Abstract
Carcinogenic DNA viruses such as high-risk human papillomaviruses (HPV) and Epstein-Barr-Virus (EBV) replicate during persistent infections as low-copy-number plasmids. EBV DNA replication is restricted by host cell replication licensing mechanisms. In contrast, copy number control of HPV genomes is not under cellular control but involves the viral sequence-specific DNA-binding E2 activator and E8∧E2C repressor proteins. Analysis of HPV31 mutant genomes revealed that residues outside of the DNA-binding/dimerization domain of E8∧E2C limit viral DNA replication, indicating that binding site competition or heterodimerization among E2 and E8∧E2C proteins does not contribute to copy number control. Domain swap experiments demonstrated that the amino-terminal 21 amino acids of E8∧E2C represent a novel, transferable DNA replication repressor domain, whose activity requires conserved lysine and tryptophan residues. Furthermore, E8∧E2C(1-21)-GAL4 fusion proteins inhibited the replication of the plasmid origin of replication of EBV, suggesting that E8∧E2C functions as a general replication repressor of extrachromosomal origins. This finding could be important for the development of novel therapies against persistent DNA tumor virus infections.
DNA viruses such as certain types of human papillomaviruses (HPV) and Epstein-Barr-virus (EBV), a member of the human herpesvirus family, are of major clinical interest because infections with these viruses have been linked to the development of human malignancies. HPV types 16, 18, 31, and 45 represent the predominant etiological agents of cervical cancer, and EBV infection has been suggested to contribute to the development of a number of lymphomas and nasopharyngeal carcinoma (28, 42, 43). Both HPV and EBV replicate as extrachromosomal elements in the nucleus of human cells. Interestingly, papillomaviruses and EBV maintain their genomes at defined copy numbers (10 to 50 virus copies per EBV-infected cell and 100 virus copies per papillomavirus-infected cell) in cultured cells over long periods of time (1, 3, 10, 16, 39, 40). This behavior in tissue culture is thought to reflect viral DNA replication during persistent infections in vivo.
In the latent phase, EBV DNA replication initiates at oriP, whereas in the lytic (productive) phase, oriLyt is responsible for the replication of EBV DNA (12, 40). In contrast to oriLyt, the activity of oriP requires the viral EBNA1 protein, and viral copy number is kept constant by host cell replication licensing mechanisms, which ensure that eukaryotic genomes are copied exactly once before cell division (1, 40, 41). Most likely, this is achieved by recruiting the human origin recognition complex to oriP (5, 8, 31).
Papillomaviruses use a different strategy to successfully maintain their DNA at defined copy numbers. In contrast to oriP, papillomavirus origins are not subject to replication licensing, but appear to use copy number control mechanisms, which are not fully understood (10). Two viral proteins are essential for the activation of papillomavirus origins of replication. The papillomavirus E1 protein is an origin-binding protein with helicase activity that interacts with host proteins to initiate DNA replication at the viral origin (32). The viral E2 protein is a sequence-specific DNA binding transcriptional modulator that interacts with E1 to enhance its DNA binding properties (23, 32). The analysis of mutant virus genomes revealed that important regulators for the copy number control of papillomavirus DNA replication in the persistent phase are E2 repressor proteins (14, 29, 34). E2 repressor proteins have been demonstrated to counteract transcriptional activation by E2 and to inhibit the E1/E2-dependent replication of papillomavirus origins in reporter assays (4, 6, 14, 15, 21, 34). All E2 repressor proteins lack the amino-terminal domain of E2 responsible for activation of transcription and DNA replication but retain the carboxy-terminal domain that mediates specific DNA recognition and dimerization among E2 proteins (23) (Fig. 1A). These observations gave rise to the model that E2 repressor proteins antagonize E2 by competing with E2 at E2 DNA binding sites (E2BS) and possibly also by the formation of heterodimers (2, 4, 15, 21, 22). This implies that the major determinant for the extent of papillomavirus genome replication is the presence of E2 proteins that are DNA binding and dimerization competent, but do not support origin replication. We have recently demonstrated that the HPV31 E2 repressor protein E8∧E2C, which is generated by alternative splicing and fuses the small E8 open reading frame (ORF) to the carboxy terminus of E2 (Fig. 1A), not only antagonizes E2 but functions on its own as a DNA-binding site-dependent, long-distance transcriptional repressor (LDTR) (36). The LDTR activity requires the E8 part of E8∧E2C, which is highly conserved among HPVs, indicating that this domain might be important for the regulation of HPV replication (36).
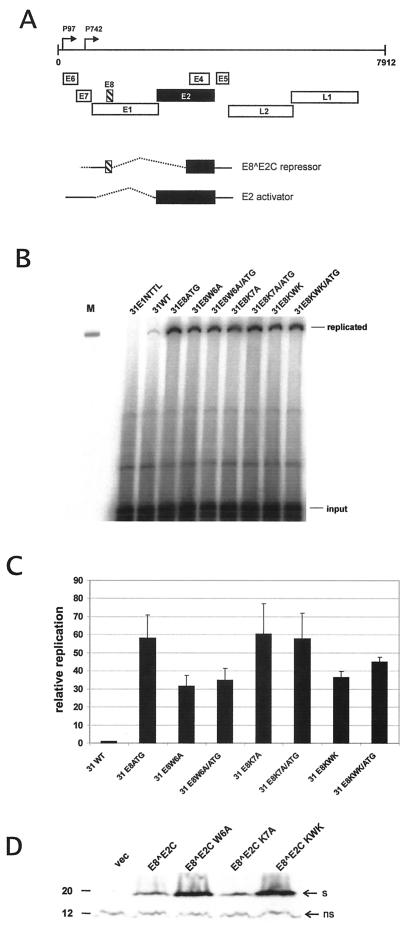
FIG. 1.
Replication properties of HPV31 E8 mutant genomes. (A) The linearized genome of HPV31 (nt 1 to 7912) with the various ORFs (E1 to E8, L1, and L2) and the positions of the major early promoter P97 and the major late promoter P742 are indicated by arrows on top. The E8 and E2 ORFs are highlighted by a striped box and black box, respectively. The structures of the spliced transcripts used to generate the E8Ê2C fusion protein and the full-length E2 protein are shown below. The carboxy terminus, which is part of both proteins, is responsible for dimerization among E2 proteins and sequence-specific DNA recognition (22). (B) Representative Southern blot of a transient replication analysis of HPV31 wild-type (31WT) and mutant genomes. The replication-deficient HPV31-E1NTTL genomeserved as a negative control. The positions of DpnI-resistant (replicated) DNA and DpnI-sensitive (input) DNA are indicated. Signal intensities of input DNA reveal similar transfection and recovery efficiencies for all constructs. One hundred picograms of EcoRI-digested HPV31 DNA was used as a size marker in lane M. (C) Graphic representation of the relative replication levels of HPV31 genomes. Signal intensities of DpnI-resistant DNA were quantified by phosphorimager analysis. The replication levels of the different HPV31 genomes are presented relative to that of wild-type HPV31 (31WT), which was set to 1. Error bars indicate standard deviations derived from data from three to five experiments. (D) Western blot analysis of E8Ê2C wild-type and mutant proteins. 293 cells were transfected with the empty expression vector pSG5 (vec) or expression vectors encoding wild-type E8∧E2C or mutant E8∧E2C W6A, K7A, or KWK proteins. A polyclonal antipeptide rabbit antiserum was used for the detection of E8∧E2C proteins. Bands representing E8∧E2C proteins are indicated by an arrow labeled “s.” A nonspecific band is also indicated (ns) and served as a loading control. A molecular mass marker (in kilodaltons) is shown on the left.
In this report, we demonstrate that the copy number of HPV31 genomes is mainly controlled by a novel activity residing in the amino-terminal 21 amino acids (aa) of E8∧E2C that is dependent on a conserved KWK amino acid motif. Domain swap experiments reveal that this 21-aa domain functions independently of DNA-binding site competition and heterodimerization with E2 to inhibit the DNA replication of the HPV31 origin. Surprisingly, the repression of replication by the E8∧E2C(1-21) domain is not restricted to the HPV31 origin, but also occurs with the EBNA1-dependent replication of oriP, which shares no similarities with the HPV31 origin. Taken together, these findings suggest that the latent and/or persistent replication of DNA viruses can be inhibited at the level of DNA replication by specific proteins.
MATERIALS AND METHODS
Plasmids.
Plasmids pBR322.HPV31, pHPV31-E1N-TTL, and pHPV31-E8ATG have been described previously (33, 34). Mutations were introduced into the HPV31 wild-type genome or the respective mutants by subcloning restriction fragments obtained by overlap-extension PCR using the appropriate synthetic primers and restriction digest. Subcloned fragments were analyzed by DNA sequencing. With exception of the HPV31 E8ATG mutation, which is silent in E1, the mutations E8W6A, E8K7A, and E8KWK also lead to amino acid changes in the overlapping E1 gene (Fig. 1A). In pHPV31-E8W6A, E1 residue 138 is changed from V to G; in pHPV31-E8K7A, E1 residue 139 is changed from E to G; and in pHPV31-E8KWK, E1 residues 137, 138, and 139 are changed from E to S, V to G, and E to S, respectively. Eukaryotic expression plasmids for HPV31 E1 (pSG-31E1), E2 (pSX-E2), E8∧E2C (pSG-E8∧E2C), and E8∧E2C KWK (pSG-E8∧E2C KWK) are all based on pSG5 (Stratagene, Amsterdam, The Netherlands) and have been described previously (9, 36). The GAL4 DNA binding domain (DBD) (aa 1 to 147) was amplified by PCR with plasmid pM (Clontech, Heidelberg, Germany) as a template and cloned into the BglII site of a modified pSG5 plasmid that contains an additional SmaI restriction site, giving rise to pSG-GAL4. N-terminal in-frame fusions between parts of E8∧E2C and the GAL4 DBD were constructed by subcloning EcoRI fragments from pSG E8∧E2C and pSG E8∧E2C KWK or by inserting double-stranded oligonucleotides encoding the respective amino acids in the EcoRI site of pSG-GAL4, giving rise to the expression vectors pSG-E8(1-12)-GAL4, pSG-E8∧E2C(1-21)-GAL4, pSG-E8∧E2C(1-37)-GAL4, and pSG-E8∧E2C(1-37) KWK-GAL4. The luciferase reporter vector pC18-SP1-4xGAL4-luc was constructed by insertion of a double-stranded oligonucleotide containing four GAL4 binding sites (4xGAL4; 5′-AGCTCGGAAGACTCTCCTCCGACGGAAGACTCTCCTCCGACGGAAGACTCTCCTCCGACGGAAGACTCTCCTCCGA-3′) in the HindIII site of plasmid pC18-SP1-luc (36). The HPV31 origin reporter pGL31URR has been described previously (33). To construct plasmid pGL31URR-4xGAL4, the 4xGAL4 binding site oligonucleotide was inserted into the HindIII site of pGL31URR. Plasmid pGL31BS2,3,4-4xGAL4 was constructed by PCR amplification of HPV31 nucleotides (nt) 7875 to 78 by using the appropriate synthetic primers and replacement of the MluI-XhoI fragment of pGL31URR-4xGAL4. The EBV oriP reporter plasmid pGL3-oriP-4xGAL4 was constructed by replacing the HPV31 origin sequence in pGL31URR-4xGAL4 with a fragment representing oriP excised from plasmid pMEP4 (Invitrogen, Karlsruhe, Germany) by digestion with AccI and XbaI digest followed by Klenow polymerase fill-in. The EBNA1 expression plasmid p1563 was kindly provided by B. Sugden.
Cell culture.
Normal human foreskin keratinocytes (NHK) and SCC13 cells, a human squamous carcinoma cell line, were cultured as described previously (33). HeLa and 293 cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal calf serum and antibiotics.
Transient luciferase expression assay.
Approximately 1.5 × 105 HeLa or SCC13 cells were seeded into 35-mm-diameter dishes the day before transfection. Cells were cotransfected with 200 ng of luciferase reporters, 30 ng of pSG5, or the respective expression vector DNA as indicated in the figure legends, and 30 ng of pSG5 or pSG-GAL4 was added to reach equal amounts of expression plasmids. Transfections were carried out with 5 μl of Lipofectamine (Invitrogen) and OptiMEM (Invitrogen, Karlsruhe, Germany) according to the manufacturer′s recommendations. Luciferase assays were carried out 48 h after transfection as described previously. The data presented in the figures are the average of at least three independent transfection experiments.
Transient DNA replication assay.
Plasmids (4 μg) containing the various HPV31 genomes were digested with EcoRI to release the viral genome from the cloning vector and then religated under dilution conditions (10 μg/ml) to facilitate intramolecular ligation. After ethanol precipitation, ligation efficiency of the products was monitored by agarose electrophoresis, and equal amounts of DNA were then transfected into 7 × 105 NHK cells grown in 60-mm-diameter dishes with 15 μl of Lipofectamine and OptiMem (Invitrogen). The next day, cells were transferred onto 100-mm-diameter dishes and were grown in the presence of mitomycin C-treated NIH 3T3 feeder cells. Low-molecular-weight DNA was isolated 96 h posttransfection by proteinase K digestion and high-salt precipitation as previously described (35). Experiments were carried out at least three times with different DNA preparations to ensure reproducibility. Transient replication experiments with HPV31 origin plasmids were carried out in SCC13 cells. Cells (5 × 105) were transfected with Lipofectamine and OptiMem and received 200 ng of the respective origin reporter plasmids, 1,000 ng of HPV31 E1 expression vector, 100 ng of HPV31 E2 expression vector, and 300 ng of expression plasmids coding for GAL4 DBD, E8∧E2C-GAL4 fusion proteins, or wild-type and mutant E8∧E2C proteins. For each transfection experiment, a mixture of the corresponding origin reporter plasmid and expression vectors for the replication activator proteins E1 and E2 was prepared to ensure equal distribution of the three plasmids among different dishes. Expression plasmids for GAL4 DBD, E8∧E2C-GAL4 fusion proteins, or wild-type E8∧E2C were then added individually to the transfection mixtures. To ensure reproducibility, experiments were carried out at least four times with different plasmid preparations. HeLa cells (5 × 105) were cotransfected with 2 μg of pGL3-oriP-4xGAL4, 200 ng of p1563 (EBNA1), and 500 ng of pSG-GAL4 or the respective expression plasmids, as indicated in the figure legends. The next day, cells were transferred into 100-mm-diameter dishes and grown for an additional 120 h. Low-molecular-weight DNA was purified as described previously (34, 35). Aliquots of the isolated DNA were analyzed by analytical agarose gels to control for DNA recovery. Equal amounts of DNA were digested with DpnI and an appropriate enzyme to linearize the replication reporter and then analyzed by Southern blot hybridization and phosphorimaging as described previously (34, 35). For quantification, only phosphorimager results that were in the linear range were used, as indicated by pixel analysis with the AIDA software package, version 2.1 (Raytest, Berlin, Germany).
RPA.
Approximately 1.5 × 106 NHK cells were lipofected with 6 μg of ligated HPV31 genomes. Total RNA was isolated with Trizol reagent (Invitrogen) 48 h after transfection. Poly(A)+ RNA was isolated from total RNA by using the Oligotex mRNA purification system (Qiagen, Hilden, Germany). Antisense riboprobe was synthesized from pRP742 as described previously (35). RNase protection analysis (RPA) was performed with the RPA III kit (Ambion, Huntingdon, United Kingdom) with 2 μg of poly(A)+ RNA and 1 × 105 cpm of 32P-labeled antisense probe per sample. Samples were separated on a 5% denaturing polyacrylamide gel and quantitated by phosphorimaging analysis.
Western blot analysis.
Transfected 293 or HeLa cells were lysed in sodium dodecyl sulfate (SDS)-polyacrylamide sample buffer by boiling to 95°C for 5 min, sonicated, and then separated in an SDS-15% polyacrylamide gel. Proteins were transferred to nitrocellulose membrane (Protran; pore size, 0.2 μm; Schleicher & Schuell, Dassel, Germany) in a buffer containing 10 mM CAPS [3-(cyclohexylamino)-1-propanesulfonic acid] (pH 10.3) and 10% methanol at 70 V for 1 h. The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline-0.1% Tween 20 (MTBST) for 1 h. The membrane was incubated overnight at 4°C with anti-GAL4 DBD antibody (sc-577; Santa Cruz Biotechnology, Heidelberg, Germany) diluted 1:3,000 in MTBST to detect GAL4 proteins, with the monoclonal antibody 1H4 diluted 1:100 in MBTST to detect EBNA1 (11), or with an affinity-purified polyclonal rabbit antiserum generated against a peptide consisting of aa 44 to 57 of the HPV31 E8∧E2C protein (Sigma Genosys, United Kingdom) diluted 1:1,000 in MBTST. Bound antibodies were detected by adding antirabbit immunoglobulin G (IgG) (GAL4, E8∧E2C) or anti rat-IgG (EBNA1) antibodies conjugated to horseradish peroxidase (Dako, Hamburg, Germany) and incubating this mixture for 1 h. Bound antibodies were detected by chemiluminescence with SuperSignal reagents (Perbio, Bonn, Germany).
RESULTS
HPV31 mutants encoding LDTR-deficient E8∧E2C proteins display an overreplication phenotype.
To gain insight into the molecular mechanisms of the inhibition of the HPV31 DNA replication by E8∧E2C, we investigated the contribution of E8∧E2C LDTR activity to viral copy number control. We made use of the fact that mutations of residues K5, W6, and K7 in E8∧E2C genetically separated the LDTR activity of E8∧E2C from sequence-specific DNA-binding activity, repression of the viral early promoter P97 through promoter-proximal E2BS, and inhibition of transcriptional transactivation by E2 (36). The corresponding mutations were introduced by site-directed mutagenesis into the HPV31 genome, giving rise to HPV31-E8W6A, HPV31-E8K7A, and HPV31-E8KWK. The HPV31 mutant genomes were transfected in normal human keratinocytes, the natural host cell for HPV, and their replication activities were compared to those of wild-type HPV31 DNA and HPV31 E8ATG mutant DNA. HPV31 E8ATG genomes carry a mutation of the E8∧E2C start codon and are therefore unable to express the E8∧E2C protein. The HPV31 E8ATG genome displays an overreplication phenotype in short-term assays both in normal and immortalized human keratinocytes (34). Low-molecular-weight DNA was isolated 96 h posttransfection, and DpnI-resistant (newly replicated) and DpnI-sensitive (input) viral DNA was detected by Southern blotting (Fig. 1B). A replication-deficient E1 mutant genome served as a negative control (Fig. 1B, 31E1NTTL) (35). In line with previously published data (34), the quantitation of several experiments revealed that the HPV31 E8ATG mutant genome showed 59-fold-higher levels of newly replicated DNA compared to wild-type HPV31 (31WT) genomes (Fig. 1C). The HPV31-E8K7A mutant displayed a 60-fold increase in DNA replication over wild-type levels, similar to the higher levels of replication seen in the HPV31 E8ATG mutant (Fig. 1C). The mutant genomes HPV31-E8W6A and HPV31-E8KWK showed 32- and 37-fold-increased replicated DNA levels (Fig. 1C). To further investigate whether the reduced replication activity of the HPV31-E8 W6A and the HPV31-E8KWK mutants relative to that of the HPV31-E8ATG and HPV31-E8K7A mutants was only due to the mutations in E8 or whether the additional mutations in the overlapping E1 gene were responsible for this (described in Materials and Methods), we constructed HPV31 E8∧E2C knockout genomes by combining the ATG mutation, which is silent in E1 (34), with the W6A, K7A, or KWK mutations (HPV31-E8W6A/ATG, HPV31-E8K7A/ATG, and HPV31-E8KWK/ATG). A comparison of the transient replication capacities of the HPV31-E8W6A/ATG and HPV31-E8KWK/ATG genomes to the HPV31-E8ATG genome revealed that the double-mutant genomes replicated to lower levels (35- and 45-fold, respectively; Fig. 1B and C). This strongly suggested that the replication capacities of the HPV31-E8W6A and HPV31-E8KWK genomes are indeed increased by the E8 mutations but reduced by the mutations in the overlapping E1 ORF, resulting in an intermediate phenotype. Examination of the protein levels of wild-type and mutant E8∧E2C proteins in transfected 293 cells by immunoblotting revealed that the E8∧E2C K7A mutant protein was expressed at wild-type levels, whereas the amounts of the E8∧E2C W6A and KWK mutant proteins were even increased (Fig. 1D). Taken together, this analysis revealed that HPV31 genomes, which encode E8∧E2C proteins that are LDTR deficient but DNA binding and dimerization competent, replicate essentially as well as E8∧E2C knockout genomes. This suggested that mainly the LDTR activity of E8∧E2C and not binding site competition and heterodimerization among E2 proteins controls viral DNA replication in the persistent phase.
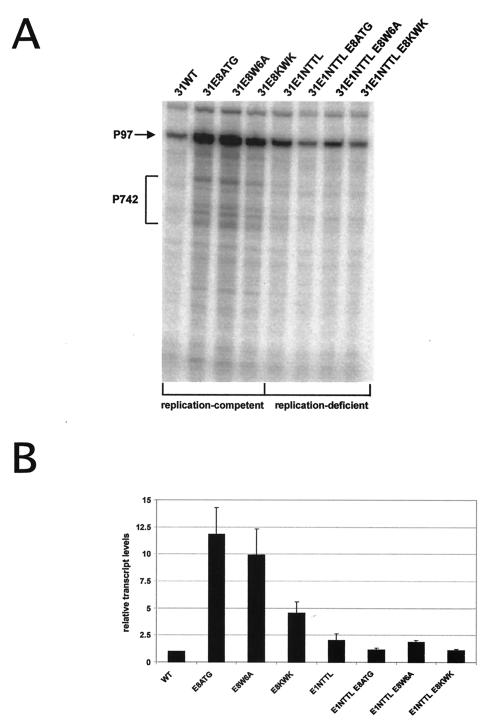
We then investigated whether HPV31 E8 LDTR mutant genomes display an increased viral gene expression due to the loss of the transcriptional repression by E8∧E2C. mRNA was isolated from normal human keratinocytes transiently transfected with wild-type HPV31 or mutant genomes (HPV31-E8ATG, HPV31-W6A, and HPV31-KWK), and the amounts of viral transcripts expressed from different HPV31 genomes were analyzed by RPA experiments. An antisense RNA probe was used that allowed simultaneous detection of viral transcripts initiated at the major early (P97) or the major late (P742) promoters. HPV31-E8ATG, HPV31-E8 W6A, and HPV31-E8 KWK mutant genomes displayed increased P97 transcript levels and also showed an induction of P742-specific transcripts compared to HPV31 wild-type genomes (Fig. 2A). The elevation in transcript levels could be a consequence of the increased viral copy number. If E8∧E2C functions primarily as a transcriptional repressor of viral promoters to restrict the amount of viral replication proteins, then HPV31 E8 LDTR mutant genomes should also display increased viral transcription in the absence of viral DNA replication. Therefore, replication-deficient E8 mutant genomes were created by introduction of a stop codon into E1 (HPV31-E1N-TTL, Fig. 1B), resulting in HPV31-E1N-TTL/E8ATG, HPV31-E1N-TTL/E8W6A, and HPV31-E1N-TTL/E8KWK (Fig. 2A). Quantification of the spliced P97 transcripts revealed that the replication-deficient HPV31-E1NTTL genome displayed slightly elevated transcript levels compared to the replication-competent HPV31 wild type. However, when comparing the replication-competent E8 mutant genomes HPV31-E8ATG, HPV31-E8W6A, and HPV31-E8KWK with their replication-deficient counterparts HPV31-E1NTTL/E8ATG, HPV31-E1NTTL/E8W6A, and HPV31-E1NTTL/E8KWK, a 5- to 10-fold drop in transcript levels was observed (Fig. 2B). The transcript levels of these mutants were similar to the amounts of transcripts produced by the HPV31-E1NTTL genome (Fig. 2B), which strongly suggested that the increase in transcript levels of the replication-competent HPV31 E8 LDTR mutant genomes was a consequence of increased DNA replication.
FIG. 2.
Analysis of viral transcripts from replication-competent and -deficient HPV31 genomes. (A) Representative RPA of mRNA isolated from normal human keratinocytes transiently transfected with different HPV31 genomes. The antisense probe spans HPV31 nt 678 to 919. The positions of transcripts initiated at the major early promoter P97 and spliced at the splice donor site at nt 877 or initiated at the major late promoter P742 are indicated by an arrow and a bracket, respectively. (B) Graphic representation of relative transcript levels initiated at P97 and spliced at nt 877. Transcript levels were quantitated by phosphorimager analysis. Levels are presented relative to HPV31 wild-type (WT) transfected cells, which were set to 1. Error bars indicate standard deviations derived from three independent experiments.
The amino-terminal 21 aa of E8∧E2C represent a transferable transcriptional repression domain.
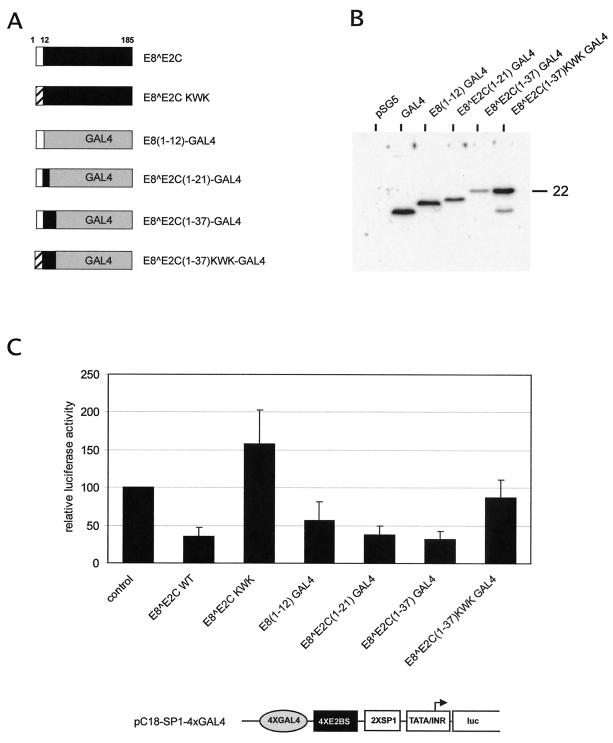
The finding that the DNA overreplication of HPV31 E8 LDTR mutant genomes was not caused by increased viral transcription pointed to the possibility that E8Ê2C directly influences DNA replication via its E8 domain. To address this experimentally, it was necessary to establish a system that allowed us to measure the influence of E8∧E2C independently of binding site competition and heterodimer formation among E2 and E8Ê2C proteins. To achieve this, we replaced the DNA recognition and dimerization domain of E8∧E2C, which is localized in the carboxy-terminal 100 aa of E2 proteins, with the DNA binding/dimerization domain (DBD) of the yeast GAL4 protein and generated fusions with the amino-terminal 12, 21, or 37 aa of E8∧E2C [E8(1-12)-GAL4, E8∧E2C(1-21)-GAL4, and E8∧E2C(1-37)-GAL4] or E8∧E2C KWK [E8∧E2C(1-37)KWK-GAL4] (Fig. 3A). First, we controlled the expression of full-length fusion proteins by Western blot analyses. 293 cells were transiently transfected with the empty expression vector pSG5 or expression vectors coding for the GAL4 DBD (pSG-GAL4) or the respective E8-GAL4 fusion proteins [pSG-E8(1-12)-GAL4, pSG-E8∧E2C(1-21)-GAL4, pSG-E8∧E2C(1-37)-GAL4], and pSG-E8∧E2C(1-37)KWK-GAL4). Protein extracts were prepared 48 h posttransfection, and GAL4 fusion proteins were detected by a GAL4 DBD-specific antiserum. This demonstrated that all fusion proteins were present at detectable levels (Fig. 3B). Next, the effects of the different E8∧E2C-GAL4 fusion proteins on transcription were evaluated by transient transfection assays (Fig. 3C). To be able to compare the activity of the E8-GAL4 fusion proteins with that of the wild-type E8∧E2C protein, a luciferase reporter plasmid was constructed, which consists of four multimerized GAL4 binding sites (4xGAL4) and four multimerized E2 binding sites (4xE2BS) upstream of SP1 binding sites (SP1) and a minimal promoter sequence from the adenovirus major late promoter (pC18-SP1-4xGAL4-luc, Fig. 3C). The reporter plasmid was cotransfected with expression vectors for GAL4-DBD, E8(1-12)-GAL4, E8∧E2C(1-21)-GAL4, E8∧E2C(1-37)-GAL4, E8∧E2C(1-37)KWK-GAL4, E8∧E2C, or E8∧E2C KWK into a human keratinocyte cell line (SCC13) and analyzed for luciferase activity (Fig. 3C). E8∧E2C(1-21)-GAL4 and E8∧E2C(1-37)-GAL4 inhibited reporter activity to a similar extent as E8∧E2C, and this was dependent on the presence of an intact KWK motif in both E8∧E2C and E8∧E2C(1-37)-GAL4. E8(1-12)-GAL4 was slightly less active and inhibited reporter activity to 64%. Taken together, these results suggested that residues 1 to 12 of E8∧E2C present a minimal transferable transcriptional repression domain whose activity is dependent on the KWK motif within E8 (Fig. 3C).
FIG. 3.
E8∧E2C-GAL4 fusion proteins act as transcriptional repressors. (A) Schematic representation of the structure of HPV31 E8∧E2C and E8∧E2C-GAL4 fusion proteins. The E8 part is depicted in white, the E2C part is depicted in black, and the GAL4 DBD is depicted in gray. The LDTR-deficient E8∧E2C KWK mutant protein was generated by changing residues 5 to 8 from KWK to AEA, and this is indicated by a striped box. Identical mutations were introduced into E8∧E2C(1-37) KWK-GAL4. Either 12, 21, or 37 amino-terminal residues of E8∧E2C or E8∧E2C KWK were fused to aa 1 to 147 of the DBD of the yeast transcription factor GAL4. (B) Western blot analysis of extracts isolated from transiently transfected 293 cells. Cells were transfected with the empty expression vector pSG5 or with plasmids expressing unfused or E8∧E2C-GAL4 proteins and analyzed with an antibody specific for the GAL4 DBD. A molecular mass marker is indicated to the right in kilodaltons. (C) SCC13 cells were cotransfected with 30 ng of expression vectors for E8∧E2C or E8∧E2C KWK, E8(1-12)-GAL4, E8∧E2C(1-21)-GAL4, E8∧E2C(1-37)-GAL4, E8∧E2C(1-37) KWK-GAL4, and 200 ng of the pC18-SP1-4xGAL4-luc luciferase reporter plasmid, respectively. In addition, 30 ng of pSG5 was included when transfecting expression vectors for GAL4 fusions, and 30 ng of pSG-GAL4 was added when E8∧E2C expression vectors were used. The average relative luciferase activities were calculated with respect to the activity of each construct in the presence of the parental pSG5 expression vector, which was set to 1. The luciferase activities are presented relative to the activity of pC18-SP1-4xGAL4-luc cotransfected withboth 30 ng of pSG5 and 30 ng of pSG-GAL4 expression vectors, which was set to 1 (control). Standard deviations are indicated by error bars. A Student′s t test analysis revealed that the observed differences between the control and E8(1-12)-GAL4 as well as between E8∧E2C(1-37)-GAL4 and E8∧E2C(1-37) KWK-GAL4 are statistically significant (P < 0.005). The structure of the promoter region of the luciferase reporter plasmid pC18-SP1-4xGAL4-luc is shown below the graph. Transcriptional control elements representing four GAL4 binding sites (4xGAL4), four E2BS (4xE2BS), two SP1 binding sites (2xSP1), the adenovirus major late promoter TATA-box/initiator element (TATA/Inr), and the RNA initiation site for the luciferase (luc) RNA are indicated.
E8∧E2C-GAL4 fusion proteins inhibit replication of the HPV31 origin independently of binding site competition or heterodimerization with E2.
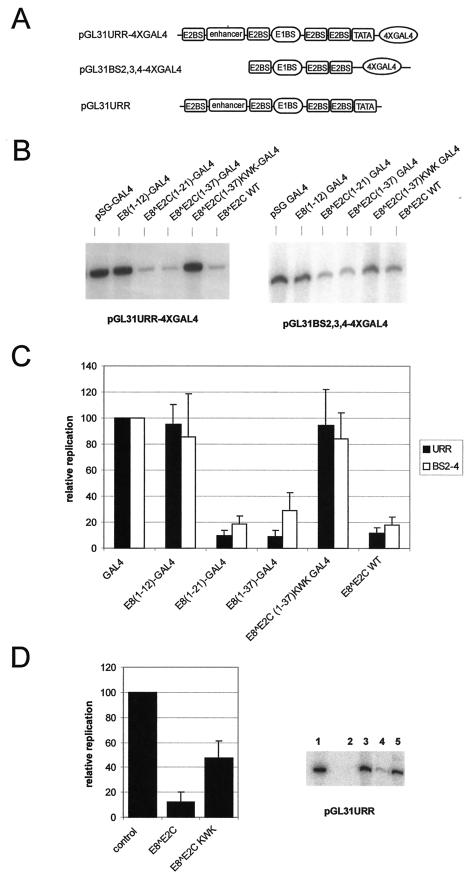
Having demonstrated that E8∧E2C and E8∧E2C-GAL4 fusion proteins are functionally equivalent in transcriptional repression assays, we next analyzed the influence of E8∧E2C-GAL4 fusion proteins on HPV31 origin replication. To target the different GAL4 fusion proteins to the HPV31 origin of replication, four GAL4 binding sites were introduced into the HPV31 replication reporter plasmid pGL31URR, giving rise to pGL31URR-4xGAL4 (Fig. 4A). The replication reporter plasmid was cotransfected with expression vectors for HPV31 E1 and E2 and the different expression vectors for GAL4 proteins into SCC13 keratinocytes, and the ability of the reporter plasmid to replicate was evaluated 72 h posttransfection by resistance to digestion with DpnI followed by Southern blotting analysis (Fig. 4B). Surprisingly, cotransfection of E8∧E2C(1-21)-GAL4 and E8∧E2C(1-37)-GAL4 reduced the E1/E2-dependent replication of the HPV31 origin to 10% compared to that of GAL4 DBD alone, and this extent was similar to the repression achieved by the wild-type E8∧E2C protein (Fig. 4C). The replication repression activity of E8∧E2C(1-37)-GAL4 required the presence of the KWK motif, because E8∧E2C(1-37)KWK-GAL4 was completely inactive. However, E8(1-12)-GAL4, despite having an intact KWK motif, displayed no replication inhibition activity (Fig. 4B and C). The replication reporter construct pGL31URR-4xGAL4 contains in addition to E1 and E2 binding sites, which are essential for origin function, transcriptional regulatory elements such as enhancer and the P97 promoter sequences. To address the question of whether the inhibitory effect of E8∧E2C-GAL4 proteins on DNA replication was mediated via transcriptional elements, a replication reporter plasmid was used that only retained the E1 binding site and the flanking E2 binding sites 2 to 4 (pGL31BS2,3,4-4xGAL4, Fig. 4A). Transient replication experiments were carried out under the same conditions as described for plasmid pGL31URR-4xGAL4. The E1/E2-dependent replication of pGLBS2,3,4-4xGAL4 was inhibited by E8∧E2C(1-21)-GAL4 and E8∧E2C(1-37)-GAL4, but not by E8(1-12)-GAL4 or E8∧E2C(1-37)KWK-GAL4, identically to the replication of pGL31URR-4xGAL4 (Fig. 4C). Next, the influence of the KWK mutation in the context of the full-length E8∧E2C protein on replication control was investigated. SCC13 cell were transfected with the HPV31 replication reporter plasmid pGL31URR (Fig. 4A), expression vectors for E1 and E2 and either the empty vector pSG5 or expression plasmids for the E8∧E2C wild-type or the E8∧E2C KWK mutant protein. Quantification of seven independent experiments revealed significant differences between E8∧E2C and E8∧E2C KWK. Whereas E8∧E2C inhibited origin replication to 12% of the control level, E8∧E2C KWK was fourfold less active in reducing replication (48% of the control; Fig. 4D). This suggested that the amino-terminal domain of E8∧E2C also plays an important role in replication control under conditions in which binding site competition and heterodimerization among E2 proteins can occur. Taken together, these data suggest that E8∧E2C residues 1 to 21 are sufficient for a novel DNA replication inhibition activity, which requires the conserved KWK motif within E8 that is also essential for the transcriptional repression activity. This suggests that the replication repression activity of E8∧E2C(1-21) functions independently of binding site competition and heterodimer formation with E2 and explains the overreplication phenotype observed with the HPV31-E8W6A, K7A, and KWK genomes
FIG.4.
E8∧E2C-GAL4 fusion proteins inhibit replication of the HPV31 origin. (A) Schematic structure of the replication origins of the different replication reporter plasmids. Binding sites for GAL4 (4xGAL4), E2 (E2BS), E1 (E1BS), and TATA-box binding (TATA) proteins, as well as the enhancer region of HPV31, which is composed of binding sites for cellular transcription factors, are shown. (B) Representative Southern blot analysis of the transient replication of the HPV31 origin of replication. SCC13 cells were transfected with either reporter plasmid pGL31URR-4xGAL4 (URR) or pGL31BS2,3,4-4xGAL4 (BS2-4) and expression vectors for HPV31 E1 and E2. In addition, cells received expression vectors for the GAL4 DBD (GAL4), the different E8∧E2C-GAL4 fusions, or wild-type E8∧E2C. (C) The levels of replicated reporter plasmids were analyzed by Southern blot analysis and quantitated by phosphorimager analysis. The replication levels of the different plasmid combinations are presented relative to the replication level of the origin reporter plasmids in the presence of E1, E2, and GAL4, which was set to 1. Error bars indicate standard deviations obtained from six independent experiments. A Student′s t test analysis revealed that the observed differences between E8(1-12)-GAL4 and E8∧E2C(1-21)-GAL4 as well as between E8∧E2C(1-37)-GAL4 and E8∧E2C(1-37) KWK-GAL4 are statistically significant (P < 0.005). (D) Southern blot analysis of the transient replication of the HPV31 origin in SCC13 cells. A representative Southern blot is shown on the right. Cells were either transfected with the reporter plasmid pGL31URR alone (lane 2) or together with expression vectors for HPV31 E1 and E2 (lanes 3 to 5). In addition, cells received expression vectors (300 ng) for wild-type E8∧E2C (lane 4) or the E8∧E2C KWK mutant (lane 5). Lane 1 received 100 pg of linearized pGLURR31 plasmid and served as a size marker. Data were quantitated by phosphorimager analysis and are presented in the graph on the right. Replication levels are presented relative to the HPV31 origin replication in the presence of E1 and E2 alone (control). Error bars indicate standard deviations obtained from seven independent experiments. Student′s t test analysis revealed that the difference between E8∧E2C and E8∧E2C KWK is statistically significant (P < 0.005).
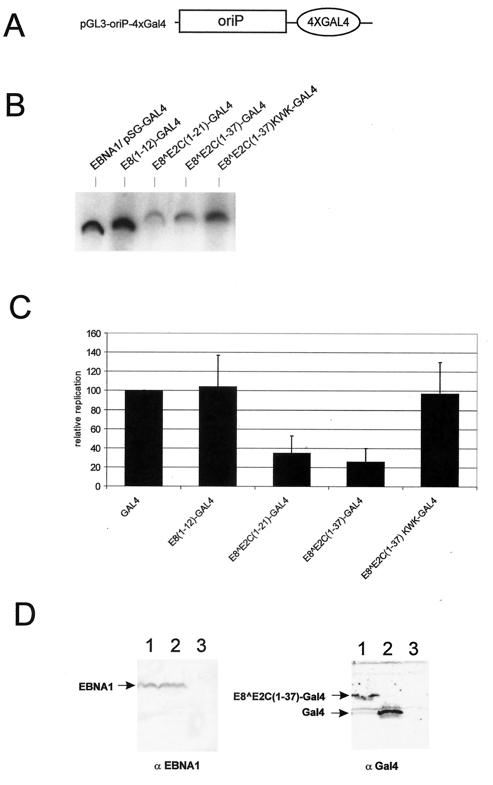
E8∧E2C-GAL4 fusion proteins inhibit replication of the EBV oriP origin.
To address the question of whether the repression of DNA replication activity by E8∧E2C is specific for the papillomavirus origin of replication, we extended our analysis to the EBV latency origin of replication, oriP. Similar to the HPV origin, oriP functions in an extrachromosomal setting and requires a virally encoded protein, EBNA1, for its activity (39, 41). However, in contrast to the HPV origin, replication at oriP initiates exactly once per cell cycle identically to cellular origins of replication, and this is most likely due to an involvement of the human origin recognition complex and MCM proteins (5, 8, 31). We constructed an oriP replication reporter plasmid, which contained four GAL4 binding sites (pGL3-oriP-4xGAL4, Fig. 5A) and analyzed its replication activity after transfection of HeLa cells together with an EBNA1 expression plasmid and different GAL4 expression plasmids (Fig. 5B). Identical to the HPV31 origin, EBNA1-dependent replication of pGL3-oriP-4xGAL4 was efficiently inhibited by E8∧E2C(1-21)-GAL4 and E8∧E2C(1-37)-GAL4, but was not repressed by GAL4 DBD, E8(1-12)-GAL4, or E8∧E2C(1-37)KWK-GAL4 (Fig. 5C). Western blot analyses demonstrated that the amounts of EBNA1 protein were similar in the presence of GAL4 and E8∧E2C(1-37)-GAL4, which suggests that the absence of newly replicated DNA is not caused by reduced amounts of EBNA1 (Fig. 5D). In summary, these data strongly suggest that we have identified a novel DNA replication repression activity of the HPV E8∧E2C protein that is able to inhibit two differentially regulated extrachromosomal origins of replication of human DNA tumor viruses.
FIG. 5.
E8∧E2C-GAL4 fusion proteins inhibit replication of the EBV oriP. (A) Schematic structure of the origin region of the replication reporter plasmid pGL3-oriP-4xGAL4. It consists of the oriP region from EBV, which is composed of multiple binding sites for EBNA1, and four binding sites for GAL4 (4xGAL4). (B) HeLa cells were transfected with the reporter plasmid pGL3-oriP-4xGAL4 and an expression vector for EBNA1. In addition, cells received expression vectors encoding the GAL4 DBD (pSG-GAL4), or the different E8-GAL4 fusions. The levels of DpnI-resistant (replicated) reporter plasmids were analyzed by Southern blot analysis and quantitated by phosphorimager analysis. (C) The replication levels of the different plasmid combinations are presented relative to the replication level of the origin reporter plasmids in the presence of EBNA1 and GAL4, which was set to 1. Error bars indicate standard deviations obtained from four independent experiments. (D) Western blot analysis of transiently transfected HeLa cells. Cells were mock transfected (lane 3) or transfected with the EBNA1 expression plasmid (lanes 1 and 2) together with pSG-GAL4 (lane 2) or pSG E8∧E2C(1-37)-GAL4. Equivalent amounts of lysate were separated by SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. Blots were probed with either the monoclonal antibody 1H4 (αEBNA1) or a polyclonal antiserum specific for the GAL4 DBD (αGal4).
DISCUSSION
It is well established that transcription is controlled both positively and negatively by DNA binding factors in eukaryotes. In contrast, sequence-specific DNA binding proteins have been mainly implicated in the activation of replication origins of viruses and of lower eukaryotes (24, 26). The data presented in this paper strongly suggest that the hitherto unknown biological function of the HPV E8∧E2C protein is the repression of viral DNA replication. The analysis of HPV31 genomes that encode mutant E8∧E2C proteins surprisingly suggested that binding site competition and heterodimerization among E2 activator and repressor proteins are not sufficient to restrict replication of the HPV31 genome. The E8 domain also contributed significantly to the inhibition of replication under experimental conditions in which binding site competition and heterodimerization can occur (Fig. 1C and 4D). In line with this new model, repression of DNA replication could also be detected when the amino-terminal 21 aa of E8∧E2C were fused to the heterologous DBD of the GAL4 transcription factor. Surprisingly, the DNA replication repression activity was not restricted to the HPV31 origin but also inhibited the replication of oriP of EBV, which is subject to replication licensing by the host cell.
The inhibition of DNA replication by transcription factors in higher eukaryotes has only been described for viral origins so far. The TATA box binding protein (TBP) inhibits in vitro DNA replication of simian virus 40 (SV40) by interacting with T antigen, thus preventing unwinding of the origin (13). The replication of HPV11 can be inhibited in a DNA binding site-independent manner by interaction of the YY1 transcription factor with the HPV11 E2 protein (17). The inhibition of both the HPV31 origin and oriP of EBV by E8∧E2C makes it unlikely that E8∧E2C directly interferes with the viral replication activator protein HPV31E1 or -E2 on one hand and EBV EBNA1 on the other, because they do not share significant sequence homologies. Also, the p53 protein has been demonstrated to inhibit replication of the bovine papillomavirus 1 (BPV1) origin in vivo and of polyomavirus DNA in vitro but not in vivo (19, 25). However, p53 does not repress replication from oriP in vivo; therefore, it is unlikely that E8∧E2C and p53 achieve origin inhibition by the same mechanism (19).
We initially characterized the E8∧E2C protein as a transcriptional repressor that, in contrast to E2, was able to inhibit transcription over a long distance (36). Interestingly, both the transcriptional and replication repression activities are dependent upon the conserved KWK motif within E8. However, several observations argue against a model in which the repression of replication is a consequence of transcriptional repression. First, replication-deficient HPV31 E8 mutant genomes displayed no changes in transcript levels (Fig. 2). Second, replication of papillomavirus origins does not require transcription control elements, such as promoter or enhancer sequences (7, 27, 30, 37). In vitro replication of BPV1 and HPV11 DNA does not require RNA polymerase II activity (38), and we demonstrated that E8∧E2C-GAL4 fusion proteins inhibited DNA replication to similar extents in the presence and absence of promoter and enhancer elements (Fig. 4). Finally, no promoter activity within the EBV oriP sequence has been described (18). Furthermore, transcriptional and replication repression activities are not completely overlapping, because E8(1-12)-GAL4 was able to inhibit transcription but not replication. Nevertheless, our data do not completely rule out the possibility that the E8∧E2C(1-21) domain regulates the transcription of cellular genes involved in replication control. However, inhibition of DNA replication occurred with both the E8∧E2C(1-21) domain in the context of the homologous DBD and fused to a heterologous DBD. Because it seems unlikely that E2BS and GAL4BS are both present in the promoter region of the same cellular gene, this model would require that cellular genes are regulated by E8∧E2C(1-21) without sequence-specific binding to DNA. So far, transcriptional repression by E8∧E2C has only been detected in the presence of E2BS (26). Thus, we conclude that the replication repression activity most likely uses a novel mechanism. For instance, E8∧E2C may directly interfere with components of the cellular DNA replication machinery that may be common to the replication of both viral episomes. Alternatively, E8∧E2C may influence the structure of the origin. It has been demonstrated that packaging of BPV1 DNA into nucleosomes prevents in vitro DNA replication, indicating that chromatin structure can repress DNA replication (20). Because the activation of replication by transcription factors also involves the modulation of chromatin structure, it seems possible that inhibition of DNA replication by E8∧E2C may occur via chromatin remodeling (24, 26).
No specific antiviral treatments for the precancers or the malignancies induced by HPV types such as HPV31 and EBV are yet available. Our results suggest that it is possible to interfere with the DNA replication of these viruses even in the presence of the respective replication activator proteins. Elucidation of the molecular mechanisms of E8∧E2C that mediate the inhibition of viral DNA replication might therefore result in novel approaches to the treatment of viral infections.
Acknowledgments
We thank B. Sugden and A. Schepers for providing reagents and E. Straub for excellent technical assistance.
This work was funded by a grant from the Deutsche Forschungsgemeinschaft to F.S. (Stu218/2-3).
REFERENCES
- 1.Adams, A. 1987. Replication of latent Epstein-Barr virus genomes in Raji cells. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barsoum, J., S. S. Prakash, P. Han, and E. J. Androphy. 1992. Mechanism of action of the papillomavirus E2 repressor: repression in the absence of DNA binding. J. Virol. 66:3941-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedell, M. A., J. B. Hudson, T. R. Golub, M. E. Turyk, M. Hosken, G. D. Wilbanks, and L. A. Laimins. 1991. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J. Virol. 65:2254-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvard, V., A. Storey, D. Pim, and L. Banks. 1994. Characterization of the human papillomavirus E2 protein: evidence of trans-activation and trans-repression in cervical keratinocytes. EMBO J. 13:5451-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choe, J., P. Vaillancourt, A. Stenlund, and M. Botchan. 1989. Bovine papillomavirus type 1 encodes two forms of a transcriptional repressor: structural and functional analysis of new viral cDNAs. J. Virol. 63:1743-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demeret, C., M. Le Moal, M. Yaniv, and F. Thierry. 1995. Control of HPV 18 DNA replication by cellular and viral transcription factors. Nucleic Acids Res. 23:4777-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 9.Frattini, M. G., and L. A. Laimins. 1994. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology 204:799-804. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, D. M., and S. N. Cohen. 1987. Bovine papilloma virus plasmids replicate randomly in mouse fibroblasts throughout S phase of the cell cycle. Cell 50:59-68. [DOI] [PubMed] [Google Scholar]
- 11.Grasser, F. A., P. G. Murray, E. Kremmer, K. Klein, K. Remberger, W. Feiden, G. Reynolds, G. Niedobitek, L. S. Young, and N. Mueller-Lantzsch. 1994. Monoclonal antibodies directed against the Epstein-Barr virus-encoded nuclear antigen 1 (EBNA1): immunohistologic detection of EBNA1 in the malignant cells of Hodgkin's disease. Blood 84:3792-3798. [PubMed] [Google Scholar]
- 12.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 13.Herbig, U., K. Weisshart, P. Taneja, and E. Fanning. 1999. Interaction of the transcription factor TFIID with simian virus 40 (SV40) large T antigen interferes with replication of SV40 DNA in vitro. J. Virol. 73:1099-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert, P. F., B. C. Monk, and P. M. Howley. 1990. Phenotypic analysis of bovine papillomavirus type 1 E2 repressor mutants. J. Virol. 64:950-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert, P. F., B. A. Spalholz, and P. M. Howley. 1987. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell 50:69-78. [DOI] [PubMed] [Google Scholar]
- 16.Law, M. F., D. R. Lowy, I. Dvoretzky, and P. M. Howley. 1981. Mouse cells transformed by bovine papillomavirus contain only extrachromosomal viral DNA sequences. Proc. Natl. Acad. Sci. USA 78:2727-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, K.-Y., T. R. Broker, and L. T. Chow. 1998. Transcription factor YY1 represses cell-free replication from human papillomavirus origins. J. Virol. 72:4911-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leight, E. R., and B. Sugden. 2000. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev. Med. Virol. 10:83-100. [DOI] [PubMed] [Google Scholar]
- 19.Lepik, D., and M. Ustav. 2000. Cell-specific modulation of papovavirus replication by tumor suppressor protein p53. J. Virol. 74:4688-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, R., and M. R. Botchan. 1994. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc. Natl. Acad. Sci. USA 91:7051-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim, D. A., M. Gossen, C. W. Lehman, and M. R. Botchan. 1998. Competition for DNA binding sites between the short and long forms of E2 dimers underlies repression in bovine papillomavirus type 1 DNA replication control. J. Virol. 72:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride, A. A., J. C. Byrne, and P. M. Howley. 1989. E2 polypeptides encoded by bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc. Natl. Acad. Sci. USA 86:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride, A. A., H. Romanczuk, and P. M. Howley. 1991. The papillomavirus E2 regulatory proteins. J. Biol. Chem. 266:18411-18414. [PubMed] [Google Scholar]
- 24.Melendy, T., and R. Li. 2001. Chromatin remodeling and initiation of DNA replication. Front. Biosci. 6:D1048-D1053. [DOI] [PubMed] [Google Scholar]
- 25.Miller, S. D., G. Farmer, and C. Prives. 1995. p53 inhibits DNA replication in vitro in a DNA-binding-dependent manner. Mol. Cell. Biol. 15:6554-6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakami, Y., and Y. Ito. 1999. Transcription factors in DNA replication. Front. Biosci. 4:D824-D833. [DOI] [PubMed] [Google Scholar]
- 27.Remm, M., R. Brain, and J. R. Jenkins. 1992. The E2 binding sites determine the efficiency of replication for the origin of human papillomavirus type 18. Nucleic Acids Res. 20:6015-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 29.Riese, D. J., II, J. Settleman, K. Neary, and D. DiMaio. 1990. Bovine papillomavirus E2 repressor mutant displays a high-copy-number phenotype and enhanced transforming activity. J. Virol. 64:944-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell, J., and M. R. Botchan. 1995. cis-Acting components of human papillomavirus (HPV) DNA replication: linker substitution analysis of the HPV type 11 origin. J. Virol. 69:651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stenlund, A. 1996. Papillomavirus DNA replication, p. 679-698. In M. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Stubenrauch, F., A. M. E. Colbert, and L. A. Laimins. 1998. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J. Virol. 72:8115-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stubenrauch, F., M. Hummel, T. Iftner, and L. A. Laimins. 2000. The E8∧E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 74:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stubenrauch, F., H. B. Lim, and L. A. Laimins. 1998. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J. Virol. 72:1071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stubenrauch, F., T. Zobel, and T. Iftner. 2001. The E8 domain confers a novel long-distance transcriptional repression activity on the E8∧E2C protein of high-risk human papillomavirus type 31. J. Virol. 75:4139-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ustav, M., E. Ustav, P. Szymanski, and A. Stenlund. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, L., R. Li, I. J. Mohr, R. Clark, and M. R. Botchan. 1991. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature 353:628-632. [DOI] [PubMed] [Google Scholar]
- 39.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yates, J. L., and N. Guan. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 42.zur Hausen, H. 1994. Human pathogenic papillomaviruses, p. 131-156. Springer-Verlag KG, Berlin, Germany.
- 43.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]