulting residual C-terminal fragments are resistant to degradation, promote Tcf-dependent gene transcription and induce expression of activity-dependent genes.
Wnt signaling is known to regulate synaptic structure and plasticity [51,52]. In Xenopus, p120ctn has recently been shown to bind Frodo, a protein that interacts with Dishevelled (Dsh), a component of the Wnt pathway [13]. Wnt signaling stabilizes p120ctn through interaction with Frodo, enhancing p120ctn interaction with the transcriptional repressor kaiso, thereby altering gene transcription. It would be very interesting to examine whether this pathway regulates synapse structure and plasticity in neurons.
Multiple functional roles
The underlying mechanisms for some functions of the cadherin–catenin complex are either unclear or can be ascribed to more than one of the above roles.
The loss of cadherin function by expression of a dominant-negative construct that competes for binding to catenins in hippocampal neurons leads to perturbation of synaptic structure and function [53], resulting in alterations in filopodia-like elongation and bifurcation of the spine head. These changes are accompanied by reductions in accumulation of pre- and postsynaptic markers and synaptic function. A similar perturbation of cadherin function reduces the accumulation of a postsynaptic marker in retinal horizontal cells [54]. Although the dominant-negative construct used in this study was originally designed to compete with β-catenin, thereby reducing cadherin-based adhesion, it is now clear that this construct also affects both cadherin-mediated interactions with p120ctn family members as well as a range of catenin-mediated interactions and functions. It therefore remains unclear which roles of the complex are required for spine morphogenesis.
Inhibition of N-cadherin, using an antagonistic peptide injected into the hippocampus of the mouse brain, inhibits long-term contextual fear memory [55]. This effect can be partly mediated by preventing phosphorylation of Erk1/2 and subsequent reduction of association of N-cadherin with IQGAP, a protein that serves as a scaffold to link the cadherin–catenin complex to the actin cytoskeleton.
In association with the extracellular domain of GluR2, N-cadherin promotes dendritic spine formation [56]. In these studies, knockdown of N-cadherin at early stages (DIV 6+12) decreased spine density; knockdown of N-cadherin at later stages (DIV 14+4) did not reduce spine density. However, knockdown of N-cadherin at later stages (DIV 14+4) does prevent extracellular domain of GluR2-induced increase in spine density and synaptic function, suggesting that the N-cadherin–GluR2 complex stimulates synapse development. Synapse formation is not perturbed in cultured N-cadherin null hippocampal neurons [57]. One possibility is that one of the other classical cadherins might compensate for loss of N-cadherin.
Activity promotes the relocalization of β-catenin from dendritic shafts to spines and enhances its association with cadherin. Association with β-catenin strengthens adhesion through stabilizing cadherins on the cell surface and through making possible associations with the cytoskeleton [58,59]. It also provides a mechanism for recruitment of signaling proteins that associate with β-catenin or α-catenin [60]. Tyrosine phosphorylation of β-catenin at residue Y654 [5] promotes dissociation from cadherin, thereby reducing association of signaling proteins. Association of β-catenin with cadherin promotes synaptic activity and an increase in size of synaptic clusters [5,61]. BDNF, a neurotrophin known to be vital for synaptic plasticity, disperses synaptic vesicle clusters. BDNF-mediated synaptic vesicle dispersion requires Y654 phosphorylation-dependent dissociation of cadherin–β-catenin interaction [62].
In neurons derived from N-cadherin null embryonic stem cells, initial synapse formation is not perturbed. However, presynaptic function, in particular the availability of vesicles for release at high frequencies, is compromised. This is accompanied by alterations in short-term plasticity and enhancement of synaptic depression [63].
In addition to N-cadherin, other cadherins are expressed in central neurons, although their functional roles are relatively less explored. Cadherin-8 is required for normal development of presynaptic terminals in hippocampal neurons; an inhibitory peptide leads to a gross reduction in the size and intensity of presynaptic puncta [64]. Cadherin-11 and -13 promote development of functional excitatory synapses in hippocampal neurons in culture [65], with data suggesting that they play somewhat different roles.
The cadherin–catenin complex in inhibitory synaptogenesis
Relatively less is known about the functional roles of the cadherin complex in the development and maintenance of inhibitory synapses. N-cadherin is excluded from inhibitory synapses during development in hippocampal neurons [21]. It is unclear whether N-cadherin is replaced by another cadherin and what the contribution of this cadherin is to inhibitory synaptogenesis. Cadherin-11 and -13 promote the formation of inhibitory synapses in cultured hippocampal neurons [65]. The roles of the catenins in inhibitory synaptogenesis remain unexplored. β-Catenin is still retained at inhibitory synapses despite loss of N-cadherin, suggesting that it is associated with other cadherins.
The cadherin–catenin complex in human neurological disease
Dendrite and spine abnormalities are associated with several human conditions, including mental retardation, normal aging process and neurodegenerative disorders. Given their wide expression pattern and their critical roles in synapse and spine morphogenesis and plasticity, it is highly likely that mutations in cell-adhesion molecules might underlie several neurological disorders.
There is currently no direct evidence for the involvement of a specific mutation of any of the components of the cadherin–catenin complex in neurological disease. Altered expression of various cadherins and catenins is associated with cancer metastasis. The genetic deletion on chromosome 5p in humans associated with the Cri du Chat syndrome [66] includes the gene encoding δ-catenin, although it is unclear whether the primary deficits arise from the lack of δ-catenin. Interestingly, mice that lack δ-catenin have severe learning deficits, including altered synaptic transmission and long-term potentiation [67]. Further studies are necessary to determine the contribution of loss of δ-catenin to Cri du Chat syndrome.
Presenilin, a gene implicated in Alzheimer's disease, promotes cleavage of N-cadherin to generate a C-terminal fragment that results in inhibition of CRE-mediated transcription [49]. Mutations in presenilin that are associated with familial Alzheimer's disease inhibit the ability of presenilin to promote this cleavage. Because CRE-regulated transcription regulates several genes, inhibition of N-cadherin cleavage might contribute to the pathology of this devastating neurodegenerative disorder.
Conclusions and future directions
It is clear that the cadherin–catenin complex is pivotal in spine and synapse morphogenesis and plasticity, with each component of the complex making a contribution. No doubt, technical advances in genomics and proteomics will greatly aid in identifying more novel interaction partners for the complex and increase our understanding of the multiple and complex functional roles of this complex in neurons.
However several questions remain. What signaling pathways are regulated by the cadherin–catenin complex in neurons? How are these roles altered during development? How are these responses coordinated in response to synaptic activity? Does the complex contribute to long-term alterations in plasticity by affecting gene expression? How does the contribution of the complex to synaptic structure and function affect neural networks and higher brain functions?
In addition, the contribution of the complex to inhibitory synapses remains unclear. No doubt advances in proteomics and imaging should make it possible to delineate the contribution of the cadherin–catenin complex to multiple aspects of synapse formation and higher brain functions.
Acknowledgements
J.A. is supported by a postdoctoral fellowship from the Larry L. Hillblom Foundation. We also thank Seung-hye Lee for comments on the manuscript and Dr. Baris Genc for assistance with the figures. Work in the authors’ laboratory is supported by the National Institutes of Health.
Footnotes
Conflict of interest statement
The authors declare no competing financial interests.
References
- 1.Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu. Rev. Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez VA, Sabatini BL. Anatomical and physiological plasticity of dendritic spines. Annu. Rev. Neurosci. 2007;30:79–97. doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- 3.Yap AS, et al. Making and breaking contacts: the cellular biology of cadherin regulation. Curr. Opin. Cell Biol. 2007;19:508–514. doi: 10.1016/j.ceb.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne J, Harris KM. Do thin spines learn to be mushroom spines that remember? Curr. Opin. Neurobiol. 2007;17:381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Schuman EM, Murase S. Cadherins and synaptic plasticity: activity-dependent cyclin-dependent kinase 5 regulation of synaptic β-catenin-cadherin interactions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:749–756. doi: 10.1098/rstb.2002.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tai CY, et al. Activity-regulated N-cadherin endocytosis. Neuron. 2007;54:771–785. doi: 10.1016/j.neuron.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Yagi T, Takeichi M. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
- 8.Shapiro L, et al. Adhesion molecules in the nervous system: structural insights into function and diversity. Annu. Rev. Neurosci. 2007;30:451–474. doi: 10.1146/annurev.neuro.29.051605.113034. [DOI] [PubMed] [Google Scholar]
- 9.Pokutta S, Weis WI. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu. Rev. Cell Dev. Biol. 2007;23:237–261. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- 10.Kobielak A, Fuchs E. α-Catenin: at the junction of intercellular adhesion and actin dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:614–625. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCrea PD, Park JI. Developmental functions of the P120-catenin sub-family. Biochim. Biophys. Acta. 2007;1773:17–33. doi: 10.1016/j.bbamcr.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Lilien J, Balsamo J. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of β-catenin. Curr. Opin. Cell Biol. 2005;17:459–465. doi: 10.1016/j.ceb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Park JI, et al. Frodo links Dishevelled to the p120-catenin/Kaiso pathway: distinct catenin subfamilies promote Wnt signals. Dev. Cell. 2006;11:683–695. doi: 10.1016/j.devcel.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 14.Drees F, et al. α-Catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada S, et al. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bekirov IH, et al. Identification and localization of multiple classic cadherins in developing rat limbic system. Neuroscience. 2002;115:213–227. doi: 10.1016/s0306-4522(02)00375-5. [DOI] [PubMed] [Google Scholar]
- 17.Elste AM, Benson DL. Structural basis for developmentally regulated changes in cadherin function at synapses. J. Comp. Neurol. 2006;495:324–335. doi: 10.1002/cne.20876. [DOI] [PubMed] [Google Scholar]
- 18.Rubio ME, et al. Assembly of the N-cadherin complex during synapse formation involves uncoupling of p120-catenin and association with presenilin 1. Mol. Cell. Neurosci. 2005;30:611–623. [PubMed] [Google Scholar]
- 19.Uchida N, et al. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J. Cell Biol. 1996;135:767–779. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fannon AM, Colman DR. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron. 1996;17:423–434. doi: 10.1016/s0896-6273(00)80175-0. [DOI] [PubMed] [Google Scholar]
- 21.Benson DL, Tanaka H. N-cadherin redistribution during synaptogenesis in hippocampal neurons. J. Neurosci. 1998;18:6892–6904. doi: 10.1523/JNEUROSCI.18-17-06892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chauvet N, et al. Distribution of p120 catenin during rat brain development: potential role in regulation of cadherin-mediated adhesion and actin cytoskeleton organization. Mol. Cell. Neurosci. 2003;22:467–486. doi: 10.1016/s1044-7431(03)00030-7. [DOI] [PubMed] [Google Scholar]
- 23.Ho C, et al. δ-Catenin is a nervous system-specific adherens junction protein which undergoes dynamic relocalization during development. J. Comp. Neurol. 2000;420:261–276. [PubMed] [Google Scholar]
- 24.Silverman JB, et al. Synaptic anchorage of AMPA receptors by cadherins through neural plakophilin-related arm protein AMPA receptor-binding protein complexes. J. Neurosci. 2007;27:8505–8516. doi: 10.1523/JNEUROSCI.1395-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sirotkin H, et al. Identification of a new human catenin gene family member (ARVCF) from the region deleted in velo-cardio-facial syndrome. Genomics. 1997;41:75–83. doi: 10.1006/geno.1997.4627. [DOI] [PubMed] [Google Scholar]
- 26.Hatzfeld M, Nachtsheim C. Cloning and characterization of a new armadillo family member, p0071, associated with the junctional plaque: evidence for a subfamily of closely related proteins. J. Cell Sci. 1996;109:2767–2778. doi: 10.1242/jcs.109.11.2767. [DOI] [PubMed] [Google Scholar]
- 27.Jontes JD, et al. In vivo trafficking and targeting of N-cadherin to nascent presynaptic terminals. J. Neurosci. 2004;24:9027–9034. doi: 10.1523/JNEUROSCI.5399-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka H, et al. Molecular modification of N-cadherin in response to synaptic activity. Neuron. 2000;25:93–107. doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 29.Bozdagi O, et al. Increasing numbers of synaptic puncta during late-phase LTP: N-cadherin is synthesized, recruited to synaptic sites, and required for potentiation. Neuron. 2000;28:245–259. doi: 10.1016/s0896-6273(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 30.Okamura K, et al. Cadherin activity is required for activity-induced spine remodeling. J. Cell Biol. 2004;167:961–972. doi: 10.1083/jcb.200406030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bamji SX, et al. Role of β-catenin in synaptic vesicle localization and presynaptic assembly. Neuron. 2003;40:719–731. doi: 10.1016/s0896-6273(03)00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perego C, et al. Mammalian LIN-7 PDZ proteins associate with β-catenin at the cell-cell junctions of epithelia and neurons. EMBO J. 2000;19:3978–3989. doi: 10.1093/emboj/19.15.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanamori M, et al. The PDZ protein tax-interacting protein-1 inhibits β-catenin transcriptional activity and growth of colorectal cancer cells. J. Biol. Chem. 2003;278:38758–38764. doi: 10.1074/jbc.M306324200. [DOI] [PubMed] [Google Scholar]
- 34.Dobrosotskaya IY, James GL. MAGI-1 interacts with β-catenin and is associated with cell-cell adhesion structures. Biochem. Biophys. Res. Commun. 2000;270:903–909. doi: 10.1006/bbrc.2000.2471. [DOI] [PubMed] [Google Scholar]
- 35.Okuda T, et al. β-Catenin regulates excitatory postsynaptic strength at hippocampal synapses. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13479–13484. doi: 10.1073/pnas.0702334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abe K, et al. Stability of dendritic spines and synaptic contacts is controlled by α N-catenin. Nat. Neurosci. 2004;7:357–363. doi: 10.1038/nn1212. [DOI] [PubMed] [Google Scholar]
- 37.Elia LP, et al. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron. 2006;51:43–56. doi: 10.1016/j.neuron.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B, et al. β-Catenin regulates acetylcholine receptor clustering in muscle cells through interaction with rapsyn. J. Neurosci. 2007;27:3968–3973. doi: 10.1523/JNEUROSCI.4691-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li XM, et al. Retrograde regulation of motoneuron differentiation by muscle β-catenin. Nat. Neurosci. 2008;11:262–268. doi: 10.1038/nn2053. [DOI] [PubMed] [Google Scholar]
- 40.Madhavan R, et al. Involvement of p120 catenin in myopodial assembly and nerve-muscle synapse formation. J. Neurobiol. 2006;66:1511–1527. doi: 10.1002/neu.20320. [DOI] [PubMed] [Google Scholar]
- 41.Yamagata K, et al. Arcadlin is a neural activity-regulated cadherin involved in long term potentiation. J. Biol. Chem. 1999;274:19473–19479. doi: 10.1074/jbc.274.27.19473. [DOI] [PubMed] [Google Scholar]
- 42.Yasuda S, et al. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2β and p38 MAP kinases. Neuron. 2007;56:456–471. doi: 10.1016/j.neuron.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis MA, et al. A core function for p120-catenin in cadherin turnover. J. Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laura RP, et al. The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of δ-catenin and ARVCF. J. Biol. Chem. 2002;277:12906–12914. doi: 10.1074/jbc.M200818200. [DOI] [PubMed] [Google Scholar]
- 45.Izawa I, et al. Densin-180 interacts with δ-catenin/neural plakophilin-related armadillo repeat protein at synapses. J. Biol. Chem. 2002;277:5345–5350. doi: 10.1074/jbc.M110052200. [DOI] [PubMed] [Google Scholar]
- 46.Ide N, et al. Interaction of S-SCAM with neural plakophilin-related armadillo-repeat protein/δ-catenin. Biochem. Biophys. Res. Commun. 1999;256:456–461. doi: 10.1006/bbrc.1999.0364. [DOI] [PubMed] [Google Scholar]
- 47.Deguchi M, et al. PAPIN. A novel multiple PSD-95/Dlg-A/ZO-1 protein interacting with neural plakophilin-related armadillo repeat protein/δ-catenin and p0071. J. Biol. Chem. 2000;275:29875–29880. doi: 10.1074/jbc.M005384200. [DOI] [PubMed] [Google Scholar]
- 48.Jaulin-Bastard F, et al. Interaction between Erbin and a catenin-related protein in epithelial cells. J. Biol. Chem. 2002;277:2869–2875. doi: 10.1074/jbc.M109652200. [DOI] [PubMed] [Google Scholar]
- 49.Marambaud P, et al. A CBP binding transcriptional repressor produced by the PS1/ε-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell. 2003;114:635–645. doi: 10.1016/j.cell.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 51.Speese SD, Budnik V. Wnts: up-and-coming at the synapse. Trends Neurosci. 2007;30:268–275. doi: 10.1016/j.tins.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ataman B, et al. Rapid activity-dependent modifications in synaptic structure and function require bidirectional Wnt signaling. Neuron. 2008;57:705–718. doi: 10.1016/j.neuron.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Togashi H, et al. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- 54.Tanabe K, et al. Cadherin is required for dendritic morphogenesis and synaptic terminal organization of retinal horizontal cells. Development. 2006;133:4085–4096. doi: 10.1242/dev.02566. [DOI] [PubMed] [Google Scholar]
- 55.Schrick C, et al. N-cadherin regulates cytoskeletally associated IQGAP1/ERK signaling and memory formation. Neuron. 2007;55:786–798. doi: 10.1016/j.neuron.2007.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saglietti L, et al. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 57.Kadowaki M, et al. N-cadherin mediates cortical organization in the mouse brain. Dev. Biol. 2007;304:22–33. doi: 10.1016/j.ydbio.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 58.Troyanovsky RB, et al. Stable and unstable cadherin dimers: mechanisms of formation and roles in cell adhesion. Mol. Biol. Cell. 2007;18:4343–4352. doi: 10.1091/mbc.E07-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamada S, Nelson WJ. Synapses: sites of cell recognition, adhesion, and functional specification. Annu. Rev. Biochem. 2007;76:267–294. doi: 10.1146/annurev.biochem.75.103004.142811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pokutta S, et al. Biochemical and structural analysis of α-catenin in cell-cell contacts. Biochem. Soc. Trans. 2008;36:141–147. doi: 10.1042/BST0360141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murase S, et al. Depolarization drives β-catenin into neuronal spines promoting changes in synaptic structure and function. Neuron. 2002;35:91–105. doi: 10.1016/s0896-6273(02)00764-x. [DOI] [PubMed] [Google Scholar]
- 62.Bamji SX, et al. BDNF mobilizes synaptic vesicles and enhances synapse formation by disrupting cadherin-β-catenin interactions. J. Cell Biol. 2006;174:289–299. doi: 10.1083/jcb.200601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jungling K, et al. N-cadherin transsynaptically regulates short-term plasticity at glutamatergic synapses in embryonic stem cell-derived neurons. J. Neurosci. 2006;26:6968–6978. doi: 10.1523/JNEUROSCI.1013-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bekirov IH, et al. Cadherin-8 and N-cadherin differentially regulate pre- and postsynaptic development of the hippocampal mossy fiber pathway. Hippocampus. 2008;18:349–363. doi: 10.1002/hipo.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paradis S, et al. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53:217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cerruti Mainardi P. Cri du Chat syndrome. Orphanet J. Rare Dis. 2006;1:33. doi: 10.1186/1750-1172-1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Israely I, et al. Deletion of the neuron-specific protein δ-catenin leads to severe cognitive and synaptic dysfunction. Curr. Biol. 2004;14:1657–1663. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]